Abstract
Mutations of the Janus family kinase JAK3 have been found to be responsible for autosomal recessive severe combined immunodeficiency (SCID) in humans. We report here the analysis of four new unrelated patients affected by JAK3-deficient SCID. The genetic defects were heterogeneous and included a large intragenic deletion as well as different point mutations, leading to missense substitutions, early stop codons, or splicing defects. We performed a series of studies of the biochemical events induced by cytokines on lymphoblastoid B-cell lines obtained from these patients. Abnormalities in tyrosine phosphorylation of JAK3 in response to interleukin-2 (IL-2) and IL-4 were present in all patients. Accordingly, IL-2–mediated phosphorylation of STAT5 was also absent or barely detectable. On the contrary, in all cases, we could show reduced but clear phosphorylation of STAT6 upon IL-4 stimulation. In one patient carrying a single amino acid change (Glu481Gly) in the JH3 domain of JAK3, we observed partially conserved IL-2 responses resulting in reduced but detectable levels of JAK3 and STAT5 phosphorylation. Interestingly, the patient bearing this mutation developed a substantial number of circulating CD4+/CD45RO+ activated T lymphocytes that were functionally impaired. In two cases, patients' cells expressed JAK3 proteins with mutations in the JH2 pseudo-kinase domain. A single cysteine to arginine substitution (Cys759Arg) in this region resulted in high basal levels of constitutive JAK3 tyrosine phosphorylation unresponsive to either downregulation by serum starvation or cytokine-mediated upregulation. The characterization of the genetic defects and biochemical abnormalities in these JAK3-deficient patients will help define the role of JAK3 in the ontogeny of a competent immune system and may lead to a better understanding of the JAK3 functional domains.
JANUS KINASES CONSTITUTE a family of recently identified protein-tyrosine kinases that play a fundamental role in cytokine receptor signal transduction pathways of several important hematopoietic cytokines.1-3 Four members of this family (JAK1, JAK2, TYK2, and JAK3) have been identified so far. One of these, JAK3, was originally cloned from natural killer (NK) cells and shown to be preferentially expressed in lymphoid and myeloid cells.4 Further studies5,6 showed the critical role played by JAK3 in multiple cytokine signal transduction pathways, in association with the common γ chain (γC ) of the receptors for interleukin-2 (IL-2), IL-4, IL-7, IL-9, and IL-15. More recently, mutations affecting the expression of JAK3 were found to be responsible for autosomal recessive severe combined immunodeficiency (SCID) in humans, a condition in which T-cell development and B-cell function are severely reduced, indicating that this kinase is also essential for the correct development of mature lymphoid lineages.7 8 Patients with SCID due to JAK3 deficiency (JAK3-SCID) have a clinical phenotype that is virtually identical to the X-linked variant of SCID which is caused by mutations of γC . Both forms of SCID are characterized by the absence of circulating mature T lymphocytes and NK cells, normal to elevated numbers of nonfunctional B lymphocytes, and marked hypoplasia of lymphoid tissues. Reconstitution of a competent immune system by allogeneic bone marrow transplantation has proven life-saving for these patients who are otherwise exposed to potentially deadly infections sustained by bacteria, viruses, and opportunistic pathogens.
JAK3-gene knock-out murine models have also been generated and differ from the human phenotype, because the JAK3-deficient mice have severe defects of both T- and B-cell development.9-11 The divergence between JAK3-deficient mice and humans seems to indicate a difference in the role played by JAK3 in different species, because there appears to be an absolute requirement for JAK3 in the ontogeny of both T and B cells in the mouse, whereas, in humans, B lymphocytes can differentiate independently of JAK3. Therefore, the study of the naturally occurring human JAK3 mutants offers a unique opportunity to better define the role of JAK3 in the development and function of the immune system and of the B-cell lineage in particular.
We report here the analysis of the molecular basis of the defect occurring in patients affected by JAK3-SCID and correlate the detected mutations with the biochemical events elicited by IL-2 and IL-4. The results obtained help define the role of JAK3 in cytokine signal transduction and may be useful to gain more insight into the functional characteristics of the Janus kinases homology domains.
MATERIALS AND METHODS
Patients
Four unrelated patients (L.E., L.P., N.K., and V.L.) with T−B+ SCID, who normally expressed the γC , were investigated as candidates for SCID due to JAK3 deficiency. Parental consanguinity was present in patients V.L. and N.K.
Clinical features included severe early onset upper and lower respiratory tract infections that were associated with failure to thrive in V.L. and N.K. and chronic diarrhea in V.L.
Laboratory investigations showed lymphopenia (700 cells/μL) in patient N.K., but a normal lymphocyte count (1,400 to 3,000 cells/μL) in the other patients. However, they showed a severely reduced proportion of peripheral blood T lymphocytes (CD3+ from <1% to 2%), with an increased proportion of B cells (CD13+, 74% to 98%). NK cells (CD16+) were undetectable in all patients, except for L.E., who had 13% CD16+ cells.
In vitro responses to phythoemoagglutinine, to anti-CD3, and to anti-CD3 + IL-2 were absent in all cases. All patients were treated by haploidentical bone marrow transplantation (BMT). Before BMT, patient L.E. developed autologous T cells. At 3 months of age, he had 41% CD3+ cells (absolute number, 3,645/μL). However, these cells had an unusual activated phenotype, in that greater than 98% of them were also CD4+ CD45RO+ and predominantly (63%) coexpressed the DR antigen. At this time, and in contrast to that observed at 1 month of life, in vitro stimulation with anti-CD3 + IL-2 resulted in a substantial proliferative response (36.2 × 103 cpm v 36.6 × 103 cpm in a normal control) as compared with that elicited by anti-CD3 alone (4.2 × 103 cpm in the patient v 39.4 × 103 cpm in the control). Furthermore, IL-2 stimulation alone also elicited a proliferative response comparable to normal (17.6 × 103 cpm in patient L.E. v 16.2 × 103 cpm in the control). Molecular typing of the purified CD4+ cells with use of the highly polymorphic markers DQ α and D1S80 showed that the circulating T cells were autologous and not of maternal origin (Brugnoni et al, manuscript submitted).
Cell Culture, DNA and RNA Extraction, and Southern Blot Analysis
Upon informed consent, lymphoblastoid cell lines (BCLs) were originated by Epstein-Barr virus immortalization of peripheral blood lymphocytes obtained from normal donors and JAK3-SCID patients. Cells were maintained in RPMI 1640 (Life Technologies, Gaithersburg, MD) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 2 mmol/L L-glutamine (Life Technologies), and 50 μg/mL gentamycin (Bio-Whittaker, Walkersville, MD), and cultured at 37°C, 5% CO2 .
Genomic DNA for Southern blot and polymerase chain reaction (PCR) amplification was extracted with the phenol-chloroform method according to standard procedures. Total cellular RNA for reverse transcription-PCR (RT-PCR) was prepared following the Chomzynski procedure.12 For Southern analysis, filters were hybridized overnight at 42°C in formamide buffer with a probe generated by random priming.13 Filters were washed twice in 2× SSC, 0.1% sodium dodecyl sulfate (SDS) at room temperature for 10 minutes and at 65°C in 1× SSC, 0.1% SDS for 10 minutes and 0.1% SSC, 0.1% SDS for 10 minutes and were finally exposed to x-ray film (DuPont NEN, Boston, MA).
PCR Amplification of the JAK3 Gene, Enzyme Digestion, Subcloning, DNA Sequencing, and Sequence Analysis
The general strategy for PCR amplification from RNA or DNA, the primers, and the genomic structure have already been published.7 14 Primers that were developed ad hoc for amplifications used in the present work are as follows: VIIIthF, GTCGCTCAGTCCCACTCAGG; XthF, TCTGATTTCTGGTTTTTCTCCC; 2260F, TCACCATGCCCATCAGTGCC; XthR, CCAGACTGAGGTATCGCCTC; 2300R, GAGTTTCTTAGCAGGATCCA; and 2440R, GAGATGAGGCTATTGAGGT.
Primer VIIIthF, located in the 8th intron, was used in conjunction with the exonic 1518R primer14 to amplify a genomic portion of patient L.P. and controls, whereas 2260F was used with 2440R for the identification of the mutation on the other allele. Primer pair 1448F/2300R was used for amplification of cDNA as well as genomic DNA from patient L.E. Primer pair 1831F/1950R was used for the amplification of cDNA from patient N.K. The primers XthF and XthR, flanking exon 10, were used for amplification of the 10th exon for single-stranded confirmation polymorphism (SSCP) analysis in patient V.L. and controls. Thirty cycles of amplification were performed. The denaturation was at 94°C for 1 minutes, the annealing step was at 60°C for 30 seconds, and polymerization was at 72°C for 30 seconds for all amplifications.
The SSCP analysis was performed loading the amplified samples onto mutation detection enhancement gel plus 5% glycerol and electrophoresed at 4°C in 0.6× TBE at 100 V for 20 hours. The gel was then stained with silver nitrate as previously described.15 Subcloning and sequencing of the PCR product and computer analysis were performed as previously described.7 14
Immunoblotting and Immunoprecipitation
For Western blot analysis, cells were lysed in buffer containing 300 mmol/L NaCl, 50 mmol/L Tris-HCL, 2 mmol/L EDTA, 0.5% Triton X-100, 2.5 μmol/L p-nitrophenyl p′-guanidino-benzoate, 10 μg/mL aprotinin, and 10 μg/mL leupeptin and centrifuged at 12,000g. One hundred micrograms of protein was then boiled, subjected to 8% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and electrotransferred onto polyvinylidene fluoride membranes (Immobilon-P; Millipore, Bradford, MA). Membranes were probed with rabbit polyclonal antibody directed against the C-terminus of JAK3 (a-JAK3)4 or a mouse monoclonal antibody (Ab) specific for JAK1 (a-JAK1; Transduction Laboratories, Lexington, KY), following previously reported procedures.5 For immunoprecipitations, cells (5 to 10 × 107) were first cultured for 4 hours in the absence of fetal bovine serum and then stimulated with 1,000 U/mL of IL-2 or IL-4 (kindly provided by Dr C. Reynolds, National Cancer Institute, Frederick, MD) for 15 minutes and lysed in the lysis buffer defined above supplemented with 200 μmol/L sodium orthovanadate (Sigma Chemical Co, St Louis, MO). After clarification by centrifugation, postnuclear supernatants were immunoprecipitated with appropriate antibodies, as reported.5 Rabbit polyclonal Ab specific for STAT5a (a-STAT5) was described previously,16 and rabbit anti-human STAT6 (a-STAT6) polyclonal antibody was purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). Immunoprecipitates were then washed, boiled, and resolved using SDS-PAGE. After electro-transfer, filters were immunoblotted with antiphosphotyrosine mouse monoclonal Ab (a-PY, 4G10; Upstate Biotechnology Inc, Lake Placid, NY) or with original immunoprecipitation antibody as described.5 Detection was then performed by enhanced chemiluminescence (ECL; Amersham Life Science, Arlington Heights, IL).
RESULTS
Characterization of the Molecular Abnormalities in JAK3-Dependent SCID Patients
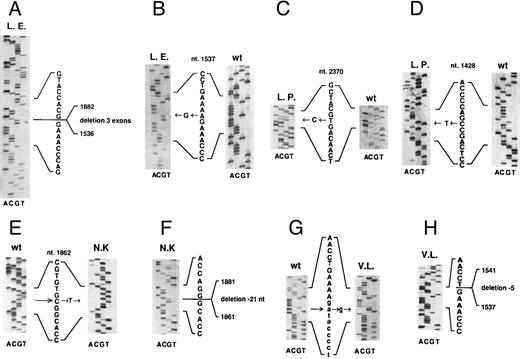
Patient L.E. Southern blot analysis of patient L.E. DNA restricted with EcoRI and hybridized to an RT-PCR product obtained with the primer pair 1448F/2140R showed the presence of a 4.2-kb band in addition to the 3.2- and 2.5-kb band present in the normal DNAs (data not shown). RT-PCR amplification of the JAK3 transcript with the exonic primers 1448F and 2300R showed a smaller band in addition to the expected one. Sequence analysis of three of these clones showed that the different size of the transcript was due to deletion of three exons (exons 10, 11, and 12, Fig 1A) that did not alter the reading frame of the downstream sequence. These results, together with the Southern findings, suggested that a genomic deletion was responsible for this abnormality.
Mutations of JAK3 gene in four unrelated patients affected by autosomal SCID. (A and B) cDNA sequences of the two alleles of patient L.E. (C and D) cDNA sequences of the two alleles of patient L.P. (E and F ) Sequence of genomic DNA of patient N.K. (E) that creates a new splice site causing a 21-bp deletion in his cDNA sequence (F ). (G and H) Sequence of genomic DNA from patient V.L. showing a point mutation in the invariant dinucleotide of the acceptor site of the 9th intron (G), causing the use of a cryptic splice site 5 bp downstream of the normal one, with a consequent 5-bp deletion in the cDNA sequence (H). All exon sequences are in bold, whereas intron sequences in (G) are indicated in lower-case letters. The numbers refer to the JAK3 cDNA sequence as present in data base. w.t., wild-type. All the sequences are shown in the sense orientation, even when performed on the reverse strand.
Mutations of JAK3 gene in four unrelated patients affected by autosomal SCID. (A and B) cDNA sequences of the two alleles of patient L.E. (C and D) cDNA sequences of the two alleles of patient L.P. (E and F ) Sequence of genomic DNA of patient N.K. (E) that creates a new splice site causing a 21-bp deletion in his cDNA sequence (F ). (G and H) Sequence of genomic DNA from patient V.L. showing a point mutation in the invariant dinucleotide of the acceptor site of the 9th intron (G), causing the use of a cryptic splice site 5 bp downstream of the normal one, with a consequent 5-bp deletion in the cDNA sequence (H). All exon sequences are in bold, whereas intron sequences in (G) are indicated in lower-case letters. The numbers refer to the JAK3 cDNA sequence as present in data base. w.t., wild-type. All the sequences are shown in the sense orientation, even when performed on the reverse strand.
To further investigate the genomic abnormality, we performed the amplification of the corresponding genomic DNA region with the same primers. Only a 2.9-kb band was seen in normal controls, including the patient mother, whereas in patient L.E. and in his father, an additional 1.4-kb band was detected (data not shown). Sequencing of the smaller band and comparison with the normal sequence that includes introns 9, 10, and 11 showed that there was a recombination between two Alu elements, the first located in the ninth intron and the second in the eleventh intron, with deletion of the intervening sequences (data not shown). Therefore, exons 10 and 11 were deleted at the genomic level, whereas apparently exon 12 is present in the genomic DNA, but it is preferentially skipped in the abnormally sized transcript.
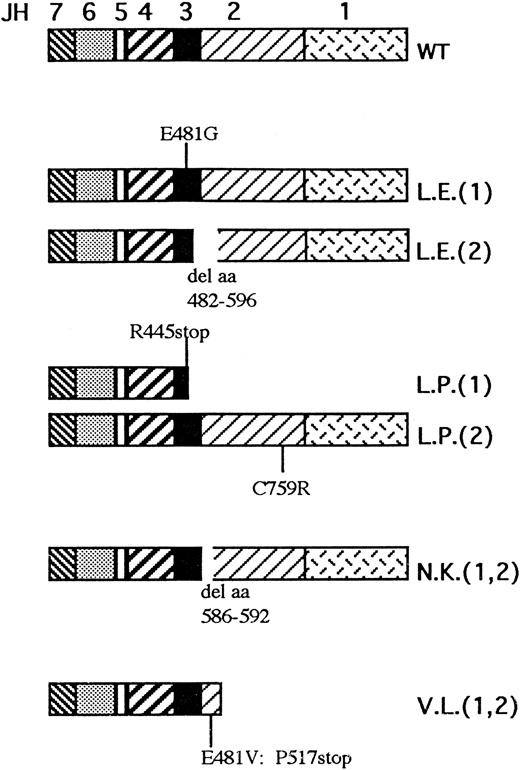
The second mutation in patient L.E. was an A to G transition at nucleotide 1537 found both in the transcript and in genomic clones (Fig 1B). This causes a missense mutation at the codon 481, causing a substitution of glutamic acid to glycine. The search for this point mutation in the maternal DNA by direct sequencing of PCR product and in 12 PCR clones did not show any abnormality, suggesting that it represents a de novo mutation. This mutation lies at the first nucleotide of exon 10, but it evidently does not affect the correct splicing of the transcript. It does not create or abolish any restriction site; therefore, we designed intronic primers around exon 10 (XthF and XthR) and performed SSCP on more than 100 normal chromosomes, none of which showed alteration in the electrophoretic migration, as opposed to that observed in the patient. Taken together, these results suggest that patient L.E. carried two different mutations, leading the first to a shorter protein with an internal deletion including portions of the JH3 and JH2 domains, but with an intact C-terminus, and the second producing a protein of normal size with a single amino acid substitution at codon 481 (Fig 2 and Table 1).
Overview of predicted protein abnormalities occurring in the four JAK3/SCID patients described in the present report. The JH1-7 domains are depicted in different patterns as indicated above the wild-type (WT) protein. The patient initials are indicated beside every mutated allele; the numbers in brackets refer to the two different alleles when the patients are compound heterozygotes.
Overview of predicted protein abnormalities occurring in the four JAK3/SCID patients described in the present report. The JH1-7 domains are depicted in different patterns as indicated above the wild-type (WT) protein. The patient initials are indicated beside every mutated allele; the numbers in brackets refer to the two different alleles when the patients are compound heterozygotes.
Nucleotide Abnormalities of JAK3 Gene in Autosomal SCID Patients
| Patient Allele . | Transcript Abnormality* . | Genomic Abnormality* . | Protein Abnormality . |
|---|---|---|---|
| L.E. 1 | nt 1537: A to G transition | Same | Missense mutation |
| L.E. 2 | Deletion of exons 10, 11, 12 | Genomic deletion | Internally deleted protein |
| L.P. 1 | nt 1428: C to T transition | Same | C-terminally truncated protein |
| L.P. 2 | nt 2370: T to C transition | Same | Missense mutation C579R |
| N.K. | Deletion of 21 nt (1861-1881) | nt 1862: C to T transition (new splice site in exon 12) | Internally deleted protein |
| V.L. | Deletion of 5 nt (1537-1541) | A to G transition at the acceptor splice site of intron 9 | C-terminally truncated protein |
| Patient Allele . | Transcript Abnormality* . | Genomic Abnormality* . | Protein Abnormality . |
|---|---|---|---|
| L.E. 1 | nt 1537: A to G transition | Same | Missense mutation |
| L.E. 2 | Deletion of exons 10, 11, 12 | Genomic deletion | Internally deleted protein |
| L.P. 1 | nt 1428: C to T transition | Same | C-terminally truncated protein |
| L.P. 2 | nt 2370: T to C transition | Same | Missense mutation C579R |
| N.K. | Deletion of 21 nt (1861-1881) | nt 1862: C to T transition (new splice site in exon 12) | Internally deleted protein |
| V.L. | Deletion of 5 nt (1537-1541) | A to G transition at the acceptor splice site of intron 9 | C-terminally truncated protein |
Nucleotide numbers refer to the published cDNA sequence (a.n. U09607).
Patient L.P. Patient L.P. is a compound heterozygote bearing two different mutations. The first is a T to C transition at nucleotide 2370 (Fig 1C), which causes a substitution of cysteine to arginine at codon 759, whereas the second is a C to T transition at nucleotide 1428, creating a stop codon at residue 445 of the cDNA (Fig 1D). Both these mutations were found in several clones and were confirmed by digestion analysis, because the first mutation introduces an Hha I site, whereas the second creates an Alu I site. With regards to the first mutation, to exclude the possibility of a rare polymorphism, we amplified the genomic region with the primer pair 2260F/2440R encompassing the mutated nucleotide. Hha I digestion of specific PCR products from more than 150 independent chromosomes showed the presence of a single band of 320 bp in all cases, whereas the patient and his mother showed three bands of 320, 250, and 70 bp, showing that the mutation was also present on the maternal allele (data not shown). To confirm the second mutation, the pertinent fragment from the genomic DNA of the patient, his parents, and controls was amplified with primers VIIIthF and 1518R and digested with Alu I (data not shown). Four bands were detected in the patient L.P. and his father (both heterozygous for this mutation), whereas a normal pattern of two bands was present in the mother's and normal DNAs. Taken together, these results show that the patient carried a stop mutation inherited by his father and a missense mutation inherited by his mother (Fig 2 and Table 1).
Patient N.K. Because this patient was born to consanguineous parents, we could anticipate that he was the carrier of a homozygous mutation. Sequencing of the RT-PCR product obtained with primer pair 1448F/2140R showed a 21-bp deletion occurring at nt 1861-1881 (inclusive) in 3 of 3 clones (Fig 1F ). To confirm the mutation and its putative homozygosity status, we amplified the transcript with primer pair 1831F/1950R from the patient, his parents, and normal controls. The patient showed a major deleted band (data not shown), whose size is compatible with the 21 nt deletion, which was shared by both parents. The deleted nucleotides are located at the 3′ boundary of exon 12,14 suggesting an abnormality of splicing mechanisms. A C to T transition at nucleotide 1862 within exon 12 was detected at the genomic level (Fig 1E). This mutation creates a new GT dinucleotide in exon 12, 21 bp upstream of the normal donor splice site, which appears to overcome the normal splice site, leading to a transcript that is translated according to the normal reading frame, but in which 7 amino acids in the JH2 domain are deleted (Fig 2 and Table 1).
Patient V.L. SSCP analysis of the region corresponding to the tenth exon amplified using the primer pairs XthF and XthR showed an abnormal migration in the DNA of patient V.L. with disappearance of the normal bands. Direct sequencing of the PCR product of this region showed an A to G transition at the nucleotide −2 of the acceptor splice site of the 9th intron (Fig 1G), which appears to be homozygous, in keeping with the SSCP findings and the known consanguinity of the parents. Sequencing of the transcript in the same region detected a 5-nt deletion, including nt 1537 to 1541 (Fig 1H). Taken together, these findings suggest that, due to the abnormality in the acceptor splice site, a cryptic splice site containing the AG dinucleotide at the exonic position 1540-1541 is used, with a consequent 5-nt deletion that leads to premature termination of the reading frame (Fig 2 and Table 1).
Analysis of JAK3 Protein Expression and Phosphorylation
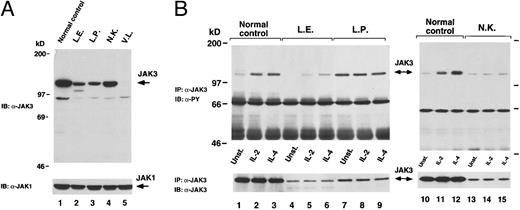
We assessed the presence of the JAK3 protein by Western blot analysis of whole cell lysates obtained from normal control and patients' BCLs by using a rabbit polyclonal antibody raised against the JAK3 C-terminus region4 (Fig 3A). In agreement with the results of the mutation analysis, we found that cells from patient L.E. (Fig 3A, lane 2) expressed two distinct forms of JAK3 protein, one of which showed a markedly reduced molecular weight and was most likely derived from the transcript lacking exons 10, 11, and 12. Of note, the truncated form of JAK3 protein was detected also in lysates of cells obtained from patient L.E.'s father (data not shown). In lysates from patients L.P. (Fig 3A, lane 3) and N.K. (Fig 3A, lane 4), we observed a single JAK3 band of apparently normal molecular weight, although we could not detect a JAK3-specific signal in patient V.L.'s cells (Fig 3A, lane 5). This last finding is not surprising, because from the mutation analysis of patient V.L. we would predict a truncated JAK3 protein lacking the C-terminus region, the region against which the JAK3 antibody was directed. Furthermore, the truncated mutant form of JAK3 protein predicted in patient L.P. cannot be detected using this particular anti-JAK3 polyclonal antibody. The finding of a JAK3 protein of apparent normal molecular weight in patient N.K., whose JAK3 mRNA carries a 21-bp deletion, may be justified by the consideration that changes in size due to a deletion of 7 amino acids can be difficult to appreciate. Of note, the levels of expression of JAK3 detectable in JAK3-SCID patients' cells were decreased in comparison to the normal control cells in all cases.
(A) Analysis of JAK3 expression. Lysates of BCLs obtained from normal control subject (lane 1) or JAK3-deficient patients cells (lanes 2 through 5) were subjected to SDS-PAGE and then immunoblotted (IB) with JAK3 antiserum (a-JAK3, top). The membrane was then stripped and reblotted with JAK1 antiserum (a-JAK1, bottom) to verify equal loading. (B) Analysis of JAK3 phosphorylation. Control (lanes 1 through 3 and 10 through 12) and JAK3-SCID patients cells (lanes 4 through 9 and 13 through 15) were stimulated with the indicated cytokine for 15 minutes, lysed, and immunoprecipitated (IP) with a-JAK3. Complexes were detected by immunoblotting (IB) with monoclonal antibody specific for phosphorylated tyrosine residues (a-PY, top). The membrane was then stripped and the presence of JAK3 in the immunoprecipitates was verified by reblotting with a-JAK3 (bottom).
(A) Analysis of JAK3 expression. Lysates of BCLs obtained from normal control subject (lane 1) or JAK3-deficient patients cells (lanes 2 through 5) were subjected to SDS-PAGE and then immunoblotted (IB) with JAK3 antiserum (a-JAK3, top). The membrane was then stripped and reblotted with JAK1 antiserum (a-JAK1, bottom) to verify equal loading. (B) Analysis of JAK3 phosphorylation. Control (lanes 1 through 3 and 10 through 12) and JAK3-SCID patients cells (lanes 4 through 9 and 13 through 15) were stimulated with the indicated cytokine for 15 minutes, lysed, and immunoprecipitated (IP) with a-JAK3. Complexes were detected by immunoblotting (IB) with monoclonal antibody specific for phosphorylated tyrosine residues (a-PY, top). The membrane was then stripped and the presence of JAK3 in the immunoprecipitates was verified by reblotting with a-JAK3 (bottom).
We then studied the tyrosine phosphorylation of JAK3 in response to cytokine stimulation. Patient V.L. was not eligible for this study because the antibody used for immunoprecipitation4 could not detect JAK3 protein in this patient's cell lysates. As shown in Fig 3B, IL-2 and IL-4 induced tyrosine phosphorylation of JAK3 in normal control cells (Fig 3B, lanes 1 through 3 and 10 through 12), whereas a reduced signal was detectable in patient L.E. cells (Fig 3B, lanes 4 through 6). This may reflect the low amount of JAK3 protein expressed in these cells. In the case of patient L.P., we found that JAK3 was constitutively tyrosine phosphorylated and unresponsive to serum starvation or cytokine stimulation (Fig 3B, lanes 7 through 9). Finally, we did not observe any increase of phosphorylation over baseline levels in N.K. patient cells upon cytokine treatment (Fig 3B, lanes 13 through 15).
Cytokine-Mediated Phosphorylation of Stat5 and Stat6
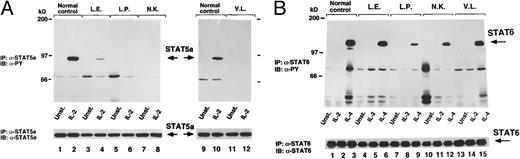
We next examined the consequences of the impairment of JAK3 expression on some of the cytokine-induced biochemical events known to be mediated by JAK3. We first assessed the tyrosine phosphorylation of STAT5 upon IL-2 stimulation. We detected a strong phosphorylation response in normal control cells (Fig 4A, lanes 1, 2, 9, and 10), whereas IL-2 did not induce any effect on the phosphorylation state of STAT5 in cells from JAK3-SCID patients L.P., N.K., and V.L. (Fig 4A, lanes 5 through 8, 11, and 12, respectively). Interestingly, and in agreement with the observed partial conservation of JAK3 phosphorylation (Fig 4A, lanes 4 through 6), we detected a minimal IL-2–induced STAT5 phosphorylation in cells from patient L.E. (Fig 4A, lanes 3 and 4).
(A) Cytokine-induced phosphorylation of STAT5. Control (lanes 1, 2, 9, and 10) and JAK3-deficient patients cells (lanes 3 through 6, 11, and 12) were stimulated with indicated cytokine for 15 minutes, lysed, and immunoprecipitated (IP) with STAT5A-specific antiserum (α-STAT5A). After transferring to a nylon membrane, complexes were immunoblotted (IB) with α-PY (top). The membrane was then stripped and reprobed with a-STAT5A (bottom). (B) Cytokine-induced phosphorylation of STAT6. Control (lanes 1 through 3) and JAK3-deficient patients cells (lanes 4 through 15) were left unstimulated or stimulated with IL-2 or IL-4, lysed, and immunoprecipitated (IP) with STAT6 antiserum (α-STAT6). After electro-transferring, immunocomplexes were immunoblotted (IB) with phosphotyrosine-specific monoclonal antibody (α-PY, top). The same membrane was then stripped and reprobed with α-STAT6 (bottom).
(A) Cytokine-induced phosphorylation of STAT5. Control (lanes 1, 2, 9, and 10) and JAK3-deficient patients cells (lanes 3 through 6, 11, and 12) were stimulated with indicated cytokine for 15 minutes, lysed, and immunoprecipitated (IP) with STAT5A-specific antiserum (α-STAT5A). After transferring to a nylon membrane, complexes were immunoblotted (IB) with α-PY (top). The membrane was then stripped and reprobed with a-STAT5A (bottom). (B) Cytokine-induced phosphorylation of STAT6. Control (lanes 1 through 3) and JAK3-deficient patients cells (lanes 4 through 15) were left unstimulated or stimulated with IL-2 or IL-4, lysed, and immunoprecipitated (IP) with STAT6 antiserum (α-STAT6). After electro-transferring, immunocomplexes were immunoblotted (IB) with phosphotyrosine-specific monoclonal antibody (α-PY, top). The same membrane was then stripped and reprobed with α-STAT6 (bottom).
In a previous study, we showed that IL-4 could induce STAT6 phosphorylation in JAK3 protein-deficient cells from JAK3-SCID patients, albeit to a lesser degree than in normal cells.17 However, to investigate whether the same would be in cells expressing defective JAK3 protein, we assessed the effects of IL-4 on the activation of STAT6 in our JAK3-SCID patients cells, and we found that IL-4–induced STAT6 phosphorylation was clearly detectable in all cases (Fig 4B, lanes 4 through 15), although the level of the response was variable and substantially lower than in normal controls (Fig 4B, lanes 1 through 3).
DISCUSSION
The aim of this work was to thoroughly investigate the biochemical consequences of mutations in JAK3 protein occurring in four patients affected by autosomal SCID (6 abnormal alleles summarized in Fig 2 and Table 1). Two abnormalities in the JAK3 gene introduce premature stop codons that should not give rise to translation products, because usually mRNA-bearing premature stop codons in internal exons are inefficiently processed and no truncated protein is produced.18 The two patients originally described by us7 produced very low or undetectable levels of JAK3 protein and can be considered null mutants. Likewise, the three knockout murine models described are null mutations, completely abrogating the production of JAK3 molecules.9-11 In contrast to these examples, four of the new mutations characterized in the present report allow for the production of abnormal JAK3 molecules that, although expressed at reduced levels, are easily detectable. In this regard, these mutants are instrumental in elucidating some of the steps involved in the JAK3-dependent signal transduction pathways and the role of Janus homology domains, whose function is not yet clear. Two of these mutants give rise to internally deleted proteins, whereas the other two code for proteins with a single amino acid change (Fig 2).
Both the 115- and 7-amino acid (aa) deleted mutants affect the JH2 domain. They are detectable by Western blot analysis, but are not efficiently tyrosine phosphorylated in response to IL-2 and IL-4 (Fig 3). The 7-aa deleted protein, which is produced in reduced, but clearly detectable amounts, is especially interesting, because it emphasizes that this small deletion in the JH2 domain can profoundly affect the function of the JAK3 kinase, rendering it unresponsive to cytokine stimulation. Although the lack of phosphorylation response could be the consequence of the adverse effect of the mutation on the JAK3 protein structure and/or its ability to cooperate with the other components (such as γC chain and JAK1) of the receptor complex activated by cytokine binding, these results also raise the intriguing possibility that the JH2 domain could be involved in the regulation of JAK3 kinase activity. This is supported by another interesting mutation, carried by patient L.P. and located in the JH2 domain, that results in the disruption of the normal mechanisms of JAK3 phosphorylation and dephosphorylation. In this case, the substitution C759R leads to the production of a JAK3 protein that is constitutively phosphorylated and apparently is neither activated by cytokine stimulation (Fig 3B) nor mediates the activation of STAT5 (Fig 4A).
The other JAK3 mutant detectable in lysates from patient L.E. cells is expressed at reduced levels, although it can be phosphorylated in response to IL-2 and IL-4. The low expression of this JAK3 mutant is compatible with a reduced stability due to the E481G substitution in the JH3 region, as shown to occur in the previously described patient G.M.7
Alternatively, the impaired phosphorylation response of this full-length JAK3 mutant could result from the interfering dominant negative effect of the truncated JAK3 protein coexpressed by patient L.E.'s cells. This would obviously have important implications for the efficacy of gene addition therapy approaches that are being developed for JAK3-SCID.19 A reduced amount of phosphorylated STAT5 is detectable in cellular extracts from L.E. patient cells, suggesting the preservation of some residual JAK3 activity. These data are in agreement with the unique immunologic phenotype of this patient who produced a substantial number of T cells (albeit characterized by an aberrant, activated phenotype) that retained, in part, the ability to proliferate in response to IL-2. Delayed development of peripheral blood T autologous cells has been recently reported20 in an X-SCID patient carrying a mutation that results in the expression of a γC chain with a truncated intracytoplasmic tail. This patient did not show IL-2–induced JAK3 phosphorylation, whereas STAT phosporylation was not investigated. As a whole, these observations raise the hypothesis that partial preservation of JAK-STAT signal pathway may allow for T-cell development or that, alternatively, a γC chain/JAK3-independent pathway of T-cell differentiation may cause delayed maturation of T cells. However, our observation and that of Morelon et al20 expand the immunologic spectrum of SCID due to a defective γC -JAK3 signalling pathway and can have important diagnostic implications.
Our findings of a conserved IL-4–mediated phosphorylation of STAT6 in all four JAK3-SCID patients (Fig 4B) confirm and expand previous observations by our group conducted on lymphoblastoid cells of a JAK3-SCID17 and on a series of BCLs derived from XSCID patients20a (Taylor et al, manuscript in preparation) and demonstrating the existence of a γC chain/JAK3-independent pathway that allows the activation of STAT6 in response to IL-4. Altogether, these data are in agreement with the observations by Matthews et al21 demonstrating the conservation of some of the biologic effects of IL-4 on B lymphocytes obtained from XSCID patients, but are in apparent contrast to other studies suggesting that γC chain22 and JAK38 are critical for IL-4–mediated STAT6 activation in human BCLs. However, a series of independent studies has shown that IL-4 can transduce signals in human vascular endothelial and colon carcinoma cells that do not express either γC or JAK3. In these cells, IL-4 is thought to signal through the activation of JAK1 and JAK2.23-27 This does not seem to be the case in B lymphocytes, because, as previously reported,17 we could not detect activation of JAK2 or TYK2 in response to IL-4 in normal control or JAK3-SCID BCLs. In light of the similar biologic effects induced by IL-4 and IL-13 on B cells, it is tempting to speculate that, in the absence of a competent γC chain/JAK3 interaction, IL-4 could inefficiently bind to and signal through the IL-13 receptor, which shares the IL-4 receptor α subunit (IL-4Rα) and comprises at least a second chain (IL-13Rα)28,29 that has not yet been associated to a known kinase of the Janus family in lymphoid cells. Alternatively, it is also conceivable that, in JAK3-SCID (and XSCID) BCLs, IL-4 could suboptimally signal through the homotypic interaction of IL-4Ra chains and JAK1 activation, a phenomenon very recently shown in transfection studies.30
Despite the large number of studies devoted to the elucidation of the mechanisms regulating the signaling of cytokines through the JAK-STAT pathways, further investigation is necessary to understand the implications of such a system in the development of a competent immune system. The findings presented in this work and the availability of cell lines carrying mutations diversely affecting the various steps along the signaling pathway will contribute to the investigation of those aspects that are still obscure and may lead to the discovery of critical links that could be the target of therapeutic intervention for immunity disorders.
ACKNOWLEDGMENT
The authors are grateful to Prof R. Dulbecco and Prof L. Rossi Bernardi for their encouragement. The technical assistance of Enrica Mira Catò and Lucia Susani is also gratefully acknowledged. We also thank Sophie Bevan for her careful typing of the manuscript.
Supported by grants from Biomed 1 (PL 1321 to L.D.N.) and from Telethon Italy (to L.D.N.). This is manuscript no. 7 of the Genoma 2000/ITBA Project funded by CARIPLO.
Address reprint requests to Anna Villa, MD, CNR-ITBA, Via Ampère 56, 20131 Milan, Italy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal