Abstract
In the course of systematic cloning of protein tyrosine kinases (PTKs) expressed in hematopoietic stem and progenitor cells, we have identified the murine homologue of human Bmx. It encodes a protein containing the five domains characteristic of the Tec family of cytoplasmic src-related PTKs: pleckstrin homology (PH), Tec homology (TH), src homology 3 and 2 (SH3 and SH2), and tyrosine kinase (TK). In adults, Bmx expression was found primarily in bone marrow and at a lower level in lung and heart. During fetal development it was also found in the spleen at late stage of gestation and in neonates. Analysis of bone marrow subpopulations showed that Bmx was expressed in the progenitor cell population and maturing hematopoietic cells of the granulo/monocytic lineage where expression increased with maturation and differentiation. At the periphery, a high level of Bmx expression was also found in neutrophils and monocytes/macrophages. Bmx expression was not detected in the primitive hematopoietic stem cell population, and cells of the B-, T-, and erythroid-lineages. It was also not detected in most of the cell lines examined. Our results indicate that Bmx is another member of the Btk/Itk/Tec PTK family, which is predominantly expressed in the granulo-monocytic lineage within the hematopoietic system.
TYROSINE phosphorylation plays a crucial role in the transduction of mitotic and differentiative signals from membrane to nucleus. Among the cytokine receptors, those that contain tyrosine kinase catalytic domains are rare. Examples of these receptors include fms, the M-CSF receptor; kit, the SCF receptor, and the less characterized flt3/flk2, tie1, and tie2 receptors. Others belong to the superfamily of cytokine receptors that is characterized by the structure of the extracellular domain and the presence of proline-rich motif in the intracytoplasmic domain. Although deprived of catalytic function, ligand binding to these receptors still triggers activation of cytoplasmic protein tyrosine kinases (PTKs). This is the starting point of multiple signal transduction pathways, based mainly on phosphorylation-dependent protein activation, as well as interactions between tyrosine-phosphorylated proteins and proteins having affinity for phospho-tyrosines through a SH2 domain.1-3
Among the cytoplasmic PTKs involved in signal transduction, several classes are commonly found. PTKs of the Jak family are directly activated by ligand bound receptors.2 However, specificity of the receptor for each particular Jak kinase is variable. For instance, the interleukin-6 (IL-6) receptor shows little specificity and activates Jak1, Jak2, and Tyk2, whereas the erythropoietin receptor activates only Jak2. Once activated, these Jak kinases phosphorylate proteins of the STAT family, again with various specificity. This triggers their dimerization and translocation to the nucleus. The src family of PTKs can also be activated following receptor stimulation.2 Their role in signal transduction is best understood in the context of the B-cell receptor (BCR).1 3 Antigen binding to the BCR first activates PTKs of the src family. Through the phosphorylation of a few other proteins, this switches on the phospholipase Cγ (PLCγ) pathway, which leads to an increase in intracellular calcium and inositoltriphosphate (IP3), the ras, and the phosphoinositide 3 kinase (PI3K) pathways.
More recently, a new class of cytoplasmic PTKs has emerged, the Tec family of PTKs. The role of the Tec family of PTKs in signal transduction has been best characterized for Btk in the context of the BCR. Btk has been identified as the gene responsible for X-linked agammaglobulinemia (XLA) in humans.4,5 In XLA patients, mutations in Btk lead to the absence of immunoglobulins in peripheral blood due to a severe blockage in early B-cell development. Btk was shown to be normally activated following BCR stimulation,6-8 due to phosphorylation by PTKs of the src family.9,10 Activated Btk is in turn involved in the activation of the PLCγ2 transduction pathway.11 Beside its role in B cells, Btk has also been shown to be involved in IL-5 receptor signaling12,13 and is also activated following stimulation of gp130, the transducing chain of IL-6 receptor.14 Other members of this PTK family may also play a role in signal transduction. Tec itself can be activated following stimulation of kit,15 gp130,14 G-CSF receptor, erythropoietin receptor, and IL-3 receptor.16,17 Another family member, Itk, also plays a role in TCR signaling similar to that of Btk for the BCR,18 as well as a role in CD2819 and FcεRI signaling.20 21
Our laboratory has been involved in the purification of hematopoietic stem and progenitor cells by fluorescence activated cell sorting (FACS) and their characterization.22 As the molecular mechanisms governing the proliferation and differentiation of these cells have not been well characterized, we attempted to gain some insight through the identification of PTKs expressed by these cells, which may play a role in hematopoietic cell development. Among three new sequences identified, one was found to be the murine homologue of human Bmx, a member of the Tec family of cytoplasmic PTKs that has not been yet fully characterized. Murine Bmx was found to be predominantly expressed in bone marrow, lung, and heart. Within the hematopoietic system, Bmx expression was observed at the level of progenitor populations, in maturing cells along the granulo-monocytic lineage, and in terminally differentiated neutrophil and macrophage populations.
MATERIALS AND METHODS
Cells
Rh123lo Lin− Kit+ Ly6A+ primitive hematopoietic stem cells and Rh123hi Lin− Kit+ Ly6A− progenitor cells were isolated from C57BL/6 bone marrow using FACS technique, as described previously.22 Briefly, this involved three major sequential steps: Rhodamine 123 staining, immuno-magnetic bead depletion of mature lineage positive cells, and staining with biotinylated anti-kit ACK2 and allophycocyanin-conjugated anti-Ly6A E13-161.7 monoclonal antibodies (MoAbs) followed by Texas Red-avidin. The MoAbs and fluorochromes used for immunodepletion and immunofluorescence labeling are all specified elsewhere.22
To obtain various subpopulations of the myelo-monocytic lineage, single cell suspension of adult bone marrow were prepared from femurs and tibia of C57BL/6 mice. After a brief incubation with 20 μg rat Ig, the cells were first stained with phycoerythrin (PE)-conjugated anti-kit 3C1 (Caltag Lab, Burlingame, CA) and either biotinylated anti-Gr-1 RB6-8C5 or anti-Mac-1 M1/70.15.11.5 MoAbs, then with FITC-avidin (Progen Industries, Ltd, Darra, Australia). FACS was performed using an unmodified FACS-Vantage instrument (Becton Dickinson Immunocytometry Systems, San Jose, CA). Gates were set as indicated on Fig 3. After completion of cell sorting, the deflected cells were centrifuged at 1,500 rpm for 5 minutes, resuspended, and counted using a hematocytometer.
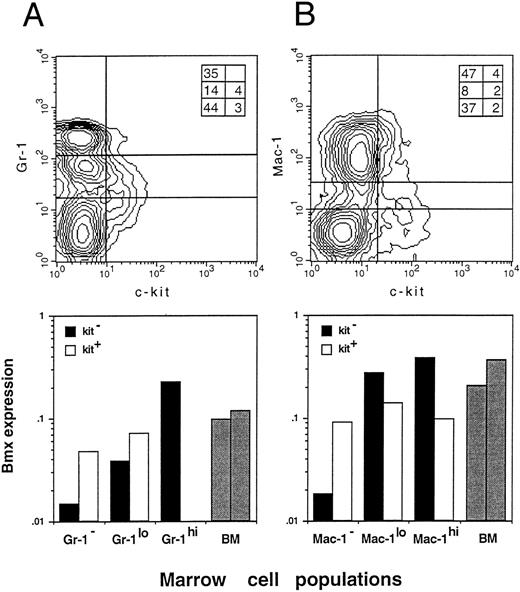
Bmx expression in Gr-1, Mac-1, and Kit sorted bone marrow cells. Bone marrow cells were sorted by FACS for the expression of Gr-1 and Kit (A) or Mac-1 and Kit (B). Populations separated by lines on the density plot (top panels) were isolated and lyzed. Inserts in the right upper corner of both density plots show the percentage of individual fractions within the normal bone marrow. Bmx expression (bottom panels) is presented as the ratio of Bmx to actin signals. Expression in total bone marrow is indicated with gray bars.
Bmx expression in Gr-1, Mac-1, and Kit sorted bone marrow cells. Bone marrow cells were sorted by FACS for the expression of Gr-1 and Kit (A) or Mac-1 and Kit (B). Populations separated by lines on the density plot (top panels) were isolated and lyzed. Inserts in the right upper corner of both density plots show the percentage of individual fractions within the normal bone marrow. Bmx expression (bottom panels) is presented as the ratio of Bmx to actin signals. Expression in total bone marrow is indicated with gray bars.
Peripheral blood of 6-week-old C57BL/6 mice was fractionated by centrifugation over a Ficoll cushion for 30 minutes at 1,800 rpm. Polymorphonuclear neutrophils were harvested from the pellet. Mononuclear cells harvested at the interphase of the Ficoll were incubated for 2 hours in Dulbecco's modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum (FCS) at 37°C and the adherent macrophage fraction was harvested. Resident peritoneal macrophages were prepared by washing the peritoneal cavity with phosphate buffered saline (PBS). Pulmonary alveolar macrophages were prepared by aspirating the lung through the treachea with PBS as described.23
Bone marrow macrophages were obtained as described.24 Briefly, 6 × 107 bone marrow cells were incubated at a density of 6 × 105 cells/mL in RPMI, 5 × 10−5 mol/L 2-mercaptoethanol, 20 mmol/L HEPES, 15% FCS, and 500 U/mL M-CSF. On day 3 of culture, nonadherent cells were harvested and incubated further at a density of 105 cells/mL in fresh medium with M-CSF. On day 9, adherent cells were harvested and lyzed for RNA preparation.
Transplantation. C57BL/6 mice at 10 to 12 weeks of age were used and kept in the Queensland Institute of Medical Research animal facility. Recipient animals were exposed to X-ray (9 Gy whole body irradiation) generated by a Toshiba X-ray machine model KXC-19-2 (Shibaura Electric Co Ltd, Tokyo, Japan) operated at 200 kV, 14 mA: half value layer was 1.8 mm copper, and a focal distance of 25 cm with full backscatter conditions was used at a dose rate of 112.5 rads/min.
Irradiated animals were transplanted with 106 nucleated bone marrow cells intravenously in 0.3 mL PBS within 3 hours of irradiation. They were sacrified at indicated time points and spleens were removed for RNA preparation. In the absence of graft, irradiated mice died within 10 to 12 days postirradiation.
RNA preparation and analysis. Total RNA from FACS-sorted and cultured cells was prepared by in situ lysis in guanidium-thiocyanate and phenol-chloroform extraction, using Total RNA Isolation kit (Advanced Biotechnologies Ltd, Surrey, UK). Total RNA from tissues and cell lines was prepared by lysis in guanidium-thiocyanate buffer and centrifugation over a caesium chloride cushion,25 and quantified by spectro-photometry. For reverse transcription polymerase chain reaction (RT-PCR) experiments, RNA from variable numbers of FACS-sorted cells or 0.5 μg RNA from tissues was reverse-transcribed using Moloney murine leukemia virus (Mo-MuLV) reverse transcriptase (GIBCO-BRL, Life Technologies, Melbourne, Australia) and random hexamers as primers in a final volume of 20 mL. One tenth of the reaction mixture was used for PCR in a final volume of 25 μL.
For systematic amplification of PTKs from hematopoietic stem and progenitor cells, RNA from 1.7 × 104 Rh123lo Lin− Kit+ Ly6A+ cells and 8 × 104 Rh123hi Lin− Kit+ Ly6A− cells were prepared. Residual DNA was digested with DNase RQ1. A 35-cycle PCR was performed with primers PTK1 (CGGGATCCAYCGNGAYYT NGC N GCNMG) and PTK2 (GGCTCTAGAYNCCRWARSWCCANACRTC). One hundredth of the reaction mixture was further amplified for 20 cycles using primers PTK1 and PTK3 (CGGAATTCYTCNRGNGCNRTCCAYTT). Both PCR reactions used 37°C as an annealing temperature. The 164- to 172-bp PCR products were gel purified, cloned into the Bluescript plasmid (Stratagene, CA), and individual clones were isolated and sequenced.
For expression analysis, Bmx and actin were amplified from adult and fetal tissues using 20 cycles of PCR. For other samples, the number of cycles was adapted so that PCR products were not visible after ethidium bromide staining of the gel. Bmx and actin were amplified from various number of bone marrow cells and from hematopoietic progenitors with 30 and 20 cycles, respectively; from spleens of irradiated mice with 20 and 10 cycles, respectively; from various bone marrow subpopulations based on Mac-1/Kit and Gr-1/Kit expression and mature hematopoietic cells with 25 and 15 cycles, respectively. Annealing temperature was 60°C. Bmx primers Pr1 (GAACTGTTTGGTG GACAG) and Pr2 (TGGAAACTTGGTTCCTACT) and actin primers Act1 (GACATG GAGAAGATCTGGCA) and Act2 (GGTCTTTACGGATGTCAACG) gave a single PCR product of 106 bp and 637 bp, respectively.
For Southern blots, 10 μL of Bmx and actin PCR products were run on 1.8% and 1.2% agarose gels, respectively. For Northern blots, 4 mg of tissue RNA was run on a formaldehyde agarose gel. Where indicated, DNA or RNA in the gel were stained with 0.5 μg/mL ethidium bromide. DNA or RNA was transferred onto Nylon membrane (HybondN; Amersham, Buckinghamshire, UK) and hybridized overnight with at least a 10-fold molar excess of probes at 65°C in 5× SSPE, 5× Denhardt's solution, 0.1 mg/mL salmon sperm DNA, and 0.5% sodium dodecyl sulfate (SDS). DNA probes were labeled by random priming using 32P-dCTP with a specific activity of 2.5 × 108 cpm/μg. The 1L4a PTK probe was from nt 1689 to 1794, the murine Bmx probe from nt 1435 to 1870, and the actin probe was a 640-bp Taq1 fragment of murine actin cDNA. Hybridization signals were quantified using PhosphorImager apparatus (Molecular Dynamics, Sunnyvale, CA). Bmx expression is given in arbitrary units as the ratio of Bmx to actin signal.
Cloning of murine Bmx. For isolation of cDNA clones, 2.6 × 106 pfu of a 15-day-old mouse embryo cDNA library (Clontech, Palo Alto, CA) were screened by PCR, using Bmx primers Pr1 and Pr2, as described.26 One clone was detected, isolated, and subcloned into the Bluescript plasmid. Sequencing was performed on both strands.
The 5′ rapid amplification of cDNA ends (RACE) technique was performed on total RNA from murine bone marrow using the Marathon cDNA Amplification kit (Clontech). Briefly, cDNA was synthesized by reverse transcription using oligo(dT) primers and ligated to an adaptor sequence. Bmx cDNA was then amplified by PCR from the Marathon library using an adaptor-specific primer AP1 and Bmx primer Pr2 for 30 cycles. The PCR product was further amplified for 30 cycles using an internal adaptor-specific primer AP2 and Bmx primer Pr3 (TCTGGGCCTCCTGAAAGA), which is located at the 5′ end of the cDNA library clone. The unique Bmx-specific PCR product was cloned into the Bluescript plasmid and sequenced. As mutations may have been introduced through these extensive amplification cycles, the sequence was recloned by performing a 30-cycle PCR directly on mouse bone marrow RNA using Bmx primers Pr4 (ACAGAAGGAGC CCAAATGGA) and Pr3. A single PCR product was obtained and cloned into the Bluescript plasmid. Four independent clones were isolated and individually sequenced on both strands.
RESULTS
In an attempt to identify PTKs involved in hematopoietic cell growth and development, an RT-PCR approach was employed using primers targeting the highly conserved tyrosine kinase domain. Two hematopoietic cell populations purified by FACS technique from bone marrow were used: primitive hematopoietic stem cells (Rh123lo Lin− c-kit+ Ly6A+) and the more mature progenitor cells (Rh123hi Lin− c-kit+ Ly6A−).22 Table 1 shows a compilation of three independent experiments with a total of 141 clones analyzed. Seventeen different sequences were encountered, including 4 known receptor-type and 10 cytoplasmic PTKs. The remaining three are novel sequences: PTK 4-20, 8-31, and 1L4a. After further analysis, PTK 4-20 (Genbank U52463) was shown to be transcribed from ψFGFR-3, a novel pseudogene of FGFR-3, in the antisense orientation compared to FGFR-3. This gene is localized on chromosome 1 and expressed primarily during fetal development.27 Sequencing of 8-31 cDNAs and expression studies indicated that it is the murine homologue of human Abelson-related gene (Arg) and it is expressed ubiquitously in both adult and fetal tissues (Genbank U40827, unpublished results). PTK 1L4a was amplified only from the progenitor cell fraction. This 160-bp sequence exhibited maximum homology with murine Btk (77%) and human Bmx (89%), two members of the Tec family of PTKs.
Sequences Identified by Systemic PCR Amplification of PTKs From Stem and Progenitor Cells
| . | Primitive Stem Cells . | Progenitor Cells . | ||||
|---|---|---|---|---|---|---|
| . | . | . | . | . | . | . |
| . | Rh123lo . | Lin− Kit+ . | Ly6A+ . | Rh123hi . | Lin− Kit+ . | Ly6A− . |
| . | . | . | ||||
| . | . | . | . | . | . | . |
| PTK . | I . | II . | III . | I . | II . | III . |
| FGFR | 0 | 1 | 4 | 0 | 0 | 0 |
| flk2/flt3 | 1 | 6 | 6 | 0 | 1 | 1 |
| kit | 0 | 0 | 1 | 0 | 1 | 2 |
| c-fms | 5 | 1 | 0 | 11 | 4 | 2 |
| c-abl | 7 | 0 | 6 | 2 | 7 | 3 |
| ferT | 3 | 3 | 3 | 3 | 3 | 3 |
| csk/ctk | 0 | 3 | 1 | 2 | 1 | 2 |
| syk | 0 | 1 | 0 | 0 | 1 | 1 |
| mushyk | 1 | 0 | 0 | 0 | 0 | 0 |
| musfak | 1 | 0 | 0 | 1 | 0 | 0 |
| pTKa | 0 | 0 | 0 | 2 | 0 | 1 |
| pTKd | 0 | 0 | 0 | 0 | 1 | 0 |
| pTKf | 0 | 0 | 0 | 2 | 0 | 0 |
| csf1 | 0 | 0 | 0 | 0 | 0 | 3 |
| 4.20 | 1 | 0 | 0 | 0 | 0 | 0 |
| 8.31 | 7 | 6 | 1 | 1 | 4 | 1 |
| 1L4a | 0 | 0 | 0 | 0 | 3 | 1 |
| Total | 26 | 21 | 22 | 24 | 26 | 22 |
| . | Primitive Stem Cells . | Progenitor Cells . | ||||
|---|---|---|---|---|---|---|
| . | . | . | . | . | . | . |
| . | Rh123lo . | Lin− Kit+ . | Ly6A+ . | Rh123hi . | Lin− Kit+ . | Ly6A− . |
| . | . | . | ||||
| . | . | . | . | . | . | . |
| PTK . | I . | II . | III . | I . | II . | III . |
| FGFR | 0 | 1 | 4 | 0 | 0 | 0 |
| flk2/flt3 | 1 | 6 | 6 | 0 | 1 | 1 |
| kit | 0 | 0 | 1 | 0 | 1 | 2 |
| c-fms | 5 | 1 | 0 | 11 | 4 | 2 |
| c-abl | 7 | 0 | 6 | 2 | 7 | 3 |
| ferT | 3 | 3 | 3 | 3 | 3 | 3 |
| csk/ctk | 0 | 3 | 1 | 2 | 1 | 2 |
| syk | 0 | 1 | 0 | 0 | 1 | 1 |
| mushyk | 1 | 0 | 0 | 0 | 0 | 0 |
| musfak | 1 | 0 | 0 | 1 | 0 | 0 |
| pTKa | 0 | 0 | 0 | 2 | 0 | 1 |
| pTKd | 0 | 0 | 0 | 0 | 1 | 0 |
| pTKf | 0 | 0 | 0 | 2 | 0 | 0 |
| csf1 | 0 | 0 | 0 | 0 | 0 | 3 |
| 4.20 | 1 | 0 | 0 | 0 | 0 | 0 |
| 8.31 | 7 | 6 | 1 | 1 | 4 | 1 |
| 1L4a | 0 | 0 | 0 | 0 | 3 | 1 |
| Total | 26 | 21 | 22 | 24 | 26 | 22 |
Results of three independent experiments (I, II, and III) are presented.
Cloning of Bmx cDNA. We next designed 1L4a-specific primers, Pr1 and Pr2, to screen a mouse bone marrow and a mouse 15-day-old embryo cDNA library following the PCR-based technique described by Israel.26 One cDNA of 1289 bp was obtained from the mouse embryo library despite the observation that 1L4a message is more abundant in bone marrow (see expression data in later section). Sequence analysis revealed that this clone contained the 3′ end of an ORF followed by a 3′ untranslated region (Fig 1A). To obtain the 5′ moiety of the ORF, a 5′RACE experiment was performed using mouse bone marrow RNA and both primers Pr 2 and Pr3, which is corresponding to the 5′ end of the cDNA clone obtained previously. A single PCR product, which contained all of the 5′ ORF region, was obtained (Fig 1A).
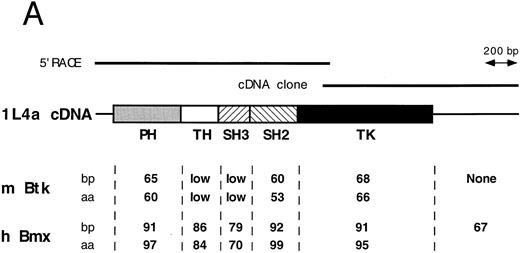
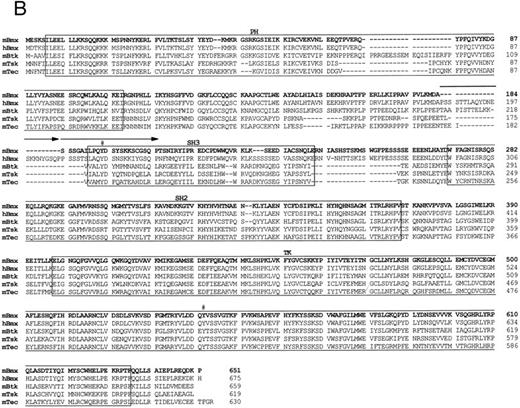
Comparison of 1L4a cDNA with members of the Btk family of PTK. (A) Schematic representation of 1L4a cDNA. Structure of 1L4a cDNA was drawn in accordance with the sequence of the 5′ RACE and the cDNA library clones. Lines and boxes represent the UTRs and ORF, respectively. Names of the protein domains encoded by the cDNA are indicated below. Homology with murine Btk and human Bmx is shown for each domain as % of identity at the bp and aa levels. (B) Alignment of the amino-acid sequences of 1L4a cDNA, human Bmx, murine Btk, Tsk, and Tec. Gaps (-) have been introduced for optimal alignment. The different protein domains have been framed. Arrows show partial repeat of the SH3 domain in human Bmx. Potentially phosphorylated tyrosines 194 and 542 are indicated with #.
Comparison of 1L4a cDNA with members of the Btk family of PTK. (A) Schematic representation of 1L4a cDNA. Structure of 1L4a cDNA was drawn in accordance with the sequence of the 5′ RACE and the cDNA library clones. Lines and boxes represent the UTRs and ORF, respectively. Names of the protein domains encoded by the cDNA are indicated below. Homology with murine Btk and human Bmx is shown for each domain as % of identity at the bp and aa levels. (B) Alignment of the amino-acid sequences of 1L4a cDNA, human Bmx, murine Btk, Tsk, and Tec. Gaps (-) have been introduced for optimal alignment. The different protein domains have been framed. Arrows show partial repeat of the SH3 domain in human Bmx. Potentially phosphorylated tyrosines 194 and 542 are indicated with #.
The compiled sequence contained a 1,980-bp long ORF starting with two inframe ATG at position 130 and 142 (Fig 1B). The second ATG was assumed to be the site for initiation of translation, as it was surrounded by a better Kozak consensus sequence. On comparison with the murine databank, this sequence presented 63% identity with Btk in the coding region and little identity in the untranslated region. The encoded protein contains the most characteristic domains of this PTK subfamily: a PH, an SH2, an SH3, and a TK domain. However, the TH domain was poorly conserved. When compared with PTKs of other species, homology was even higher with human Bmx,28 with 89% identity in the ORF and 67% in the 3′ untranslated region. At the amino acid level, highest identity was seen in the PH (97%), SH2 (99%), and TK (95%) domains. We therefore concluded that 1L4a was the murine homologue of human Bmx and renamed as murine Bmx.
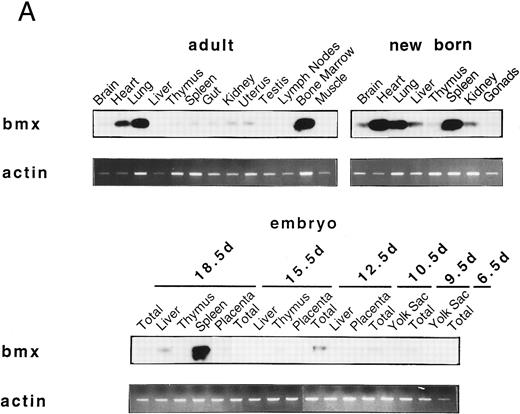
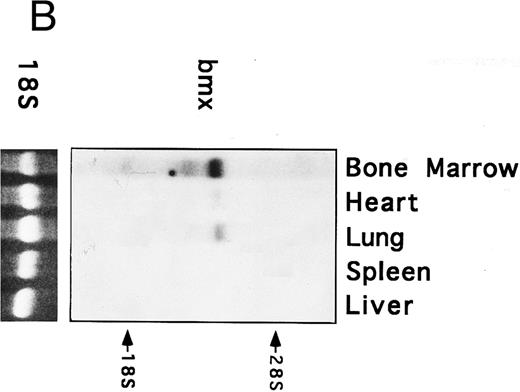
Bmx expression in murine tissues and cell lines. To investigate the expression pattern of Bmx, equal amounts of RNA from various adult, newborn, and fetal mouse tissues were subjected to RT-PCR using primers Pr1 and Pr2. These primers specifically amplified a 106-bp or a 2-kb fragment using cDNA or genomic DNA as a template, respectively (data not shown). This indicated the presence of at least one intron in this region and enabled us unambiguously to identify the amplified cDNA. During fetal development, Bmx expression was very low in most of the tissues examined, including hematopoietically active tissues such as yolk sac or fetal liver (Fig 2A). High level of Bmx expression was first seen in the spleen at day 18.5 of gestation. In newborns, abundant Bmx message was found in the spleen, heart, and lung and at a lower level in kidney, liver, and brain. In adult, its expression was primarily detected in the bone marrow and to a lesser extent lung and heart. Bmx was not detected in any of the lymphoid tissues examined, such as thymus, lymph nodes, and spleens. Northern blot analysis was also performed on a range of the adult tissues (Fig 2B). Bmx was expressed as a 3.5-kb message, and its relative abundance in bone marrow, lung, and heart was similar to previous results obtained using RT-PCR technique.
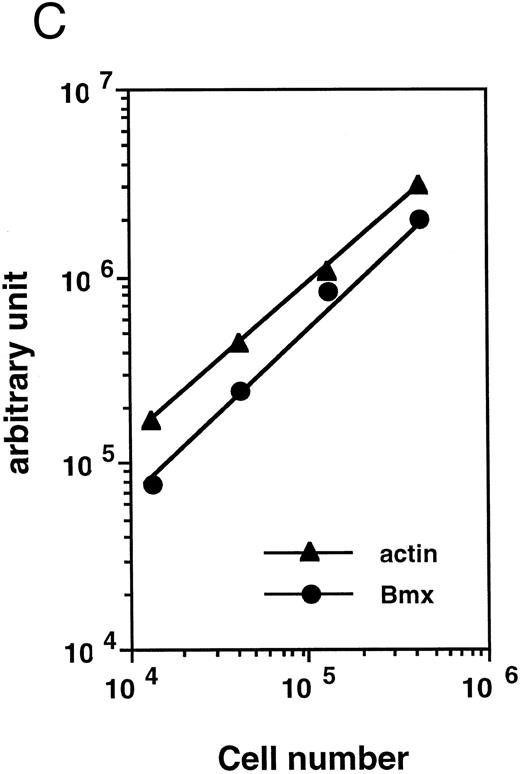
Bmx expression in tissues. (A) Analysis of Bmx expression in adult and fetal tissues by RT-PCR. RNA purified from indicated tissues were amplified by RT-PCR using Bmx primers Pr1 and Pr2. PCR products were transferred onto Nylon membrane after separation on agarose gel and hybridized with 32P-labeled PTK 1L4a probe (top panel). As a control of cDNA quality, actin was amplified from the same cDNA samples using Act1 and Act2 primers. PCR products were stained with ethidium bromide after separation on agarose gel (bottom panel). (B) Northern blot analysis of Bmx expression. RNA purified from indicated adult tissues were transferred onto Nylon membrane after separation on agarose formaldehyde gel and hybridized with 32P-labeled murine Bmx probe. Position of rRNA is indicated. Ethidium bromide-stained 18S RNA on the membrane is shown below. (C) Quantitation of Bmx and actin RT-PCR products as a function of the number of bone marrow cells. Signals were quantified with PhosphorImager.
Bmx expression in tissues. (A) Analysis of Bmx expression in adult and fetal tissues by RT-PCR. RNA purified from indicated tissues were amplified by RT-PCR using Bmx primers Pr1 and Pr2. PCR products were transferred onto Nylon membrane after separation on agarose gel and hybridized with 32P-labeled PTK 1L4a probe (top panel). As a control of cDNA quality, actin was amplified from the same cDNA samples using Act1 and Act2 primers. PCR products were stained with ethidium bromide after separation on agarose gel (bottom panel). (B) Northern blot analysis of Bmx expression. RNA purified from indicated adult tissues were transferred onto Nylon membrane after separation on agarose formaldehyde gel and hybridized with 32P-labeled murine Bmx probe. Position of rRNA is indicated. Ethidium bromide-stained 18S RNA on the membrane is shown below. (C) Quantitation of Bmx and actin RT-PCR products as a function of the number of bone marrow cells. Signals were quantified with PhosphorImager.
We next performed both Northern blot and RT-PCR to assess the expression of Bmx in a panel of murine cell lines: a pre-T lymphocytic BW5147; a cytotoxic T lymphocytic CTLL-2; a pre-B lymphocytic BAF-3; a B lymphocytic WEHI231; four myeloid FDCP-1, M1, WEHI-3BD+, G32D; a macrophage RAW309; a mast cell MC9; three stromal PA6, K062 and BAd and two fibroblastic cell lines NIH3T3 and Balb/c3T3. Apart from the PA6 murine stromal cell line, we did not detect Bmx message in any of the cell lines examined. In PA6 cells, the level of Bmx expression was about fivefold less than that of total bone marrow cells (data not shown).
Bmx expression in adult bone marrow subpopulations. We next examined the expression of Bmx, by RT-PCR, in different hematopoietic subpopulations within the marrow. To quantitate Bmx expression between different cell populations, expression of both Bmx and actin was measured and expressed as Bmx/actin ratio. Figure 2C shows a cell dose-dependent detection of both Bmx and actin PCR products over 104 − 5 × 105 marrow cells, thus validating the quantitation of Bmx expression as the ratio of Bmx to actin signal.
Results from three independent experiments showed that Bmx expression was fivefold to ninefold lower in progenitor population compared to total bone marrow and was not detected in the primitive stem cell population (data not shown). As the progenitor cell population represent only 4% to 7% of total bone marrow cells,22 these results indicate that hematopoietic progenitors are not the only cell population that express Bmx within the marrow.
To obtain further insight into which other cell populations may express Bmx in the marrow, individual myeloid, erythroid, and lymphoid populations were isolated by FACS, based on the presence of lineage-specific markers at their surface, and analyzed by RT-PCR. Expression was observed mainly in individual B220−, Ter119−, Gr-1+, and Mac-1+ fractions, suggesting that Bmx is predominantly expressed in the granulo-monocytic lineage (data not shown). As expression of Gr-1 antigen in the marrow is directly correlated with granulocytic differentiation and maturation,29 while kit is mainly expressed in stem and progenitor cells,22,30 31 we then used Gr-1– and kit-specific antibodies to subfractionate the bone marrow population (Fig 3A, upper panel). The proportion of individual fractions isolated for the expression study is shown in the insert at the upper right corner. The kit+ population (either Gr-1− or Gr-1lo) expressed moderate level of Bmx (Fig 3A, lower panel). The level of Bmx expression seen in the kit+ Gr-1− population was similar to that observed in the Rh123hi Lin− c-kit+ Ly6A− progenitor cell population. In agreement with previous observations, the kit− Gr-1− population, which contains mainly lymphoid and erythroid cells in the marrow, showed little Bmx expression. High level of Bmx expression was observed in the kit− Gr-1hi population, which contains mainly mature neutrophils upon microscopic examination, whereas the kit− Gr-1lo maturing granulocytic population expressed intermediate levels of Bmx.
Similar experiments were performed using the Mac-1– and kit-specific antibodies for the fractionation. Mac-1 is expressed on both granulocytes and monocytes/macrophages, though it is commonly referred as a cell surface marker for the latter cell population.32 33 The results are shown in Fig 3B with the insert in the upper panel showing the percent of individual fractions in the marrow. Similar to the results obtained from previous experiments, moderate level of Bmx expression was seen in all three kit+ subpopulations. The kit− Mac-1− population expressed very low level of Bmx and high level of Bmx expression was seen in both kit− Mac-1lo and kit− Mac-1hi populations.
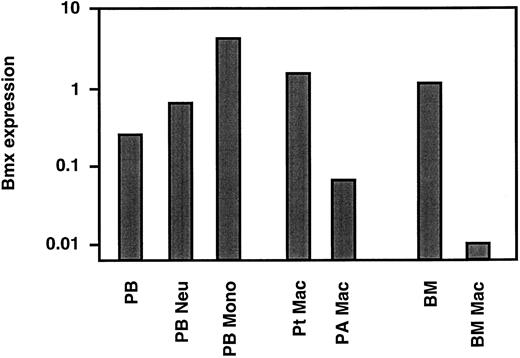
Bmx expression in mature neutrophils and monocyte/macrophages. The above studies showed that in the marrow Bmx was expressed mainly in cells of the granulo-monocytic lineage and increased with differentiation and maturation. We next investigated Bmx expression in mature neutrophils and monocytes in the circulation. Neutrophil and monocyte populations were isolated from peripheral blood as described in Materials and Methods. RT-PCR analysis showed that, in comparison to the total peripheral nucleated blood cells, Bmx expression was approximately 2.5- and 17-fold higher in neutrophil and monocyte fractions, respectively (Fig 4). Bmx expression was also detected in resident macrophages isolated from the peritoneal cavity. In contrast, pulmonary alveolar macrophages and primary cultures of bone marrow macrophages expressed little Bmx (Fig 4).
Bmx expression in mature hematopoietic cells. Bmx expression was measured in total nucleated peripheral blood cells (PB), peripheral blood neutrophils (PB Neu), peripheral blood monocytes (PB Mono), peritoneal macrophages (Pt Mac), pulmonary alveolar macrophages (PA Mac), total bone marrow (BM) and bone marrow derived macrophages in cultures (BM Mac), as for Fig 3.
Bmx expression in mature hematopoietic cells. Bmx expression was measured in total nucleated peripheral blood cells (PB), peripheral blood neutrophils (PB Neu), peripheral blood monocytes (PB Mono), peritoneal macrophages (Pt Mac), pulmonary alveolar macrophages (PA Mac), total bone marrow (BM) and bone marrow derived macrophages in cultures (BM Mac), as for Fig 3.
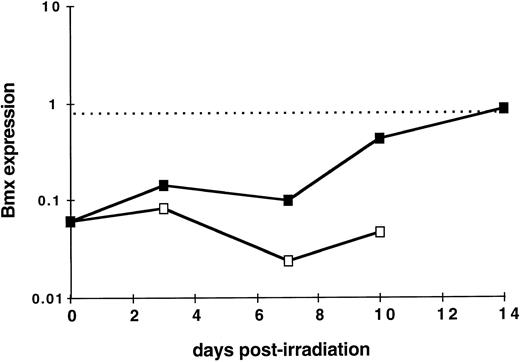
Bmx expression during hematopoietic reconstitution in vivo. As Bmx expression was seen increasing with expression of Gr-1 and Mac-1 differentiation markers, we next investigated whether it would be correlated with in vivo hematopoietic activity. We took advantage of the observation that normal adult spleen showed little expression of Bmx but it will become hematopoietically active when the mice receive exogenous bone marrow cells after lethal irradiation. Bone marrow transplantation experiments were performed on lethally-irradiated mice. At days 3, 7, 10, and 14, mice were sacrified, RNA were prepared from the spleens, and Bmx expression was measured. Figure 5 shows that in normal adult mice, Bmx expression detected in the spleen was at least 13-fold lower than that in the marrow. After receiving bone marrow graft, Bmx expression in the spleen remained low for up to day 7, then increased steadily until day 14 where it reached a level similar to that observed in normal adult marrow.
Kinetics of Bmx expression in the spleen of irradiated mice after bone marrow graft. Mice were irradiated and grafted with 106 bone marrow cells (▪) or no cells (□). At indicated days postirradiation, mice were sacrified, RNA was prepared from the spleens, and Bmx expression was measured as for Fig 3. Mean Bmx expression in normal bone marrow is indicated as a dashed line.
Kinetics of Bmx expression in the spleen of irradiated mice after bone marrow graft. Mice were irradiated and grafted with 106 bone marrow cells (▪) or no cells (□). At indicated days postirradiation, mice were sacrified, RNA was prepared from the spleens, and Bmx expression was measured as for Fig 3. Mean Bmx expression in normal bone marrow is indicated as a dashed line.
DISCUSSION
In an attempt to identify PTKs involved in the hematopoietic cell development, we have cloned a novel murine PTK of the Tec family. As its cDNA was 79% to 92% identical to human Bmx over the whole ORF and 67% identical over its 3′ untranslated region, we concluded that it is the murine homologue of human Bmx. Like human Bmx, the predicted murine Bmx protein contains all of the characteristic features shared by members of the Tec family of PTKs: it has a PH, a TH, an SH3, an SH2, and a TK domain but lacks myristylation signals and C-terminal tyrosine residue corresponding to Y527 of c-src. Interestingly, the partial duplication of the SH3 domain present in human Bmx is absent in murine Bmx. A peculiarity of both murine and human Bmx compared to other members of the family is that only the 5′ moiety of the TH domain is conserved while the proline-rich region present in the 3′ moiety is absent. This 3′ region contains motifs that have previously been shown to be involved in interactions with SH3 containing proteins.15,34,35 In the case of Btk, two tyrosines have been shown to be crucial during activation, Y551 and Y223. The former tyrosine residue is in the TK domain and can be transphosphorylated by Lyn kinase.10 The resulting activated Btk then autophosphorylates Y223 within its SH3 domain.36 Interestingly, both tyrosine residues have been conserved in Bmx and may also play a regulatory role in its activation.
All PTKs belonging to the Tec family are abundantly expressed in hematopoietic tissues, though some are also found in other tissues. For example, Tec is expressed in a wide range of hematopoietic cells, but it is also found in liver, heart, kidney, and ovary.37 Btk is expressed mostly in B cells at all stages of development,4 but is also found in monocytes, lung, and pancreas.5,38 So far, Itk and Txk expression have only been reported in T cells.39-42 Our current study on murine Bmx confirms and extends previous observations with human Bmx28,43 in that its expression in hematopoietic cells is found predominately in cells of granulo-monocytic lineage but not in cells of the B-, T-, and erythroid lineages. We further show that murine Bmx is not detected in the primitive hematopoietic stem cells but in the more mature progenitor cells (Rh123hi Lin− Kit+ Ly6A−, Kit+ Gr-1−, Kit+ Gr-1lo, Kit+ Mac-1−, Kit+ Mac1lo ) and in the kit− Gr-1+, kit− Mac-1+ granulocyte-monocyte populations. In contrast to human, both mature neutrophils and monocytes express Bmx in mice whereas human Bmx is not detected in the monocyte preparation.43 In addition, murine Bmx is only seen in spleen taken from neonates and at late stage of gestation (eg, day 18.5) but not from normal adult mice. These observations indicate that murine Bmx is a myeloid cell specific member of the Tec-family of PTKs.
Studies in human have indicated that Bmx expression could also be detected in muscle-forming tissues such as skeletal muscle and heart, and in umbilical vein endothelial cells. We also found murine Bmx expression in nonhematopoietic tissues such as newborn and adult heart and lung (see Fig 2) and in PA6 murine stromal cells. We have further excluded the broncho-alveolar macrophages as Bmx-expressing cells in the lung (see Fig 4). The cell types that express Bmx in these two tissues remain yet to be identified.
During fetal development, Bmx expression was first detected in the spleen at late stage of gestation and in newborns and was not detected in most of other fetal tissues examined including yolk sac and fetal liver. Spleen has been well described as a transitory hemopoietic organ around birth when hematopoiesis shifts from fetal liver to adult marrow. In particular, granulopoiesis predominates over erythropoiesis after day 17 of gestation.44 Expression of Bmx found in spleens at this transitional stage may thus be related to this activity. This is also consistent with the expression detected in spleens of adult irradiated mice after bone marrow transplantation, a situation where spleen becomes the major hematopoietic organ during early phase of hematopoietic regeneration. The absence of Bmx expression in fetal liver raises the possibility that myeloid cells from fetal liver and fetal spleen may be of different ontogeny or that the two fetal hematopoietic organs provide different microenvironment for the expression of Bmx. Investigation into these possibilities are in progress.
In contrast to most other cytoplasmic PTKs, Bmx expression is absent or low in most of the cell lines examined. Though Bmx expression can be readily detected in freshly isolated progenitor cells, it could not be detected in their progeny after 7 to 10 days of cultures in the presence of various colony-stimulating factors where extensive proliferation and granulo-monocytic differentiation occur (data not shown). Thus, Bmx seems not to be required for ligand-receptor mediated mitogenic/differentiation signaling pathways for the myeloid cells in vitro. However, transplantation experiments following marrow ablation demonstrate that Bmx expression is closely associated with the proliferation and differentiation of hematopoietic stem and progenitor cells in vivo. The function of Bmx in signal transduction during granulo-monocytic differentiation remains to be elucidated.
ACKNOWLEDGMENT
We thank Professor A. Boyd for critical review of the manuscript, Drs D. Dumenil and M. Lagranderie for helpful discussions, S. Liu and G. Chojnowski for expert technical assistance in cell preparation and flow cytometry, and J. Kaye, a summer vacation scholar, for the isolation of PTK 1L4a clone.
Supported by the Leukaemia Foundation of Queensland; the University of Queensland, Queensland; National Health and Medical Research Council, Canberra (project Grant No. 941036); and AMRAD Corporation, Melbourne, Australia. D.W. has been supported by the French CNRS and a postdoctoral fellowship of the University of Queensland. S.I.S. is the recipient of a Queensland Cancer Fund John Earnshaw/PhD research scholarship.
Bmx sequence has been registered in Genbank under accession number U88091.
Address reprint requests to Chung Leung Li, PhD, Queensland Insititue of Medical Research, Post Office, Royal Brisbane Hospital, 4029 Queensland, Australia.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal