Abstract
Using simultaneous Hoechst 33342 (Hst) and Pyronin Y (PY) staining for determination of DNA and RNA content, respectively, human CD34+ cells were isolated in subcompartments of the G0 /G1 phase of the cell cycle by flow cytometric cell sorting. In both bone marrow (BM) and mobilized peripheral blood (MPB) CD34+ cells, primitive long-term hematopoietic culture-initiating cell (LTHC-IC) activity was higher in CD34+ cells isolated in G0 (G0CD34+ cells) than in those residing in G1 (G1CD34+ cells). However, as MPB CD34+ cells displayed a more homogeneous cell-cycle status within the G0 /G1 phase and a relative absence of cells in late G1 , DNA/RNA fractionation was less effective in segregating LTHC-IC in MPB than in BM. BM CD34+ cells belonging to four subcompartments of increasing RNA content within the G0 /G1 phase were evaluated in functional assays. The persistence of CD34 expression in suspension culture was inversely correlated with the initial RNA content of test cells. Multipotential progenitors were present in G0 or early G1 subcompartments, while lineage-restricted granulomonocytic progenitors were more abundant in late G1 . In vitro hematopoiesis was maintained for up to 6 weeks with G0CD34+ cells, whereas production of clonogenic progenitors was more limited in cultures initiated with G1CD34+ cells. To test the hypothesis that primitive LTHC-ICs would reenter a state of relative quiescence after in vitro division, BM CD34+ cells proliferating in ex vivo cultures were identified from their quiescent counterparts by a relative loss of membrane intercalating dye PKH2, and were further fractionated with Hst and PY. The same functional hierarchy was documented within the PKH2dim population whereby LTHC-IC frequency was higher for CD34+ cells reselected in G0 after in vitro division than for CD34+ cells reisolated in G1 or in S/G2 + M. However, the highest LTHC-IC frequency was found in quiescent PKH2bright CD34+ cells. Together, these results support the concept that cells with distinct hematopoietic capabilities follow different pathways during the G0 /G1 phase of the cell cycle both in vivo and during ex vivo culture.
THE CELL-CYCLE STATUS and responsiveness to in vitro cytokine stimulation is critical in defining strategies for hematopoietic progenitor/stem cell expansion1-3 and somatic gene therapy.4,5 It is generally believed that primitive hematopoietic progenitor cells (HPCs) lie dormant within the bone marrow (BM) microenvironment.6-8 Supporting this hypothesis is evidence that primitive HPCs, unlike committed progenitors, display a latency period before they respond to cytokine-driven cell-cycle activation9-11 and that they are resistant to exposure to antimitotic drugs like 5-fluorouracil.12-14 Cell-cycle analysis and sorting by direct measurement of DNA content with Hoechst 33342 (Hst) has been used to compare the hematopoietic function of cells in G0 /G1 versus those in S/G2 + M phases.15 However, such a static “snapshot” measurement does not distinguish quiescent G0 cells from those in G1 that may represent cell populations with distinct hematopoietic capabilities. Although the suicide techniques using 3H-thymidine16-18 or cytosine arabinoside19 are dynamic procedures capable of determining the relative proportion of resting versus cycling HPCs, they rely on selective killing of actively cycling cells, therefore preventing their functional characterization. This prompted us to evaluate the use of multiparameter flow cytometry methods applicable to viable CD34+ cells and potentially capable of discriminating between subcompartments of the G0 /G1 phase of the cell cycle.
The ability to correlate the DNA content of individual cells to other cell constituents such as total RNA or proteins has led to the description of different kinetic compartments within the traditional phases of the cell cycle. In particular, subcompartments of the G0 /G1 phase were described based on simultaneous DNA/RNA determination.20-23 Cells with low RNA content maintain a state of deep dormancy in G0 . As cells enter G1 , they accumulate RNA, mainly in the form of ribosomal RNA, until they reach S phase. In a variety of cell systems, the rate of progression of individual cells through the cell cycle was found to be correlated with RNA content.24 Although Lajtha25 originally coined the term G0 to describe dormant hepatic cells responding to liver insult, other cell types residing in quiescent mitotic state characterized by a lower RNA content than G1 cells have been referred to as G1Q or G0 cells.22 In a recent study from our laboratory, simultaneous DNA/RNA staining of BM CD34+ cells was used successfully to isolate, by flow cytometry, quiescent and primitive HPCs in the G0 phase of the cell cycle.26 The possibility to isolate viable HPCs with distinct DNA/RNA content was used for the present study to compare hematopoietic function of CD34+ cells isolated in subcompartments of the cell cycle.
We have previously shown10 that cytokine nonresponsive (CNR) cells, which resist initial cytokine stimulation and survive in culture for up to 7 days, are enriched for primitive HPCs, while those undergoing in vitro proliferation appear to gradually lose their hematopoietic potential.27 We hypothesized that if proliferating cells were to maintain primitive function, they would reenter after cell division into a quiescent state characterized by minimal RNA content. Experiments were performed to investigate whether primitive HPCs could be reisolated from ex vivo expansion cultures by cell-cycle fractionation.
In the present studies, we assayed the hematopoietic function of CD34+ cells isolated from distinct subcompartments of the G0 /G1 phase of the cell cycle by DNA/RNA staining. Such CD34+ cell subsets were evaluated in steady-state BM, in mobilized peripheral blood (MPB), and in the context of ex vivo expansion. Our results demonstrate that major functional differences could be attributed to the position of these cells in the G0 /G1 phase, and suggest that primitive and committed progenitor cells follow different pathways in the G0 /G1 phase of the cell cycle.
MATERIALS AND METHODS
CD34+ cell purification. BM and MPB samples were obtained from healthy adult volunteers according to guidelines established by the Human Investigation Committee of the Indiana University School of Medicine. Mobilization was achieved by daily G-CSF administration at 5 μg/kg (maximum, 480 μg/d) for 4 consecutive days. MPB cells were collected by apheresis on day 5. Low-density mononuclear cells from both tissues were isolated by centrifugation over Ficoll-Paque (Pharmacia Fine Chemicals, Piscataway, NJ) and were enriched for CD34+ cells by immunomagnetic selection as previously described.10 All reagents for the immunomagnetic separation procedure were a generous gift from Baxter Healthcare (Santa Ana, CA). Immunomagnetically selected CD34+ cells were labeled with fluorescein isothiocyanate (FITC)-conjugated anti-CD34 antibody (Becton Dickinson Immunocytometry Systems [BDIS], San Jose, CA) or in some cases with allophycocyanin (APC)-conjugated anti-CD34 (Caltag, San Francisco, CA) to allow for further staining with FITC-conjugated anti–Ki-67 antibody. Total BM or MPB CD34+ cells were isolated on a FACStar Plus flow cytometer (BDIS). For some of the MPB samples in which CD34+ cell purity was greater than 90% after immunomagnetic selection, the flow cytometry purification step was omitted.
Cell-cycle fractionation with Hst and Pyronin Y. Total CD34+ cells were resuspended at 1 to 2 × 106 cells in 1.5 mL 1.67-μmol/L solution of Hst (Molecular Probes, Eugene, OR) in Hst buffer. Hst buffer consisted of Hanks' balanced salt solution (Biowhittaker, Walkersville, MD), 20 mmol/L HEPES (Biowhittaker), 1 g/L glucose, and 10% fetal calf serum ([FCS] Hyclone, Logan, UT). After incubation at 37°C for 45 minutes, Pyronin Y ([PY] Sigma, St Louis, MO) prepared in Hst buffer was added at a final concentration of 1 μg/mL and the cells were further incubated for another 45 minutes at 37°C. The cells were washed once, resuspended in Hst buffer, and sorted on a FACStar Plus equipped with an argon laser providing the 488-nm excitation for PY and a krypton laser providing the 350-nm excitation for Hst. The PY signal was selected with a 575 ± 13 nm band-pass filter, and Hst was detected with a 424 ± 22 nm band-pass filter. Sorting windows were constructed as depicted in Fig 1A. Since, by definition, RNA staining with PY yields a continuous histogram without demarcation between positive and negative cells, an arbitrary sorting window comprising 10% to 15% of cells displaying minimal PY staining and 2n DNA content was used to sort G0CD34+ cells in all experiments (Fig 1A). Viability and purity of sorted cells always exceeded 98% and 90%, respectively.
General method of cell-cycle fractionation using DNA and RNA analysis with Hst and PY. (A) Dual-parameter dot plot showing analysis of BM CD34+ cells stained with Hst and PY. Cells residing in G0 (low PY uptake) appear at the bottom of the G0 /G1 region identified by low Hst staining. Cells in G1 have a brighter PY signal within the G0 /G1 peak. Cells in S/G2 + M display both high Hst and high PY staining. (B) Postsort analysis of BM CD34+ cells sorted in G0 . (C) Hst/PY fluorescence distribution of G0 CD34+ cells shown in B after 24 hours under IL-3-IL-6-SCF stimulation, showing G0-G1 progression. (D) Same sample after 72 hours in culture, depicting cells moving into S/G2 + M phase.
General method of cell-cycle fractionation using DNA and RNA analysis with Hst and PY. (A) Dual-parameter dot plot showing analysis of BM CD34+ cells stained with Hst and PY. Cells residing in G0 (low PY uptake) appear at the bottom of the G0 /G1 region identified by low Hst staining. Cells in G1 have a brighter PY signal within the G0 /G1 peak. Cells in S/G2 + M display both high Hst and high PY staining. (B) Postsort analysis of BM CD34+ cells sorted in G0 . (C) Hst/PY fluorescence distribution of G0 CD34+ cells shown in B after 24 hours under IL-3-IL-6-SCF stimulation, showing G0-G1 progression. (D) Same sample after 72 hours in culture, depicting cells moving into S/G2 + M phase.
Short-term culture and cell-cycle analysis. CD34+ cells used in these experiments were initially sorted with APC-conjugated anti-CD34 (Caltag) followed by Hst/PY fractionation. Cells were plated in culture medium consisting of IMDM, 10% FCS, 2 mmol/L L-glutamine, and antibiotics, referred to hereafter as complete medium, supplemented with interleukin-3, (IL-3), IL-6, and stem cell factor (SCF ) at 100 ng/mL each. After 24 to 144 hours in culture, cells were harvested and subjected to high-resolution cell-cycle analysis as recently described by Jordan et al,28,29 with minor modification. Cells were washed and resuspended in 1 mL phosphate-buffered saline (PBS) + 0.4% formaldehyde. After 30 minutes at 4°C, 1 mL PBS + 0.2% Triton X-100 was added, and the cells were left overnight at 4°C. The cells were then washed twice in PBS + 1% bovine serum albumin (BSA) and stained with FITC-conjugated anti–Ki-67 (clone MIB-1; Immunotech, Westbrook, ME) for 60 minutes at 4°C. Isotype controls were stained in parallel. Finally, cells were washed and resuspended in PBS + 1% BSA containing 5 μg/mL 7-aminoactinomycin-D ([7-AAD] Sigma). After a 3-hour incubation on ice, samples were run on a FACScan flow cytometer (BDIS) using FL-1 and FL-3 channels for Ki-67 and 7-AAD, respectively. Alternatively, conventional cell-cycle analysis by DNA staining with propidium iodide was used as previously described.3
Long-term culture of CD34+ subpopulations and progenitor assays. CD34+ cells isolated in G0 /G1 subcompartments were seeded in 24-well plates at a density of 104 cells/mL in complete medium supplemented with IL-3, IL-6, and SCF, each at 100 ng/mL. Every week, half of the cells were removed, followed by replacement with fresh medium and cytokines. Aliquots of harvested cells were assayed for progenitor cell content in 1.3% methylcellulose, 30% FCS, 100 ng/mL SCF, 10 ng/mL IL-3, 10 ng/mL IL-6, 5 ng/mL GM-CSF, and 2 U/mL erythropoietin (EPO). Hematopoietic colonies were scored 2 weeks later according to standard criteria.
Persistence of CD34 expression in culture. After 2 weeks of long-term culture, aliquots of cultured cells were harvested, washed, and resuspended in 100 μL mouse serum (Sigma). After a 10-minute incubation at room temperature, FITC-conjugated anti-CD34 antibody or an isotype-matched control (BDIS) were added to the cell suspension. Staining was performed for 20 minutes on ice, after which cells were washed and analyzed on a FACScan flow cytometer. CD34 expression was measured only on cells with low forward- and side-scatter properties, and was defined as cells displaying a fluorescence greater than 99% of the isotype control.
Analysis of ex vivo expansion with PKH2 cell tracking and cell-cycle fractionation. A total of 0.5 to 1 × 106 CD34+ cells were labeled with the membrane dye PKH2 (Sigma) for cell tracking per the manufacturer's instructions. Although PKH26 (emission at 575 nm) has been reported to be a more reliable indicator of cell proliferation, PKH2 (emission at 530 nm) was used in this study to allow simultaneous use with PY. After staining, cells were counted and plated in 24-well plates at a density of 2 × 105 cells/mL in complete medium supplemented with IL-3, IL-6, and SCF, all at 100 ng/mL. An aliquot of PKH2-stained cells was fixed in 1% paraformaldehyde and kept at 4°C for determination of day 0 PKH2 fluorescence. Cultures were fed with cytokines every 48 hours. Care was taken to avoid cell densities higher than 5 × 105 cells/mL by frequent splitting of the cultures to minimize dye transfer between cells. After 7 days, subpopulations of CD34+ cells were sorted in two steps as follows.
Cultured cells were counted, washed, and stained with biotinylated anti-CD34 monoclonal antibody (Caltag) followed by streptavidin (SA)-APC (Molecular Probes). Controls consisted of biotinylated isotype-matched control IgG followed by SA-APC. Cells were washed and resuspended in PBS + 1% BSA and sorted into two groups. The first consisted of CD34+PKH2bright cells, designating those that had not divided in culture, and the second contained CD34+PKH2dim cells, representing those that had divided and therefore lost part of their original PKH2 fluorescence. Sorted CD34+PKH2dim cells were further stained with Hst and PY as already described, and were sorted in a second step into cells residing in G0 , G1 , or S/G2 + M.
Limiting dilution analysis of long-term hematopoietic culture-initiating cells. LTHC-IC frequencies were determined by a stroma-free limiting dilution analysis as described previously.10 LTHC-IC frequencies were calculated using Poisson statistics.30
Growth factors. Human recombinant IL-3, IL-6, SCF, and GM-CSF were a kind gift from Amgen (Thousand Oaks, CA). Human recombinant EPO was obtained from Amgen.
Statistics. Comparisons of cell populations isolated in various phases of the cell cycle were made by analysis of variance (ANOVA). If P (ANOVA) was less than .05, pairwise multiple comparisons were made using Student-Newman-Keuls (SNK) tests. When only two groups were to be compared, Student's t-tests were used. All P values are two-sided. SigmaStat 2.0 software (Jandel, San Rafael, CA) was used for all calculations.
RESULTS
Fractionation of CD34+ cells into subcompartments of the G0/G1 phase of the cell cycle. Traditional cell-cycle analysis with a DNA probe such as Hst distinguishes between cells in the S/G2 + M phase and those in the G0 /G1 phase. We have recently described the successful isolation of CD34+ cells in the G0 phase of the cell cycle by simultaneous DNA/RNA staining.26 In this method, cells determined to be in the G0 /G1 phase based on Hst fluorescence distribution can be further fractionated into subcompartments of varying cellular RNA content by staining with PY. Quiescent cells, in G0 , have a low RNA content.22 As cells progress through G1 , they accumulate RNA and finally move to the S/G2 + M phase during which Hst staining increases (Fig 1).
To assess the validity of Hst/PY cell-cycle fractionation, we compared it with a recently described method of high-resolution cell-cycle analysis, ie, the Ki-67/7-AAD method.28,29 In this procedure, a DNA histogram is generated by 7-AAD and plotted against expression of the nuclear antigen Ki-67, which is present in cycling cells but not in G0 cells.31 Consecutive measurements of Hst/PY and Ki-67/7-AAD were performed. MPB CD34+ cells were sorted in either G0 or G1 as defined by Hst/PY staining, plated in short-term cultures, and analyzed at different time intervals with Ki-67/7-AAD (Fig 2). Most of the cells (87% in the example shown in Fig 2) recovered from cultures initiated with G0CD34+ cells were Ki-67–negative 24 hours after initiation. Expression of Ki-67 was progressively upregulated while cells entered G1 and S/G2 + M stages at 72 and 144 hours, but a significant percentage of cells, 14.3%, were still Ki-67–negative after 144 hours in culture. In G1CD34+ cell–initiated cultures, the vast majority of cells entered into active phases of the cell cycle immediately after cytokine stimulation and were Ki-67–positive (>93%) at 24 hours after initiation of short-term culture. Interestingly, whereas only 3% of G0CD34+ cells traversed into active phases of the cell cycle 24 hours after exposure to cytokines, almost 50% of initial G1CD34+ cells were detected in S/G2 + M at the same time point. A similar delayed expression of Ki-67 and late onset of DNA synthesis in cultures initiated with G0CD34+ cells compared with cultures initiated with G1CD34+ cells was observed for BM CD34+ cells (data not shown). These results demonstrate that CD34+ cell subsets possessing different rates of cell-cycle activation by cytokines were identified and isolated by Hst/PY staining.
Serial high-resolution cell-cycle analysis of cytokine-stimulated G0 and G1 CD34+ cells. MPB G0 and G1 CD34+ cells were plated in complete medium supplemented with IL-3-IL-6-SCF. At indicated times, cells were harvested and analyzed for cell-cycle status with 7-AAD and Ki-67. (A) Dual-parameter plot showing fractionation of CD34+ cells in G0 and G1 phases with Hst and PY. (B to D) Cell-cycle analysis of cells harvested from G1 CD34+ cell–initiated culture at 24, 72, and 144 hours, respectively. (E to G) Cell-cycle analysis of cells harvested from G0CD34+ cell–initiated culture at 24, 72, and 144 hours, respectively. The percentage of cells in G0 , G1 , and S/G2 + M (abbreviated as G2 ) is indicated. Cells in the lower right quadrant were not considered.
Serial high-resolution cell-cycle analysis of cytokine-stimulated G0 and G1 CD34+ cells. MPB G0 and G1 CD34+ cells were plated in complete medium supplemented with IL-3-IL-6-SCF. At indicated times, cells were harvested and analyzed for cell-cycle status with 7-AAD and Ki-67. (A) Dual-parameter plot showing fractionation of CD34+ cells in G0 and G1 phases with Hst and PY. (B to D) Cell-cycle analysis of cells harvested from G1 CD34+ cell–initiated culture at 24, 72, and 144 hours, respectively. (E to G) Cell-cycle analysis of cells harvested from G0CD34+ cell–initiated culture at 24, 72, and 144 hours, respectively. The percentage of cells in G0 , G1 , and S/G2 + M (abbreviated as G2 ) is indicated. Cells in the lower right quadrant were not considered.
Assessment of LTHC-IC activity after cell-cycle fractionation of BM and MPB CD34+ cells. The ability of Hst/PY staining to achieve fractionation of primitive HPCs was compared for BM and MPB. Reports from several groups18,32 and from our laboratory33 indicated that MPB CD34+ cells consist of a population of noncycling cells, as compared with BM CD34+ cells. These observations were confirmed in the present study by standard propidium iodide DNA staining, such that 13.0% ± 2.0% of freshly isolated BM CD34+ cells were in S/G2 + M phase versus 0.81% ± 0.66% for MPB CD34+ cells (P < .0001). We extended these previous findings by examining the relative abundance of PYlow (G0 ) and PYbright (G1 ) cells within the G0 /G1 peak in both tissues. Representative dot plots in Fig 1A (BM) and Fig 2A (MPB) show that MPB CD34+ cells are enriched for cells displaying low RNA staining within the G0 /G1 phase, suggesting that relative to BM CD34+ cells, MPB CD34+ cells are not only devoid of cells in S/G2 + M but also cells in late G1 phase. To compare the efficiency of Hst/PY staining to separate primitive HPCs in BM and MPB, G0 and G1 CD34+ cells were sorted from both sources and assayed for LTHC-IC activity (Fig 3). For these experiments, G0 and G1 CD34+ cells were defined for both tissues as having minimal and maximal PY staining within the G0 /G1 peak, respectively. A statistical difference was observed between the LTHC-IC frequency of G0CD34+ cells and G1CD34+ cells for both tissues. However, although a ninefold difference in the frequency of LTHC-IC between G0CD34+ cells and G1CD34+ cells was documented for BM (P = .009), only a twofold difference (P = .02) was demonstrated for the same two fractions of MPB cells, indicating that increased homogeneity in the cell-cycle status of MPB CD34+ cells versus BM CD34+ cells was associated with a more pronounced functional homogeneity. In addition, G1CD34+ cells isolated from BM had a lower LTHC-IC frequency (0.7% ± 0.2%) than their counterparts isolated from MPB (5.7% ± 2.0%, P < .01), whereas no statistical difference was observed between G0CD34+ cells isolated from these two sources (P = .17). This suggests that a specific subpopulation of CD34+ cells residing in late G1 in the BM microenvironment, and whose primitive HPC function is largely compromised, is not recovered after mobilization into the PB.
LTHC-IC frequencies of G0 and G1 CD34+ cells isolated from BM and MPB. Frequencies are given per 100 cells as the mean ± SEM, n = 8 BM and 7 MPB. Samples were compared with t tests. *P = .009 v BM G1CD34+ cells; **P = .02 v MPB G1CD34+ cells; ***P = .01 v BM G1CD34+ cells.
LTHC-IC frequencies of G0 and G1 CD34+ cells isolated from BM and MPB. Frequencies are given per 100 cells as the mean ± SEM, n = 8 BM and 7 MPB. Samples were compared with t tests. *P = .009 v BM G1CD34+ cells; **P = .02 v MPB G1CD34+ cells; ***P = .01 v BM G1CD34+ cells.
Hematopoietic function of BM CD34+ cells isolated in subcompartments of the G0/G1 phase of the cell cycle. Additional experiments were performed to examine in more detail the functional heterogeneity of BM CD34+ cells as a function of their position within the prereplicating phase of the cell cycle. Using Hst/PY staining, we arbitrarily defined four subcompartments of increasing RNA content in the G0 /G1 phase of the cell cycle, termed hereafter G0 , G1a , G1b , and G1c (Fig 4). Sorting windows for G1a , G1b , and G1c were constructed to contain the same percentage of cells as contained in the sorting window for G0 cells defined as depicted in Fig 1A. BM CD34+ cells isolated in these subcompartments were used for subsequent functional assays.
Fractionation of steady-state BM CD34+ cells into 4 G0 /G1 subcompartments using DNA/RNA analysis with Hst and PY. Sorting gates defining four subcompartments of G0 /G1 phase are indicated.
Fractionation of steady-state BM CD34+ cells into 4 G0 /G1 subcompartments using DNA/RNA analysis with Hst and PY. Sorting gates defining four subcompartments of G0 /G1 phase are indicated.
The relative contribution of each of the CD34+ cell subsets to late and primitive progenitor populations was assessed by colony-forming unit (CFU) and LTHC-IC assays, respectively. Overall CFU frequency was not significantly different between cell subsets, and was 15.2% ± 3.4% in G0CD34+ cells, 18.3% ± 5.8% in G1aCD34+ cells, 17.4% ± 3.8% in G1bCD34+ cells, and 8.3% ± 3.8% in G1cCD34+ cells (n = 3, P > .05). However, different types of progenitors were detected in these cell populations such that G0CD34+ cells had a significantly higher frequency (P < .05) of multipotential progenitors (CFU-MIX) than G1a , G1b , and G1c CD34+ cell populations (Fig 5A), but a significantly lower frequency (P < .05) of myeloid committed progenitors (CFU-GM) than G1a and G1b CD34+ cells.
Functional characterization of steady-state BM CD34+ cells isolated in subcompartments of G0 /G1 phase. (A) Progenitor cell assay. The number of hematopoietic colonies are given per 100 cells plated. Data shown above are the mean ± SEM of three experiments performed in duplicate. BFU-E, burst-forming unit–erythroid; CFU-GM, colony-forming unit–granulocyte/macrophage; CFU-MIX, CFU-mixed. *P < .05 v G1a , G1b , and G1c CD34+ cells; **P < .05 v G1a and G1b CD34+ cells. (B) LTHC-IC assay. Frequencies are given per 100 cells as the mean ± SEM, n = 4. ***P < .05 v G1b and G1c CD34+ cells. Statistical analysis was made by ANOVA followed by SNK pairwise comparisons.
Functional characterization of steady-state BM CD34+ cells isolated in subcompartments of G0 /G1 phase. (A) Progenitor cell assay. The number of hematopoietic colonies are given per 100 cells plated. Data shown above are the mean ± SEM of three experiments performed in duplicate. BFU-E, burst-forming unit–erythroid; CFU-GM, colony-forming unit–granulocyte/macrophage; CFU-MIX, CFU-mixed. *P < .05 v G1a , G1b , and G1c CD34+ cells; **P < .05 v G1a and G1b CD34+ cells. (B) LTHC-IC assay. Frequencies are given per 100 cells as the mean ± SEM, n = 4. ***P < .05 v G1b and G1c CD34+ cells. Statistical analysis was made by ANOVA followed by SNK pairwise comparisons.
LTHC-IC frequencies were measured in our stroma-free limiting dilution analysis assay as previously described (Fig 5B). LTHC-IC frequencies were inversely correlated with the initial RNA content of test cells. The highest LTHC-IC content was found in the G0CD34+ cell subset. G1aCD34+ cells had 50% less LTHC-IC than G0CD34+ cells (P > .05), while CD34+ cells isolated in late G1 phases, G1b and G1c CD34+ cells, had low or nearly undetectable LTHC-IC frequencies (P < .05 v G0CD34+ cells). Altogether, these results indicate that in steady-state hematopoiesis, functionally distinct populations of HPCs can be identified along the G0 /G1 pathway of the cell cycle. Multipotential CFU and CFU precursors reside in G0 and to a lesser degree in early G1 phase, while late G1 subcompartments appear to contain mainly committed progenitors.
We next determined the ability of G0 /G1 subcompartments of BM CD34+ cells to maintain in vitro hematopoiesis in long-term cultures. CD34+ cells isolated with varying RNA content within the G0 /G1 phase (Fig 4) were plated in stroma-free suspension cultures and stimulated by a combination of IL-3, IL-6, and SCF. At weekly intervals, aliquots of cells were replated in progenitor cell assays to determine CFU production (Fig 6). CFUs were detectable up to 6 weeks in cultures initiated with both G0CD34+ cells and G1aCD34+ cells, while CFU production was exhausted after 2 weeks in cultures of CD34+ subsets isolated in G1b or G1c . Total CFU output was maximal in G0CD34+ cell–initiated cultures, and decreased in other cell subsets in inverse relationship to the initial RNA content of the test cells (Fig 6). The persistence of CD34 expression among cultured cells was monitored on day 14 in similar long-term cultures (Table 1). The percentage of CD34+ cells in vitro correlated with CFU production and was highest in cultures initiated with CD34+ cells displaying low RNA content (P < .05). It is interesting that after 2 weeks in LTC, the absolute number of CD34+ cells had expanded threefold in cultures initiated with G0CD34+ cells, while it had remained stable in G1aCD34+ cells and decreased approximately 80% and 88% in G1bCD34+ and G1cCD34+ cell–initiated cultures, respectively.
Time course of CFU production in long-term cultures initiated with BM CD34+ cells isolated in subcompartments of G0 /G1 phase. Data are from one representative experiment of a total of three. Cells with indicated phenotype were cultured for 7 weeks in complete medium supplemented with IL-3-IL-6-SCF. Every week, CFUs were enumerated in HPC assays. The number of CFUs (×103) per 104 initial cells plated are shown above for each time point.
Time course of CFU production in long-term cultures initiated with BM CD34+ cells isolated in subcompartments of G0 /G1 phase. Data are from one representative experiment of a total of three. Cells with indicated phenotype were cultured for 7 weeks in complete medium supplemented with IL-3-IL-6-SCF. Every week, CFUs were enumerated in HPC assays. The number of CFUs (×103) per 104 initial cells plated are shown above for each time point.
Persistence of CD34 Expression in Cultures Initiated With BM CD34+ Cells Isolated in Subcompartments of the G0/G1Phase of the Cell Cycle
| Cultures Initiated on Day 0 With . | % CD34+ Cells on Day 14 . | Total No. of CD34+ Cells on Day 14‡ . |
|---|---|---|
| G0CD34+ | 7.83 ± 0.85* | 29,383 ± 14,756 |
| G1aCD34+ | 4.31 ± 0.39† | 10,667 ± 2,462 |
| G1bCD34+ | 1.21 ± 0.57 | 6,167 ± 1,999 |
| G1cCD34+ | 0.53 ± 0.23 | 1,869 ± 1,255 |
| Cultures Initiated on Day 0 With . | % CD34+ Cells on Day 14 . | Total No. of CD34+ Cells on Day 14‡ . |
|---|---|---|
| G0CD34+ | 7.83 ± 0.85* | 29,383 ± 14,756 |
| G1aCD34+ | 4.31 ± 0.39† | 10,667 ± 2,462 |
| G1bCD34+ | 1.21 ± 0.57 | 6,167 ± 1,999 |
| G1cCD34+ | 0.53 ± 0.23 | 1,869 ± 1,255 |
A total of 104 BM CD34+ cells isolated from G0/G1 subcompartments (Fig 4) were plated in complete medium supplemented with IL-3-IL-6-SCF. After 14 days, the persistence of CD34 expression was determined by FACS analysis of cultured cells. Data are the mean ± SEM, n = 3. Statistical analysis was by ANOVA followed by SNK pairwise comparisons.
P < .05 v cultures initiated with G1a , G1b , and G1c CD34+ cells.
P < .05 v cultures initiated with G1b and G1c CD34+ cells.
Total number of CD34+ cells calculated by multiplying the percentage of CD34+ cells by the total cell number present in each culture.
Maintenance of hematopoietic function in ex vivo–expanded BM CD34+ cells isolated in different phases of the cell cycle.In a subsequent step, we reasoned that upon completion of cell division, the most primitive progenitors may reenter transiently into a state of quiescence while more committed progenitors remain mitotically active. Previous studies from our laboratory using PKH2 cell tracking10 had already pointed out that a significant proportion of LTHC-ICs contained in ex vivo expansion cultures belonged to a group of quiescent cells surviving in the absence of cell division, termed CNR cells. To distinguish nondividing LTHC-ICs from those reentering G0 after in vitro proliferation, we combined PKH2 cell tracking with DNA/RNA fractionation (Fig 7). Following ex vivo expansion for 7 days, CD34+ cells were first separated into PKH2bright and PKH2dim subsets as quiescent and proliferating cells, respectively (Fig 7A and B). PKH2dim cells were further fractionated by Hst/PY staining (Fig 7C) into three subpopulations, namely G0 , G1 , and S/G2 + M. All cell populations were assayed for LTHC-IC content.
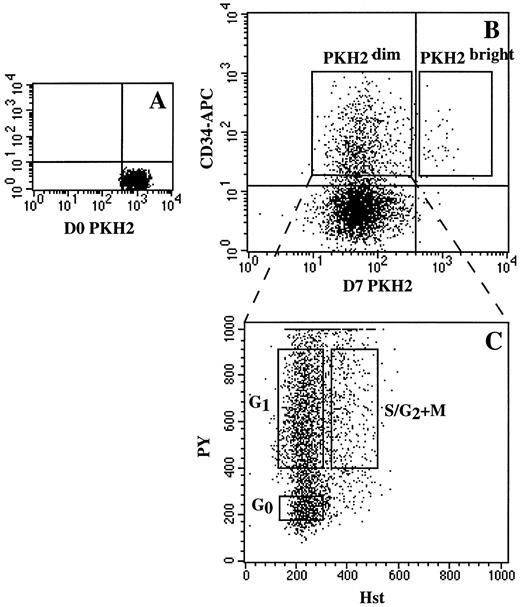
Sequential sorting of ex vivo–expanded CD34+ cells by cell tracking with PKH2 and cell-cycle fractionation with Hst and PY. Fresh sorted CD34+ cells were stained with PKH2 on day 0 (A). Cells were then cultured for 7 days in complete medium supplemented with IL-3-IL-6-SCF (100 ng/mL each). On day 7, harvested cells were stained with CD34-Biotin/streptavidin-APC and analyzed with PKH2 (B). Placement of the vertical cursor was adjusted with reference to day 0 PKH2, and the horizontal cursor with reference to the isotype control. Sorting windows including CD34+PKH2bright and CD34+PKH2dim are indicated. In the second step, sorted CD34+PKH2dim cells were stained with Hst and PY and were further fractionated by DNA/RNA analysis into G0 , G1 , and S/G2 + M phases of the cell cycle (C).
Sequential sorting of ex vivo–expanded CD34+ cells by cell tracking with PKH2 and cell-cycle fractionation with Hst and PY. Fresh sorted CD34+ cells were stained with PKH2 on day 0 (A). Cells were then cultured for 7 days in complete medium supplemented with IL-3-IL-6-SCF (100 ng/mL each). On day 7, harvested cells were stained with CD34-Biotin/streptavidin-APC and analyzed with PKH2 (B). Placement of the vertical cursor was adjusted with reference to day 0 PKH2, and the horizontal cursor with reference to the isotype control. Sorting windows including CD34+PKH2bright and CD34+PKH2dim are indicated. In the second step, sorted CD34+PKH2dim cells were stained with Hst and PY and were further fractionated by DNA/RNA analysis into G0 , G1 , and S/G2 + M phases of the cell cycle (C).
In accordance with our previous studies,10 33 the total number of LTHC-ICs (Table 2) slightly decreased over a 7-day period in suspension cultures supported with IL-3, IL-6, and SCF. Analysis of PKH2 fluorescence showed that approximately 30% of total LTHC-ICs recovered from ex vivo cultures did not respond to the cytokine stimulation used in these studies, and were still in the CD34+PKH2bright subset after 7 days in ex vivo expansion cultures.
Absolute Number of LTHC-ICs Detected Among Freshly Isolated and Ex Vivo–Expanded CD34+ Cells
| . | Phenotype . | No. of LTHC-ICs* . | Percent Cells† . |
|---|---|---|---|
| Day 0 | Total CD34+ | 3,750 ± 1,440 | |
| Day 7 | Total CD34+ | 3,481 ± 617 | |
| CD34+ PKH2bright | 1,092 ± 764 | 3.1 ± 2.3 | |
| CD34+ PKH2dim | 2,389 ± 558 | 14.9 ± 1.9 |
| . | Phenotype . | No. of LTHC-ICs* . | Percent Cells† . |
|---|---|---|---|
| Day 0 | Total CD34+ | 3,750 ± 1,440 | |
| Day 7 | Total CD34+ | 3,481 ± 617 | |
| CD34+ PKH2bright | 1,092 ± 764 | 3.1 ± 2.3 | |
| CD34+ PKH2dim | 2,389 ± 558 | 14.9 ± 1.9 |
Simultaneous CD34/PKH2 cell sorting on day 7 was performed as illustrated in Fig 7B. Paired data from 4 BM samples are reported as the mean ± SEM.
Total number of LTHC-ICs normalized per 105 initial CD34+ cells.
Percent cells with indicated phenotype given relative to the total number of cells in culture on day 7.
Within the CD34+PKH2dim cell population (Fig 8), cells reentering G0 retained a significantly higher frequency of LTHC-ICs (1.9 ± 0.2) compared with cells reisolated in G1 or S/G2 + M (0.8 ± 0.25 and 0.3 ± 0.2, respectively, P < .05), indicating that reacquisition of minimal RNA content after in vitro division was a distinct property of primitive hematopoietic cells assayed as LTHC-ICs. However, the LTHC-IC frequency of CD34+ cells reentering G0 after a 7-day ex vivo expansion was significantly lower than that detected among nonproliferating CD34+PKH2bright cells, harvested from the same cultures (4.1 ± 0.5, P < .05; Fig 8), adding further evidence that a prolonged quiescent state was associated with primitive hematopoietic function.10
LTHC-IC frequencies of ex vivo–expanded CD34+ cells isolated in different phases of the cell cycle. CD34+ cell subsets were sorted as described in Fig 7. LTHC-IC frequencies are given per 100 cells of each phenotype indicated. Paired data from four BM samples are reported as the mean ± SEM. Statistical analysis was made by ANOVA followed by SNK pairwise comparisons. *P < .05 v G0 , G1 , and S/G2 + M CD34+ PKH2dim cells; **P < .05 v G1 and S/G2 + M CD34+PKH2dim cells.
LTHC-IC frequencies of ex vivo–expanded CD34+ cells isolated in different phases of the cell cycle. CD34+ cell subsets were sorted as described in Fig 7. LTHC-IC frequencies are given per 100 cells of each phenotype indicated. Paired data from four BM samples are reported as the mean ± SEM. Statistical analysis was made by ANOVA followed by SNK pairwise comparisons. *P < .05 v G0 , G1 , and S/G2 + M CD34+ PKH2dim cells; **P < .05 v G1 and S/G2 + M CD34+PKH2dim cells.
The apparent difference in the LTHC-IC content of freshly isolated G0CD34+ cells (6.5% ± 2.0%, Fig 3) versus cultured CD34+ cells reisolated with minimal RNA content (1.9% ± 0.2%; Fig 8) prompted us to compare the relative level of cell-cycle dormancy of these two groups of cells to investigate the relationship between cell-cycle quiescence and primitive hematopoietic function. The rate of cell-cycle activation of G0CD34+ cells isolated before and after ex vivo expansion was measured in short-term cultures. Suspension cultures from paired BM samples were initiated with day 0 G0CD34+ cells and 1 week later with day 7 G0CD34+PKH2dim cells reisolated with minimal RNA content after ex vivo expansion. Both cultures were stimulated with the IL-3-IL-6-SCF combination. Cells from paired cultures were subjected to cell-cycle analysis at initiation and at 24-hour intervals (Table 3). Day 7 G0CD34+ cells quickly progressed into active phases of the cell cycle (14.24% of cells in S/G2 + M at 24 hours) compared with freshly isolated G0CD34+ cells (2.61% of cells in S/G2 + M, P < .05), indicating that after in vitro division under our culture conditions, G0CD34+ cells not only had a reduced frequency of LTHC-ICs but were unable to reacquire their initial degree of cell-cycle dormancy.
Proliferation Rate of G0CD34+ Cells Isolated Before and After Ex Vivo Expansion
| Cell Phenotype . | % Cells in S/G2 + M at Indicated . | ||
|---|---|---|---|
| . | Time (h) After Isolation . | ||
| . | 0 . | 24 . | 48 . |
| G0CD34+ | 1.45 ± 0.523-150 | 2.61 ± 1.41† | 12.73 ± 3.383-150 |
| G0CD34+PKH2dim | 2.55 ± 0.873-150 | 14.24 ± 5.36† | 11.31 ± 2.373-150 |
| Cell Phenotype . | % Cells in S/G2 + M at Indicated . | ||
|---|---|---|---|
| . | Time (h) After Isolation . | ||
| . | 0 . | 24 . | 48 . |
| G0CD34+ | 1.45 ± 0.523-150 | 2.61 ± 1.41† | 12.73 ± 3.383-150 |
| G0CD34+PKH2dim | 2.55 ± 0.873-150 | 14.24 ± 5.36† | 11.31 ± 2.373-150 |
Cultures were initiated with freshly isolated BM G0CD34+ cells and G0CD34+PKH2dim cells at day 7 isolated from paired samples as illustrated in Fig 7. Both cultures were supported with 100 ng/mL each of IL-3, IL-6, and SCF in complete medium. At initiation and at 24-hour intervals, samples were assayed for cell-cycle analysis by the propidium iodide method. Results are expressed as the mean ± SEM, n = 3.
P > .05, † P < .05: paired t-tests.
DISCUSSION
In this study, we used simultaneous DNA/RNA staining and flow cytometric cell sorting to isolate and characterize fresh CD34+ cells in different subcompartments of the G0 /G1 phase of the cell cycle, and to examine the relationship between reacquisition of mitotic quiescence and maintenance of primitive hematopoietic potential among proliferating ex vivo–expanded CD34+ cells. Conventional cell-cycle analysis based on measurement of DNA content can classify cells in G0 /G1 or S/G2 + M phases of the cell cycle as noncycling and cycling cell populations, respectively.15,32 This kind of static measurement categorizes all cells containing 2n DNA as G0 /G1 , and as such fails to address the kinetics of cell-cycle progression or heterogeneity of the cell-cycle rate within a given cell population. In addition, since the vast majority of primitive HPCs isolated in steady-state hematopoiesis reside in the G0 /G1 phase,1 the simple distinction between S/G2 + M and G0 /G1 cells is of limited interest. Data obtained with simultaneous DNA/RNA staining allow for further fractionation of G0 /G1 cells and can provide additional insight into the cell-cycle kinetics of hematopoietic cells. Our studies demonstrate that CD34+ cells within the G0 /G1 phase of the cell cycle can be separated into distinct subpopulations based on RNA content. In kinetic experiments reported here (Fig 2), we demonstrate the ability of our staining procedure to discern between mitotically dormant cells (G0CD34+ cells) and those in which cell-cycle activation had already been triggered (G1CD34+ cells).
Discrimination between G0 and G1 phases is a controversial subject. Our definition of G0 cells as those displaying minimal PY staining may appear arbitrary. In fact, we made no attempt to determine absolute counts of CD34+ cells in the G0 phase, but rather assayed the functional properties of cell subsets isolated relative to one another using combined DNA/RNA staining. While expression of other markers such as Ki-6728 or D-type cyclins34 may be more absolute criteria for G0 /G1 discrimination, staining for these markers requires cell permeabilization and therefore prevents functional characterization of cells isolated in separate cell-cycle subcompartments. Since the studies of Darzynkiewicz and Traganos,24 total RNA measurements have been used to describe the progression of cells from a resting state, ie, G0 , to an active prereplicating state, G1 . In addition, the appearance of cell-cycle stage-specific markers such as Ki-67 or D-type cyclins requires RNA synthesis, an event that may be detectable with DNA/RNA staining before the actual appearance of these moieties.
In steady-state BM CD34+ cells, the relationship between position within the G0 /G1 phase and primitive hematopoietic function is striking. Whereas late committed progenitors belonged to immediate prereplicating phases, CD34+ cells isolated in G0 or early G1 were enriched for multipotential CFUs and for LTHC-ICs. These observations support the concept that in steady-state hematopoiesis, the turnover of BM primitive hematopoietic cells is low compared with that of more committed progenitors, and that a direct relationship exists between the rate of cycling and the degree of lineage commitment. These observations are in line with a recent report35 showing a correlation between the proportion of cells in S/G2 + M and the proportion of committed progenitors in murine BM.
The same method of cell-cycle fractionation was less effective in separating primitive LTHC-ICs when applied to MPB CD34+ cells, thus confirming previous reports demonstrating the cell-cycle status homogeneity of MPB CD34+ cells, ie, the small percentage of cells in G281 and the absence of cells in S/G2 + M,18,32 33 compared with BM CD34+ cells. The comparison of correlated LTHC-IC activity and cell-cycle status in BM and MPB reported here (Fig 3) extends these findings and suggests that a subgroup of BM CD34+ cells residing in late G1 phase and whose primitive hematopoietic function is largely compromised are not recovered in the MPB of G-CSF–treated donors.
The application of membrane dye cell tracking coupled with DNA/RNA fractionation allowed us to examine for the first time whether primitive hematopoietic cells (LTHC-ICs) proliferating in vitro are capable of reentering into a G0 state. A relationship between cycling status and hematopoietic function similar to that observed among freshly isolated CD34+ cells was demonstrated for ex vivo–expanded CD34+ cells belonging to different phases of the cell cycle. Indeed, CD34+ cells reisolated with minimal RNA content (G0 ) were enriched for LTHC-IC activity compared with CD34+ cells reisolated in G1 or S/G2 + M. However, when comparing the level of quiescence of freshly isolated G0CD34+ cells with that of G0CD34+ cells recovered after cell division in ex vivo expansion cultures, our data showed that CD34+ cells reentering G0 have a shorter prereplicating phase than freshly isolated G0CD34+ cells. These findings suggest that under our ex vivo culture conditions, once HPCs have started to proliferate in vitro, they can no longer return to the same initial dormancy state, possibly as a result of a progressive loss of developmental capacity. Indeed, our group has recently demonstrated a correlation between cell division and loss of primitive hematopoietic function.27 Together, these observations may be interpreted to suggest that primitive cells reenter a state of transient quiescence after in vitro division, thereby allowing their reisolation in G0 , while less primitive HPCs may bypass the G0 phase and return immediately to a prereplicating stage. These results lend support to a hypothesis proposed by Ogawa8 that “the self-renewal process is associated with renewed dormancy in the cell cycle, while the differentiation process is characterized by continuous cell doubling.”
The initial cell-cycle position before ex vivo expansion of LTHC-ICs reselected in G0 after in vitro proliferation was not directly determined in the present study. Our data (Figs 3 and 5B) show that the majority of LTHC-ICs originate in G0 . However, a significant proportion of these primitive progenitor cells arise from early G1 . Therefore, the possibility exists that a cycling G1 LTHC-IC would reenter G0 after expansion culture. However, the higher generation of CD34+ cells (Table 1) and CFUs (Fig 6) in identical expansion cultures by G0 LTHC-ICs suggests that secondary LTHC-ICs detected in the PKH2dim fraction belong initially to the G0 fraction.
The findings in this communication shed new light on previous data reported by our laboratory. We previously demonstrated in both the human10,27 and the murine11 systems that the primitive hematopoietic potential of primitive HPCs undergoing proliferation in vitro was inferior to that of cells remaining quiescent in expansion cultures, referred to as CNR cells. The present results indicate that although the frequency of LTHC-ICs among proliferating cells was lower than that of CNR cells, the residual primitive hematopoietic activity of in vitro proliferating cells could be mostly attributed to a specific identifiable subgroup of these cells, namely that composed of cells that had reentered into a relative quiescent state. It remains to be established whether different culture conditions such as coculture with stromal layers or addition of cytokines such as Flt3 ligand36-38 or thrombopoietin,39 40 which have been shown to affect activation and maintenance of primitive cells, would modify the distribution of LTHC-ICs among the different fractions isolated after ex vivo expansion as described here.
When viewed together, our results can be used to propose a model of the cell cycle of hematopoietic cells similar to that proposed by Gerdes et al31 for PHA-stimulated peripheral blood lymphocytes. In this model (Fig 9), primitive LTHC-ICs, initially residing in G0 in the BM microenvironment, can enter the cell cycle via start sequences leading to G1 . In the event of a self-renewal cycle, cells may reenter into the initial low RNA dormancy state after mitosis, whereas during the process of differentiation and lineage commitment, cells remain in cycle with a shorter prereplicating phase. Furthermore, our results demonstrate that ex vivo–expanded CD34+ cells isolated with minimal RNA content have a higher degree of responsiveness to cytokine stimulation than their freshly isolated counterparts selected according to the same criteria. Since a direct relationship could be demonstrated between the hematopoietic potential of CD34+ cells and the level of mitotic quiescence both in vivo and during ex vivo culture, it is possible that the loss of LTHC-IC activity in cultured G0CD34+ cells is a reflection of the higher turnover rate of these cells relative to similar cells in vivo. These findings may have important implications in designing strategies for ex vivo expansion and genetic transduction of primitive hematopoietic cells.
A model of the cell cycle of hematopoietic cells showing different pathways of the G0 /G1 phase followed by primitive and committed progenitor cells in vivo and during ex vivo culture.
A model of the cell cycle of hematopoietic cells showing different pathways of the G0 /G1 phase followed by primitive and committed progenitor cells in vivo and during ex vivo culture.
ACKNOWLEDGMENT
The authors thank Ryan Cooper for expert technical assistance in selecting CD34+ cells from MPB.
Supported by National Institutes of Health Grant No. R01-HL55716, National Institutes of Health Centers of Excellence in Molecular Hematology Grant No. P50-DK49218, National Institutes of Health Grant No. P01-HL53586, a research award from the Phi Beta Psi Sorority (E.F.S.), a fellowship from the Fonds National de la Recherche Scientifique (FNRS, Brussels, Belgium; A.G.), and a travel grant from the Centre Anticancéreux près l'ULg (University of Liège, Belgium; A.G.).
Address reprint requests to Edward F. Srour, PhD, Indiana University School of Medicine, 1044 W Walnut St, R4-202, Indianapolis, IN 46202-5121.









This feature is available to Subscribers Only
Sign In or Create an Account Close Modal