Abstract
We have recently established a clonal culture system that supports the growth of immature natural killer (NK) cells from murine fetal thymocytes. We now describe a culture system for mixed NK cell colony formation from single lymphohematopoietic progenitors. When Sca-1+c-kit+ fetal liver cells were cultured in methylcellulose media with interleukin (IL)-2, IL-7, IL-11, and steel factor (SF ), we found mixed colonies consisting of diffuse small round cells characteristic of immature NK cells and other types of cells. The single cell origin of the mixed colonies was established by micromanipulation. Individual mixed colonies derived from single cells were characterized by flow cytometric analysis and May-Grünwald Giemsa staining. All mixed colonies contained Thy-1+B220− cells, which can differentiate to mature NK cells in fetal thymus organ culture. Most of the colonies contained B220+ B-lineage cells and macrophages, and some contained mast cells. IL-1α and IL-3, which have previously been shown to inhibit the T- and B-cell potentials of blast colonies, suppressed the formation of mixed NK cell colonies. The clonal culture assay presented here may be useful in analysis of the developmental pathway and commitment of NK cells from multipotential progenitors.
NATURAL KILLER (NK) cells comprise a small population of lymphocytes, which are distinct from T and B lymphocytes.1 NK cells exert cytolytic activity against certain tumor cells or transplanted allogenic cells and are thought to play important roles in various immune responses.2,3 Although the functions of NK cells have been studied extensively, their ontogeny and regulation of development are largely unknown. Several reports suggest that NK cells are closely related to T lymphocytes in their developmental pathway.4 NK/T common progenitors have been identified in human fetal thymus.5 In mice, a small subset of thymocytes has been shown to have the potential to develop into both NK and T cells6 7 in vivo, suggesting that NK/T common precursors also exist in murine thymus. In contrast to the developmental relationship between NK and T cells, the lineage relationship of NK cells with B and myeloid cells largely remains to be examined.
To clarify the pathway and regulation of NK cell development, it is necessary to establish a clonal cell culture system, which supports the development of NK cells from multipotential hematopoietic progenitor cells. Recently, we established the culture system that supports the growth of immature NK cell colonies from fetal thymocytes. The colonies appeared as a diffuse collection of small round cells showing no central cores. The unique morphology of these colonies and constituent cells provided an opportunity to identify multilineage colonies containing NK cells in cultures of extrathymic tissues. Here, we report that purified progenitors from fetal liver cells can form mixed colonies consisting of immature NK cells, B lineage cells, macrophages and/or mast cells in the presence of interleukin (IL)-2, IL-7, IL-11, and steel factor (SF, kit ligand). We determined the single cell origin of the mixed colonies by micromanipulation of the progenitors. We also show that IL-1α and IL-3, cytokines previously shown to exert inhibitory effects on T- and B-cell development,8 9 negatively regulate the development of NK cells. The culture system, which enables the development and growth of NK progenitor cells from uncommitted progenitors, will be useful for studies of the developmental pathway and commitment of NK cells.
MATERIALS AND METHODS
Cell preparation. Fetuses at day 14 of gestation were obtained from mice with timed pregnancy as described previously.10 Pregnant mice were killed and thymi and livers of the fetuses were harvested. Cell suspensions of the two organs were prepared by gently pressing the thymic lobes or minced livers between two slide glasses followed by repeated pipeting.
Cytokines. Human IL-1α was provided by Y. Hirai (Otsuka Pharmaceutical, Tokushima, Japan). Purified murine recombinant IL-2 and IL-3 were purchased from R&D system (Minneapolis, MN). Purified recombinant human IL-6 was a gift from M. Naruto of Toray Industries Inc (Kamakura, Japan). Recombinant human IL-7 was provided by Sanofi Winthrop, Inc (Malvern, PA). Recombinant murine SF was a gift from Elaine K. Thomas (Immunex, Seattle, WA). Recombinant human IL-11, human IL-12, and human erythropoietin was provided by Genetics Institute (Cambridge, MA). Recombinant human granulocyte colony-stimulating factor (G-CSF ) was a gift from A. Shimosaka of Kirin Brewery, Co, Ltd (Maebashi, Japan). Concentrations of cytokines used were as follows: IL-1α, 1.0 ng/mL; IL-2, 20 ng/mL; IL-3, 10 ng/mL; IL-7, 200 U/mL (20 ng/mL); IL-11, 100 ng/mL; IL-12, 10 ng/mL; SF, 100 ng/mL; G-CSF, 100 ng/mL; erythropoietin, 2 U/mL.
Monoclonal antibodies. Anti-B220 (14. 8)11, anti-Gr–1 (RB6-8C5)12, anti-Mac–1 (M1/70)13, anti-CD4 (GK1.5)14, anti-CD8 (53-6.72),15 and anti-TER–11916 were used as lineage-specific antibodies in negative immunomagnetic selection. Monoclonal antibodies used for flow cytometric analysis and sorting were as follows: phycoerythrin (PE)-conjugated–anti-CD3 (145-2C11)17 (Pharmingen, San Diego, CA), PE-conjugated–anti-NK1.1 (PK136)18 (Pharmingen), PE-conjugated–anti-CD25 (PC61.5.3)19 (Caltag Laboratories, San Francisco, CA), PE-conjugated–anti-B220 (RA3-6B2),11 PE-conjugated–anti-Sca–1 (E13-161.7)20 (Pharmingen), fluorescein isothiocyanate (FITC)-conjugated–anti-Thy–1 (30-H12)15 (Pharmingen), FITC-conjugated–anti-Ly5.1 (A20-1.7, kindly provided by Dr H. Flemming, Emory University, Atlanta, GA), FITC-conjugated–anti-c–kit (2B8)21 (Pharmingen).
Purification of progenitors from fetal livers. Purification of progenitors was performed by using the methods described by Shih et al22 and Fujimoto et al23 with slight modifications. First, cells with a density between 1.0000 and 1.0770 were collected with discontinuous density gradients of Nicodenz (Accurate Chemical & Scientific Corp, Westbury, NY). The cells were further enriched for progenitors by negative immunomagnetic selection with a cocktail of lineage-specific monoclonal antibodies, and the lineage-negative cells were sorted for Sca-1–and c-kit–positive cells using FACS Vantage (Becton Dickinson, Mountain View, CA).
Flow cytometric analysis and cell sorting. Basic methods of cell staining for flow cytometric analysis were described previously.10 Briefly, cells were incubated with appropriately diluted monoclonal antibodies for 20 minutes on ice. In all experiments, cells stained with isotype-matched control immunoglobulins were also prepared as negative controls. Cells were washed once with phosphate-buffered saline supplemented with 1% deionized bovine serum albumin (BSA), analyzed, and/or sorted by using FACS Vantage.
Cultures for mixed NK cell colonies. Cells were cultured in the medium consisting of α-medium (Flow Laboratories, Rockville, MD), 1.2% 1500-centipoise methylcellulose (Shinetsu Chemical, Tokyo, Japan), 5% fetal calf serum (FCS, Intergen, Purchase, NY), 1% deionized fraction-V BSA (Sigma, St Louis, MO), 0.1 mmol/L 2-mercaptoethanol (Sigma) and designated cytokines. Dishes were incubated for 18 days at 37°C in a humidified atmosphere of 5% CO2 . Colonies were scored on an inverted microscope using criteria described previously.10,24-26 Single cell manipulation was performed as described previously.27 Briefly, cells were first plated in 1 mL of methylcellulose medium at the concentration of 2,000/mL. Transfer of a single cell to cytokine containing methylcellulose medium was accomplished by using a fine Pasteur pipet (with a diameter of approximately 30 μm) attached to a micromanipulator (H-10M, Hacker Instruments, Fairfield, NJ) under direct microscopic visualization.
Fetal thymus organ culture. Fetal thymus organ culture was performed as described previously.10 Briefly, fetal thymi were first incubated with 4 × 104 test cells in hanging drop culture for 20 hours. Thymic lobes were then transferred onto filter membranes (Costar, Cambridge, MA; pore size, 8 μm) and cultured for 11 days. Cells were recovered from the thymic lobes, stained with antibodies, and analyzed by flow cytometry.
RESULTS
Formation of mixed NK cell colonies from fetal liver progenitors. We recently described a clonal culture assay for immature NK cells from fetal thymocytes.10 The colonies were very diffuse without central core, and most of the cells were small round cells. Because of the unique morphology, the immature NK cell colonies could be easily distinguished from other types of colonies in situ on an inverted microscope. In an effort to identify similar colonies from extrathymic tissues, we found that Sca-1+ c-kit+ cells of fetal liver can form mixed colonies containing immature NK cells when cultured in the presence of IL-2, IL-7, SF, and IL-11 (Table 1 and Fig 1). The colonies consist of diffuse small round cells characteristic of immature NK cells and other types of cells constituting a central core (Fig 1). An average of seven cells of 50 Sca-1+c-kit+ fetal liver cells formed the mixed colonies. Pure NK cell colonies consisting of only diffuse small round cells were also observed, although the frequency was low (Table 1). About 15 of 50 fetal liver Sca-1+ c-kit+ cells formed colonies, which apparently lacked immature NK cells. These colonies were identified as pre-B cell, mast cell, macrophage, and mixed mast cell-macrophage colonies in situ using criteria described previously.10,24-26 Table 1 shows the results of a representative study of the colony forming ability of fetal liver Sca-1+c-kit+ cells of day 14 gestation. In this table, mast cell, macrophage, and mixed mast cell-macrophage colonies were collectively designated as myeloid colonies. The data shown in Table 1 indicate that a cytokine combination of IL-2, IL-7, and SF, which supports the growth of pure NK cell colonies from fetal thymocytes, fails to support the formation of mixed NK cell colonies from fetal liver. We also examined the colony forming ability of the fetal liver Sca-1+c-kit+ cells on day 13 and 15 of gestation. The frequencies of mixed NK cell colonies were similar to that of day 14. In Table 1, the colony forming abilities of fetal thymocytes are also shown. Colony forming ability of CD25−c-kit+ thymocytes was examined along with unfractionated fetal thymocytes because this population of cells has been previously shown to have T, B, and NK cell potential.28 In contrast to Sca-1+c-kit+ cells of fetal liver, neither unpurified thymocytes nor purified CD25−c-kit+ fetal thymocytes formed mixed NK cell colonies even in the presence of IL-2, IL-7, SF, and IL-11.
Effects of Addition of IL-11 on the Colony Formation From Fetal Liver and Fetal Thymus Cells
| Cell Plated . | Cytokines . | No. of Colonies . | ||||
|---|---|---|---|---|---|---|
| Type . | No./Dish . | . | Mixed-NK . | Pure-NK . | Pre-B . | Myeloid . |
| Sca-1+c-kit+ FL | 50 | IL-2, IL-7, SF | 0 | 1 ± 1 | 3 ± 1 | 9 ± 1 |
| IL-2, IL-7, IL-11, SF | 7 ± 1 | 2 ± 1 | 4 ± 1 | 9 ± 2 | ||
| Unfractionated FT | 1000 | IL-2, IL-7, SF | 0 | 11 ± 1 | 0 | 2 ± 1 |
| IL-2, IL-7, IL-11, SF | 0 | 13 ± 1 | 0 | 3 ± 2 | ||
| CD25−c-kit+FT | 300 | IL-2, IL-7, SF | 0 | 22 ± 3 | 0 | 1 ± 1 |
| IL-2, IL-7, IL-11, SF | 0 | 18 ± 2 | 0 | 2 ± 1 | ||
| Cell Plated . | Cytokines . | No. of Colonies . | ||||
|---|---|---|---|---|---|---|
| Type . | No./Dish . | . | Mixed-NK . | Pure-NK . | Pre-B . | Myeloid . |
| Sca-1+c-kit+ FL | 50 | IL-2, IL-7, SF | 0 | 1 ± 1 | 3 ± 1 | 9 ± 1 |
| IL-2, IL-7, IL-11, SF | 7 ± 1 | 2 ± 1 | 4 ± 1 | 9 ± 2 | ||
| Unfractionated FT | 1000 | IL-2, IL-7, SF | 0 | 11 ± 1 | 0 | 2 ± 1 |
| IL-2, IL-7, IL-11, SF | 0 | 13 ± 1 | 0 | 3 ± 2 | ||
| CD25−c-kit+FT | 300 | IL-2, IL-7, SF | 0 | 22 ± 3 | 0 | 1 ± 1 |
| IL-2, IL-7, IL-11, SF | 0 | 18 ± 2 | 0 | 2 ± 1 | ||
Sca-1+c-kit+FL cells were prepared as described in the Materials and Methods section. CD25−c-kit+FT cells were prepared by FACS sorting. Cells were cultured in methylcellulose media in the presence of designated cytokines. Colonies were scored after 18 days of incubation. Data represent mean ± SD of quadruplicate cultures.
Abbreviations: Mixed-NK, mixed NK colonies; Pure-NK, NK cell colonies; Pre-B, Pre-B cell colonies; Myeloid, colonies consisting of mast cells, macrophages or both; FL, fetal liver; FT; fetal thymus.
A photomicrograph of a mixed NK cell colony. One quarter of a representative mixed NK cell colony seen on an inverted microscope is shown. Original magnification × 40.
A photomicrograph of a mixed NK cell colony. One quarter of a representative mixed NK cell colony seen on an inverted microscope is shown. Original magnification × 40.
Minimal cytokine requirements for the growth of the mixed NK cell colony. We next examined the minimal cytokine requirement for the formation of the mixed NK cell colony. Sca-1+c-kit+ cells of fetal liver were cultured in methylcellulose media with cytokines designated in Table 2. The results indicate that IL-7, IL-11, and SF are necessary for the growth of mixed NK cell colony. IL-2 is not essential for the colony formation, but increases the frequency of colonies.
Minimal Cytokine Requirement of Mixed NK Cell Colonies
| Cytokines . | No. of Colonies . | |||
|---|---|---|---|---|
| . | Mixed-NK . | Pure-NK . | Pre-B . | Myeloid . |
| SF | 0 | 0 | 0 | 4 ± 1 |
| IL-2 | 0 | 0 | 0 | 0 |
| IL-7 | 0 | 0 | 0 | 0 |
| IL-11 | 0 | 0 | 0 | 0 |
| SF, IL-2 | 0 | 0 | 0 | 4 ± 1 |
| SF, IL-7 | 0 | 0 | 2 ± 1 | 4 ± 1 |
| SF, IL-11 | 0 | 0 | 0 | 10 ± 2 |
| IL-2, IL-7 | 0 | 0 | 0 | 0 |
| IL-2, IL-11 | 0 | 0 | 0 | 0 |
| IL-7, IL-11 | 0 | 0 | 0 | 0 |
| SF, IL-2, IL-7 | 0 | 0 ± 1 | 0 ± 1 | 5 ± 2 |
| SF, IL-2, IL-11 | 0 | 0 | 0 ± 1 | 10 ± 2 |
| SF, IL-7, IL-11 | 2 ± 1 | 1 ± 1 | 3 ± 1 | 9 ± 3 |
| IL-2, IL-7, IL-11 | 0 | 0 | 0 ± 1 | 0 |
| SF, IL-2, IL-7, IL-11 | 8 ± 1 | 0 ± 1 | 4 ± 2 | 6 ± 1 |
| Cytokines . | No. of Colonies . | |||
|---|---|---|---|---|
| . | Mixed-NK . | Pure-NK . | Pre-B . | Myeloid . |
| SF | 0 | 0 | 0 | 4 ± 1 |
| IL-2 | 0 | 0 | 0 | 0 |
| IL-7 | 0 | 0 | 0 | 0 |
| IL-11 | 0 | 0 | 0 | 0 |
| SF, IL-2 | 0 | 0 | 0 | 4 ± 1 |
| SF, IL-7 | 0 | 0 | 2 ± 1 | 4 ± 1 |
| SF, IL-11 | 0 | 0 | 0 | 10 ± 2 |
| IL-2, IL-7 | 0 | 0 | 0 | 0 |
| IL-2, IL-11 | 0 | 0 | 0 | 0 |
| IL-7, IL-11 | 0 | 0 | 0 | 0 |
| SF, IL-2, IL-7 | 0 | 0 ± 1 | 0 ± 1 | 5 ± 2 |
| SF, IL-2, IL-11 | 0 | 0 | 0 ± 1 | 10 ± 2 |
| SF, IL-7, IL-11 | 2 ± 1 | 1 ± 1 | 3 ± 1 | 9 ± 3 |
| IL-2, IL-7, IL-11 | 0 | 0 | 0 ± 1 | 0 |
| SF, IL-2, IL-7, IL-11 | 8 ± 1 | 0 ± 1 | 4 ± 2 | 6 ± 1 |
A total of 50 Sca-1+c-kit+ fetal liver cells were plated in methylcellulose media in the presence of designated cytokines. Colonies were scored after 18 days of incubation. Data represent mean ± SD of quadruplicate cultures.
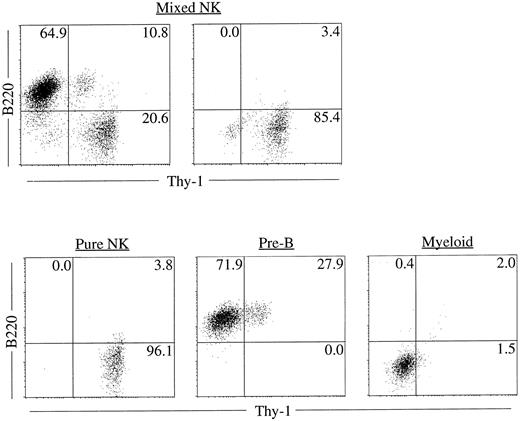
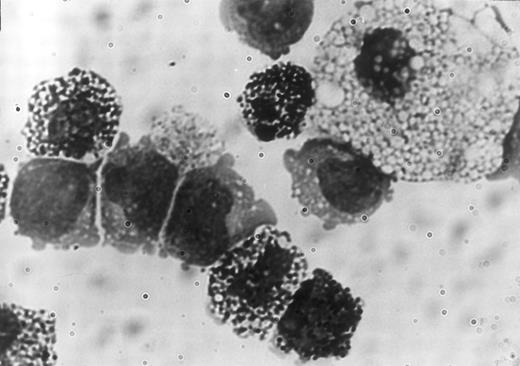
Clonality and characterization of mixed NK colonies. To establish the clonal origin of the mixed colonies, we next plated individual Sca-1+c-kit+ fetal liver cells by micromanipulation into methylcellulose media containing IL-2, IL-7, IL-11, and SF. The results of two experiments are shown in Table 3. Twenty-seven of 196 and 24 of 193 cells formed mixed NK colonies in experiments 1 and 2, respectively. These results established the single cell origin of the mixed colonies. We then picked individual mixed colonies from the methylcellulose media and analyzed their size. The mean ± standard deviation (SD) of the cell number of the mixed colonies was 3 × 104 ± 1.1 × 104 cells. One aliquot of the remaining cells was examined for the expression of B220 and Thy-1 by flow cytometry. As shown in Table 4, all colonies identified in situ contained Thy-1+B220− cells. This is consistent with our previous observation10 that the cells in the immature NK colonies express Thy-1 at a high level, but not B220. Most of the colonies (25 of 27 colonies and 20 of 24 colonies in experiment 1 and experiment 2, respectively) showed B220+Thy-1− cells indicating the presence of B-lineage cells. Representative fluorescence-activated cell sorting (FACS) profiles of individual colonies are shown in Fig 2. The upper left and right panels in Fig 2 represent mixed NK colonies with and without B220+ cells, respectively. Consistent with earlier observations,24 26 some of the B220+ cells in the pre-B cell and mixed NK cell colonies expressed Thy-1. The other aliquot of the cells was subjected to May-Grünwald-Giemsa staining for examination of myeloid lineage cells. As shown in Table 4, most of the colonies contained macrophages, and some contained mast cells. Photomicrograph of the May-Grünwald Giemsa-stained smear of a representative mixed colony is shown in Fig 3. We also performed differential cell counting of the mixed NK cell colony. One hundred Sca-1+c-kit+ cells were individually plated by micromanipulation in methylcellulose media containing IL-2, IL-7, IL-11, and SF. Ten mixed NK cell colonies were observed 18 days later. Each colony was picked and subjected to May-Grünwald Giemsa staining. As shown in Table 5, the majority of the cells constituting the mixed colonies were lymphoid lineage cells, but the proportions of the lymphoid cells, macrophages, and mast cells varied among colonies.
NK Cell Colony Formation From Individually Plated Cells
| Exp. # . | No. of Cells Plated . | No. of Colonies . | |||
|---|---|---|---|---|---|
| . | . | Mixed-NK . | Pure-NK . | Pre-B . | Myeloid . |
| Exp. 1 | 196 | 27 | 4 | 13 | 43 |
| Exp. 2 | 193 | 24 | 5 | 18 | 36 |
| Exp. # . | No. of Cells Plated . | No. of Colonies . | |||
|---|---|---|---|---|---|
| . | . | Mixed-NK . | Pure-NK . | Pre-B . | Myeloid . |
| Exp. 1 | 196 | 27 | 4 | 13 | 43 |
| Exp. 2 | 193 | 24 | 5 | 18 | 36 |
Sca-1+c-kit+ cells purified from fetal liver were individually plated into methylcellulose media by micromanipulation. Cells were cultured in the presence of IL-2, IL-7, IL-11, and SF. Colonies were scored after 18 days of incubation.
B Cell and Myeloid Expression of Individual Mixed NK Cell Colonies
| Thy-1+B220− . | B220+ . | Macrophage . | Mast Cell . | No. of Colonies . | |
|---|---|---|---|---|---|
| . | . | . | . | Exp. 1 . | Exp. 2 . |
| + | + | + | + | 4 | 4 |
| + | + | + | 20 | 16 | |
| + | + | 2 | 4 | ||
| + | + | 1 | 0 | ||
| Thy-1+B220− . | B220+ . | Macrophage . | Mast Cell . | No. of Colonies . | |
|---|---|---|---|---|---|
| . | . | . | . | Exp. 1 . | Exp. 2 . |
| + | + | + | + | 4 | 4 |
| + | + | + | 20 | 16 | |
| + | + | 2 | 4 | ||
| + | + | 1 | 0 | ||
Mixed NK cell colonies derived from single cells were harvested from methylcellulose media, divided into two aliquots, and subjected to two-color flow cytometry and May-Grünwald Giemsa staining.
Representative FACS profiles of colonies derived from single cells. Sca-1+c-kit+ cells were purified from fetal liver and were plated individually by micromanipulation into methylcellulose media containing IL-2, IL-7, IL-11, and SF. After 18 days of culture, colonies were identified in situ and picked individually. Cells from each colony were stained with PE-anti–B220 and FITC-anti–Thy-1 and analyzed by FACS Vantage using Cell Quest software. Upper left: A mixed NK colony with B lineage cells. Upper right: A mixed NK colony without B lineage cells. FACS profile of a typical pure NK, pre-B or myeloid colony is also shown for comparison.
Representative FACS profiles of colonies derived from single cells. Sca-1+c-kit+ cells were purified from fetal liver and were plated individually by micromanipulation into methylcellulose media containing IL-2, IL-7, IL-11, and SF. After 18 days of culture, colonies were identified in situ and picked individually. Cells from each colony were stained with PE-anti–B220 and FITC-anti–Thy-1 and analyzed by FACS Vantage using Cell Quest software. Upper left: A mixed NK colony with B lineage cells. Upper right: A mixed NK colony without B lineage cells. FACS profile of a typical pure NK, pre-B or myeloid colony is also shown for comparison.
Photomicrograph of a portion of the May-Grünwald Giemsa-stained smear of a mixed NK cell colony showing a macrophage, mast cells, and 5 lymphoid cells.
Photomicrograph of a portion of the May-Grünwald Giemsa-stained smear of a mixed NK cell colony showing a macrophage, mast cells, and 5 lymphoid cells.
Differential Counts of Mixed NK Cell Colonies Derived From Single Progenitors
| Colony No. . | Differential Cell Counts . | ||
|---|---|---|---|
| . | Lymphoid . | Macrophage . | Mast Cell . |
| 1 | 88 | 12 | 0 |
| 2 | 98 | 2 | 0 |
| 3 | 86 | 11 | 3 |
| 4 | 91 | 9 | 0 |
| 5 | 98 | 2 | 0 |
| 6 | 65 | 35 | 0 |
| 7 | 78 | 22 | 0 |
| 8 | 45 | 51 | 4 |
| 9 | 95 | 5 | 0 |
| 10 | 83 | 17 | 0 |
| Colony No. . | Differential Cell Counts . | ||
|---|---|---|---|
| . | Lymphoid . | Macrophage . | Mast Cell . |
| 1 | 88 | 12 | 0 |
| 2 | 98 | 2 | 0 |
| 3 | 86 | 11 | 3 |
| 4 | 91 | 9 | 0 |
| 5 | 98 | 2 | 0 |
| 6 | 65 | 35 | 0 |
| 7 | 78 | 22 | 0 |
| 8 | 45 | 51 | 4 |
| 9 | 95 | 5 | 0 |
| 10 | 83 | 17 | 0 |
100 Sca-1+c-kit+ cells were individually plated in methylcellulose media containing IL-2, IL-7, IL-11, and SF. Ten mixed NK cell colonies were observed 18 days later. The colonies were individually harvested and subjected to May-Grünwald Giemsa staining.
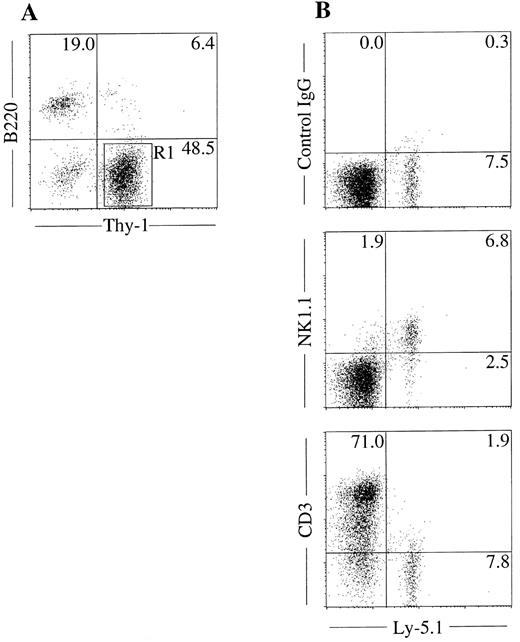
NK cell potential of Thy-1+B220− cells in mixed NK cell colonies. The unique in situ appearance of the colonies and the presence of Thy-1+B220− cells indicated that mixed colonies contain immature NK cells. To confirm this, we next examined the NK cell potential of the cells in the mixed colonies using fetal thymus organ culture. We have shown previously that this culture method supports the maturation of immature NK cells from fetal thymocytes.10 We picked mixed NK cell colonies individually, pooled and prepared Thy-1+B220− cells by FACS sorting as shown in Fig 4A. Cells were then plated in the fetal thymus organ culture for identification of NK1.1+ cells. As shown in Fig 4B, Ly5.1+ donor cells cultured in the presence of thymic lobes of Ly5.2+ mice expressed NK1.1, but not CD3. This result confirmed that the Thy-1+B220− cells in the mixed colonies have the ability to develop into NK cells. We also examined the possible presence of the clonogenic cells in the mixed NK cell colonies by replating the pooled mixed NK cell colonies. Cells were cultured in methylcellulose media containing IL-3, IL-11, and erythropoietin for analysis of the myeloid potential and in media containing SF and IL-7, for analysis of B-cell potential. Microscopic observation was performed every 2 days during 10 days of culture. There was no colony formation indicating that hematopoietic progenitors are not present in the mixed NK cell colonies.
NK cell potential of Thy-1+ cells present in mixed colonies. Sca-1+c-kit+ cells were purified from C57BL/6-Ly5.1/C57BL/6-Ly5.2 F1 (Ly5.1/Ly5.2) mice. Cells were cultured in the methylcellulose media containing IL-2, IL-7, IL-11, and SF. After incubation for 18 days, mixed NK cell colonies were individually picked, pooled, stained with PE-anti–B220 and FITC-anti–Thy-1, and analyzed by flow cytometry (A). Thy-1+B220− cells (R1) were sorted and incubated in hanging drop for 20 hours with fetal thymus lobes from BDF1 (Ly5.2) mice. The lobes were transferred to filter membranes and cultured for 11 days. Cells recovered from lobes were stained with FITC-anti–Ly5.1 and PE-anti–NK1.1, PE-anti–CD3 or mixture of PE-hamster–IgG and PE-mouse IgG2a and analyzed by FACS Vantage (B).
NK cell potential of Thy-1+ cells present in mixed colonies. Sca-1+c-kit+ cells were purified from C57BL/6-Ly5.1/C57BL/6-Ly5.2 F1 (Ly5.1/Ly5.2) mice. Cells were cultured in the methylcellulose media containing IL-2, IL-7, IL-11, and SF. After incubation for 18 days, mixed NK cell colonies were individually picked, pooled, stained with PE-anti–B220 and FITC-anti–Thy-1, and analyzed by flow cytometry (A). Thy-1+B220− cells (R1) were sorted and incubated in hanging drop for 20 hours with fetal thymus lobes from BDF1 (Ly5.2) mice. The lobes were transferred to filter membranes and cultured for 11 days. Cells recovered from lobes were stained with FITC-anti–Ly5.1 and PE-anti–NK1.1, PE-anti–CD3 or mixture of PE-hamster–IgG and PE-mouse IgG2a and analyzed by FACS Vantage (B).
Effects of IL-1α and IL-3. We have shown previously that IL-1α and IL-3 have inhibitory effects on the development of B and T cells from lymphohemopoietic cells.9,23 24 We, therefore, examined the effects of IL-1α and IL-3 on the mixed NK cell colony formation because it has been suggested that NK, B, and T cells share common progenitors at certain developmental stages. Sca-1+c-kit+ cells were purified from the fetal liver and cultured in methylcellulose with cytokines designated in Table 6. In the presence of IL-1α or IL-3, neither NK nor mixed NK cell colonies formed. This observation suggested that IL-1α and IL-3 negatively regulate the development of immature NK cells.
Effect of IL-1α and IL-3 on the Formation of NK Cell Colonies From Fetal Liver Cells
| Cytokines . | No. of Colonies . | |||
|---|---|---|---|---|
| . | Mixed-NK . | Pure-NK . | Pre-B . | Myeloid . |
| IL-2, IL-7, IL-11, SF | 7 ± 1 | 1 ± 1 | 2 ± 1 | 9 ± 3 |
| IL-1α, IL-2, IL-7, IL-11, SF | 0 | 0 | 0 | 14 ± 2 |
| IL-3, IL-2, IL-7, IL-11, SF | 0 | 0 | 0 | 18 ± 4 |
| Cytokines . | No. of Colonies . | |||
|---|---|---|---|---|
| . | Mixed-NK . | Pure-NK . | Pre-B . | Myeloid . |
| IL-2, IL-7, IL-11, SF | 7 ± 1 | 1 ± 1 | 2 ± 1 | 9 ± 3 |
| IL-1α, IL-2, IL-7, IL-11, SF | 0 | 0 | 0 | 14 ± 2 |
| IL-3, IL-2, IL-7, IL-11, SF | 0 | 0 | 0 | 18 ± 4 |
Sca-1+c-kit+ cells were purified from fetal liver. A total of 50 cells was plated in methylcellulose media in the presence of designated cytokines. Colonies were scored after 18 days of incubation. Data represent mean ± SD of quadruplicate cultures.
Effects of early-acting cytokines. Previously, we have reported that IL-11, IL-6, IL-12, and G-CSF in combination with SF support the growth and survival of murine lymphohematopoietic progenitors.24,29,30 We next examined the effects of these early-acting cytokines on the colony formation supported by IL-2, IL-7, and SF. The results presented in Table 7 indicate that IL-11, IL-6, IL-12, and G-CSF can individually support the growth of mixed-NK cell colonies in the presence of IL-2, IL-7, and SF. Consistent with previous data obtained by using bone marrow progenitors as materials,29 31 IL-12 exhibited the least activity among these cytokines, and the colonies developing in the presence of IL-12 were smaller than those supported by IL-11, IL-6, or G-CSF (data not shown).
Effects of Early-Acting Cytokines on the Formation of NK Cell Colonies
| Cytokines . | No. of Colonies . | |||
|---|---|---|---|---|
| . | Mixed-NK . | Pure-NK . | Pre-B . | Myeloid . |
| IL-2, IL-7, SF | 0 ± 1 | 1 ± 1 | 5 ± 1 | 8 ± 2 |
| IL-11, IL-2, IL-7, SF | 9 ± 1 | 1 ± 1 | 2 ± 1 | 11 ± 3 |
| IL-6, IL-2, IL-7, SF | 5 ± 1 | 0 ± 1 | 1 ± 1 | 8 ± 2 |
| G-CSF, IL-2, IL-7, SF | 6 ± 1 | 1 ± 1 | 3 ± 1 | 7 ± 2 |
| IL-12, IL-2, IL-7, SF | 3 ± 1 | 0 | 4 ± 1 | 8 ± 1 |
| Cytokines . | No. of Colonies . | |||
|---|---|---|---|---|
| . | Mixed-NK . | Pure-NK . | Pre-B . | Myeloid . |
| IL-2, IL-7, SF | 0 ± 1 | 1 ± 1 | 5 ± 1 | 8 ± 2 |
| IL-11, IL-2, IL-7, SF | 9 ± 1 | 1 ± 1 | 2 ± 1 | 11 ± 3 |
| IL-6, IL-2, IL-7, SF | 5 ± 1 | 0 ± 1 | 1 ± 1 | 8 ± 2 |
| G-CSF, IL-2, IL-7, SF | 6 ± 1 | 1 ± 1 | 3 ± 1 | 7 ± 2 |
| IL-12, IL-2, IL-7, SF | 3 ± 1 | 0 | 4 ± 1 | 8 ± 1 |
Sca-1+c-kit+ cells were purified from fetal liver. A total of 50 cells was plated in methylcellulose media in the presence of designated cytokines. Colonies were scored after 18 days of incubation. Data represent mean ± SD of quadruplicate cultures.
DISCUSSION
Although much effort has been made to clarify the pathway and regulation of NK cell development, there are still many questions to be resolved in NK cell ontogeny. One of the major problems that hampered the study of NK cell development is the lack of a clonal culture, which supports the growth and development of NK cells from uncommitted progenitor cells. Here, we report the development of a culture system, which supports the growth of immature NK, B, macrophages, and/or mast cells from a single progenitor.
Accumulating data support the concept that NK, T, and B cells arise from common progenitors.7,28,32-34 However, because of the lack of a clonal culture, which supports the growth of multiple lymphoid lineage cells simultaneously, common progenitors for lymphocytes have not been identified at the clonal level. The culture system presented here will greatly facilitate identification of common lymphoid progenitors because this culture supports simultaneous growth of NK and B lineage cells. It has been reported that a small subset of thymocytes can develop to yield both NK and B cells. These progenitors are thought to contain common lymphoid progenitors.7 28 It is noteworthy that fetal thymocytes could not form mixed colonies containing NK and B lineage cells. This may indicate that fetal thymi contain committed NK and B cell progenitors, but not common NK/B progenitors. Alternatively, very few thymocytes may have potential to develop to both NK and B cells, and our culture condition may not be sensitive enough to detect the common progenitors. In each case, a careful clonal analysis must be used for examining multipotency of a certain population of cells.
Multiple roles of IL-11 in hematopoiesis have been documented.35-37 We have previously reported that IL-11 supports the growth and/or survival of lymphohemopoietic progenitors.24-30 Consistent with this observation, the data presented here suggest that IL-11 supports the proliferation of NK, B and myeloid common progenitors of fetal liver. It is unlikely that IL-11 stimulated the growth of NK cells in a lineage-specific manner because IL-11 did not support the formation of pure NK cell colonies (Table 1). It is more likely that IL-11 acted on early progenitors before commitment to NK cell lineage. The exact developmental stages of the progenitors whose growth IL-11 supports, however, remain to be clarified further.
It has been shown that IL-1α and IL-3 can individually abrogate the T- and B-cell potential of colonies derived from murine bone marrow and fetal liver cells.8,9 23 The data presented in Table 4 indicate that IL-1α and IL-3 also negatively regulate the growth and differentiation of NK cells. It is possible that these cytokines suppress the proliferation or differentiation of lymphoid common progenitors, which have a T, B, and NK potential. Alternatively, they may negatively regulate T, B, and NK-committed progenitors. Regardless, the observations may have significant impact on the in vitro manipulation of lymphohematopoietic stem cells and progenitors.
ACKNOWLEDGMENT
We thank Dr Haiqun Zeng for assistance in cell sorting and Dr Pamela N. Pharr and Anne G. Leary in preparation of this manuscript.
Supported by National Institutes of Health Grants No. DK32294 and DK/HL48714, Office of Research and Development, Medical Research Service, Department of Veterans Affairs.
Address reprint requests to Makio Ogawa, MD, PhD, Ralph H. Johnson Medical Center, 109 Bee St, Charleston, SC 29401-5799.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal