Abstract
The α2β1 integrin, a collagen/laminin receptor, is expressed by a variety of cell types, including epithelial cells, mesenchymal cells, and hematopoietic cells. To understand the molecular mechanisms that regulate expression of the α2β1 integrin in cells with megakaryocytic differentiation, we characterized the 5′ flanking region of the α2 integrin gene and identified three distinct regulatory regions, including a core promoter, a silencer, and megakaryocyte enhancers in the distal 5′ flank (Zutter et al, Blood 96:3006, 1995 and Zutter et al, J Biol Chem 269:463, 1994). We now focus on the core promoter of the α2 integrin gene located between bp −30 and −92 that is required for transcriptional activity of the α2 integrin gene. Sequence analysis identified two Sp1 consensus sites and a potential AP2 site. Gel retardation assays showed that nuclear proteins from uninduced K562 cells and K562 cells induced to become megakaryocytic bound specifically to the core promoter region (bp −30 to bp −92) producing two DNA-protein complexes. In addition, nuclear extracts from cells induced along the megakaryocyte lineage produced a selective increase in the slower migrating complex. Site-directed mutagenesis of the 5′, the 3′, or both Sp1 binding sites suggested that both Sp1 binding sites are required for full promoter activity and for DNA-protein complex formation. DNA footprinting also showed specific protection of the 5′ Sp1 site by nuclear extracts from uninduced K562 cells and protection of both the 5′ and the 3′ Sp1 sites by nuclear extracts from induced K562 cells. Sp1 protein-DNA complex formation was dependent on Sp1 phosphorylation. The faster migrating DNA-protein complex was enhanced by dephosphorylation; the slower migrating DNA-protein complex was diminished or lost.
THE INTEGRIN FAMILY of adhesion receptors has been implicated in a number of developmental processes, including cell adhesion, migration, differentiation, and organogenesis.1-5 Accordingly, expression of the distinct integrin subunits is exquisitely regulated. The α2β1 integrin, a collagen/laminin receptor, is expressed by many distinct cell types, including certain epithelial, mesenchymal, and hematopoietic cells.6-12 Expression of the α2 integrin subunit in cells of hematopoietic origin is limited to megakaryocytes and platelets, where it serves as a collagen receptor essential for normal platelet hemostatic function.13,14 During megakaryocytic differentiation, the α2 integrin subunit is expressed at a late stage in megakaryocyte maturation and appears later than the αIIbβ3 integrin.15-17 Consequently, regulation of these two essential megakaryocyte/platelet proteins has similarities but distinct differences.
To understand the regulation of the α2 integrin expression during hematopoietic differentiation, we have used several models of megakaryocytic differentiation. K562 cells, when induced with phorbol esters, develop megakaryocytic features and express functional α2β1 integrin and other megakaryocyte-specific proteins.17 We showed that the increased expression of the α2β1 integrin was due to transcriptional activation of the α2 integrin gene.18 Subsequent studies led to the identification of the transcription start site and putative promoter and enhancer elements within the 5′ flanking region of the gene.19 The promoter region lacks TATA and CAAT boxes but contains an abbreviated initiator sequence. Three distinct regions of the α2 integrin gene promoter are required for expression of the α2 integrin gene in hematopoietic cells with megakaryocytic features.20 These regions include a core promoter region between bp −30 and −92, silencer element(s) between bp −92 and −351, and cell-type–specific enhancers in the distal 5′ flanking region.
The core promoter between bp −30 and −92 of the α2 integrin gene is required for basal promoter activity in cells that express the α2 integrin gene, but not in cells that do not express the integrin.19,20 Sequence analysis of this 62-bp region showed a GC-rich region with two consensus binding sites for Sp1.19 Overlapping the two consensus Sp1 sites is a region with potential for binding the transcription factor AP2. We have now undertaken a detailed molecular analysis of the 62-bp core promoter using the well-characterized model of megakaryocyte differentiation. Using gel-mobility shift, DNA footprinting, and reporter gene assays, we have determined the role of specific transcription factors in mediating function of the core promoter region in α2 integrin gene regulation.
MATERIALS AND METHODS
Cell cultures and transfection assays.Dami and K562 cell lines obtained from the American Type Culture Collection (Rockville, MD) were propagated in RPMI 1640 medium. Megakaryocytic differentiation of K562 was induced by the addition of 40 nmol/L phorbol dibutyrate in dimethyl sulfoxide, as previously described.17
The K562 and Dami cell lines were transfected by electroporation using a BTX Electro Cell Manipulator 600 (BTX Inc, San Diego, CA).21 Approximately 1.0 × 107 cells were transfected in RPMI medium containing 100 μg/mL salmon sperm DNA, 30 μg of plasmid DNA, and 3 μg of RSV-luciferase DNA by electroporation at 275 V and 600 μF.
Cell extracts were harvested after 48 hours. Luciferase activity produced by 10 μL of cell lysate in 190 μL of assay buffer (10 mmol/L Mg[OAc]2 , 50 mmol/L Tris-MES, pH 7.8, and 2 mmol/L ATP) was analyzed using a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA), as described,22 and was used to normalize for transfection efficiency. Cell extracts containing identical amounts of luciferase activity were then assayed for chloramphenicol acetyl transferase (CAT) activity using the standard method of Gorman et al.23 Acetylation of [14C] chloramphenicol was quantified by thin-layer chromatography.
Gel mobility shift analysis.Nuclear extracts were obtained from uninduced and phorbol-induced K562 and Dami cells by isolating nuclei, as described.24 The cells were washed with phosphate-buffered saline and lysed in 0.5% Triton X-100 in 10 mL of STKM buffer (30% sucrose, 40 mmol/L Tris, pH 7.5, 37 mmol/L KCl, 12 mmol/L MgCl2 ). After pelleting, the nuclei were washed in TNM buffer (100 mmol/L Tris, pH 7.5, 10 mmol/L NaCl, 30 mmol/L MgCl2 ). The proteins were extracted from the nuclei using a buffer containing 20 mmol/L HEPES, 20% glycerol, 0.35 mol/L KCl, 0.1 mmol/L EGTA, 0.5 mmol/L EDTA, 1% aprotinin, and 10 μg/mL leupeptin. After incubation with gentle rotation for 1 hour at 4°C, the sample was centrifuged at 100,000g for 2 hours. The supernatant was collected and the concentration of nuclear proteins in the extracts was determined by Bradford assay (Bio-Rad Laboratory, Hercules, CA). Gel-purified double-stranded DNA fragments isolated from the described plasmid were end-labeled with 32P-dCTP using Klenow DNA polymerase.25 Approximately 1 to 2 μg of nuclear protein extract was added with 40,000 counts per minute (cpm) of labeled DNA in 15 μL of binding buffer (10% glycerol, 25 mmol/L Tris, pH 7.5, 100 mmol/L KCl, 5 mmol/L spermidine, 5 mmol/L EDTA, 1 mmol/L dithiothreitol, 2 mmol/L phenylmethylsulfonyl fluoride) and incubated on ice for 15 minutes. In reactions using competitor DNA or oligonucleotide, a molar excess (as designated) of the indicated unlabeled competitor DNA fragment was used. Double-stranded oligonucleotides containing the consensus binding site for Egr-1 or Sp1 were used in competition experiments (Santa Cruz Biotech, Inc, Santa Cruz, CA). In antibody inhibition experiments, the indicated amount of monoclonal anti-Sp1, anti-AP2, or anti-Egr-1 antibody (Santa Cruz Biotech, Inc) was preincubated with the nuclear extract for 18 hours at 4°C before the addition of 32P-labeled probe. The reactions were then analyzed by polyacrylamide gel electrophoresis using 6% acrylamide/Bis (19:1) in Tris, pH 8.8. Experiments requiring dephosphorylation of nuclear extracts were conducted as described.26 Dephosphorylation was performed in 25 mmol/L HEPES, pH 7.5, 34 mmol/L KCl, 50 mmol/L MgCl2 at 33°C for 5 minutes and then for 15 minutes on ice with calf intestinal alkaline phosphatase (1 U/50 μg of nuclear extract). To stop the reaction, phosphatase inhibitors to a final concentration of 10 nmol/L NaF, 10 nmol/L sodium vanadate, 10 nmol/L potassium pyrophosphate, and 5 nmol/L sodium phosphate were added. For the dephosphorylation controls, phosphatase inhibitors were added to the extracts before incubation at 33°C.
Gel mobility shift experiments using recombinant Sp1 protein (Promega, Madison, WI) or recombinant Egr-1 protein (a generous gift from Dr Jeffrey Milbrandt, Washington University School of Medicine, St Louis, MO) were performed in an identical manner as experiments with crude nuclear extracts.
DNA footprinting.The DNA probe was prepared by digesting the 5′ flanking region of the α2 integrin genomic sequence extending from bp −961 to bp +109 cloned into pBS-SK plasmid using Xma III (at bp −92) and Kpn I (within the pBS-SK polylinker) restriction enzymes and subsequently end-labeled with 32P-dCTP by Klenow reaction (only the 5′ end was labeled).25 Twenty thousand cpm of probe were incubated with 25 μg of nuclear extract in the presence of 25 mmol/L Tris, pH 7.9, 0.5 mmol/L EDTA, 50 mmol/L KCl, 0.5 mmol/L dithiothreitol, 10% glycerol, and 2 mg of poly(dI-dC) to a total volume of 50 μg for 15 minutes on ice. After 2 minutes at room temperature, 50 μL of DNase buffer (10 mmol/L MgCl2 , 5 mmol/L CaCl2 ) was added and incubated at room temperature for 1 minute. One unit of DNase I was then added at room temperature for 1 minute. The reaction was terminated using 100 μL of stop solution (20 mmol/L NaCl, 20 mmol/L EDTA, 1% sodium dodecyl sulfate [SDS], and 250 μg/mL yeast tRNA) and 1 μg of proteinase K followed by incubation at 37°C for 1 hour. Two phenol/chloroform extractions and one chloroform extraction were subsequently performed, and the samples were precipitated and counted. Ten thousand cpm of each sample was run on a polyacrylamide-urea sequencing gel, and the results were analyzed by autoradiography.
α2CAT fusion constructs.The pα2 92-CAT and pα2 30-CAT constructs were generated by restriction enzyme digestion of the original pα2 961-CAT construct19 at the restriction enzyme sites Xma III (−92) and Pst I (−30), respectively. The pα2 92-CAT construct was then used as a template for polymerase chain reaction (PCR) reactions to generate the 5′ deletion mutant pα2 65-CAT and constructs containing site-specific mutations that disrupt the two Sp1 binding sites pα265Δ5′Sp1-CAT, pα265Δ3′Sp1-CAT, and pα265Δdbl Sp1-CAT.27-29 The following are 5′ primers used for each construct: pα265-CAT (5′-GGAGGGGCGGGCCGGGGCGG-3′); pα265Δ5′Sp1-CAT (5′-GGAGGGAAAGGCCGGGGCGG-3′); pα265Δ3′Sp1-CAT (5′-GGAGGGGCGGGCCGGGAAAGGCCCTCG-3′); and pα265ΔdblSp1-CAT (5′-GGAGGGAAAGGCCGGGAAAGGCCCTCG-3′).
A common 3′ primer extending from the Ava II site at +110 to +90 was used in all PCR reactions (5′-GACCGTAGTTGCGCTGGGTT-3′). PCR products were blunted with T4 DNA polymerase and treated with T4 DNA-kinase and cloned into pCAT-BASIC at the blunted Sal I site. All constructs were completely sequenced by the dideoxynucleotide chain termination method of Sanger et al30 to insure that the site-specific mutations were incorporated with no errors.
Immunoblot analysis.For immunoblot analysis, cell cultures were lysed in SDS-sample buffer (10% glycerol, 62.5 mmol/L Tris [pH 6.8], 2% SDS, and 2% BME; β-mercaptoethanol). Equivalent amounts of protein lysates were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto Immobilon-P transfer membrane (Millipore, Bedford, MA). Immunoblots were incubated with an appropriate dilution of anti-Sp1 polyclonal antibody (Santa Cruz Biotech, Inc) and secondary horseradish peroxidase-conjugated donkey antirabbit IgG (Amersham Life Science, Arlington Heights, IL). The ECL Chemiluminescence System (Amersham Life Science) was used for visualization.
RESULTS
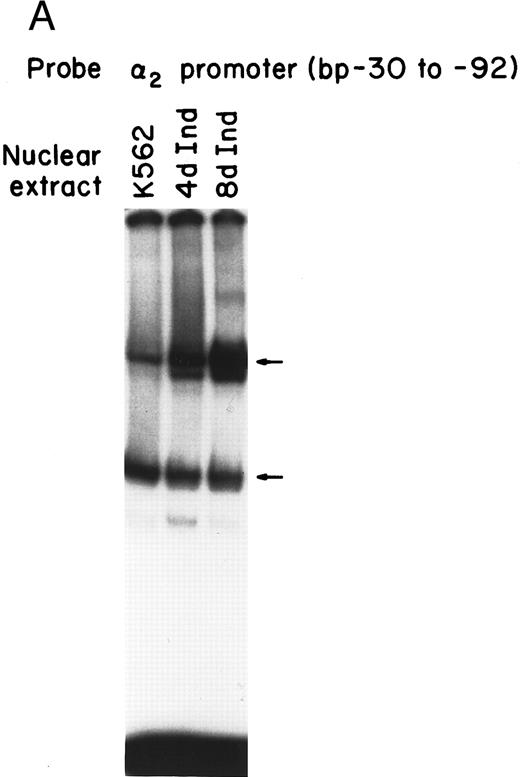
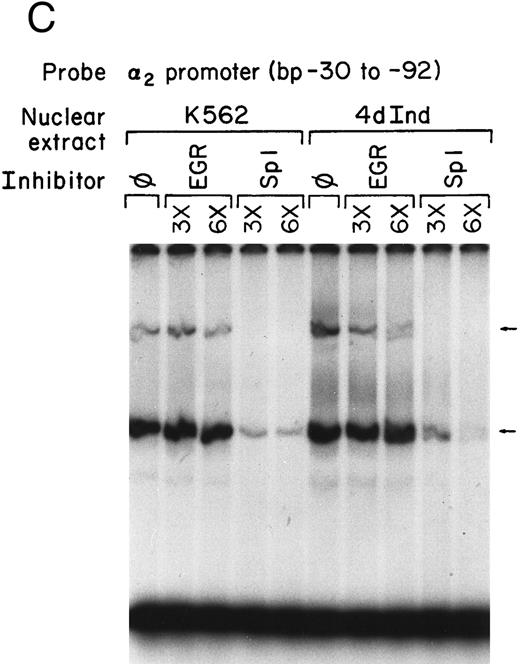
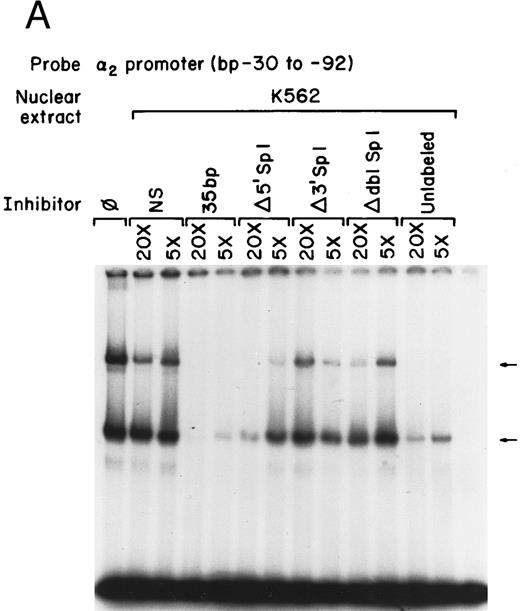
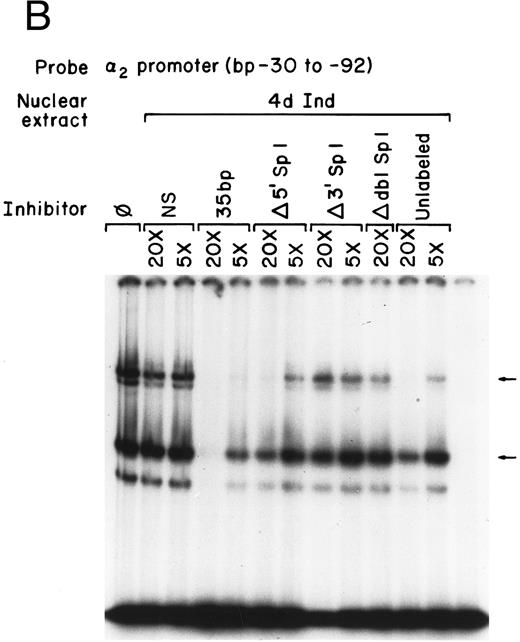
Nuclear proteins bind specifically to adjacent Sp1 consensus sequences in the core promoter.Nuclear proteins from uninduced K562 cells and from K562 cells induced to differentiate along the megakaryocyte pathway for 4 or 8 days with phorbol dibutyrate bound the 62-bp core promoter fragment (bp −30 to −92) in a gel mobility shift analysis. Nuclear extracts from both uninduced and induced K562 cells produced two major, specific DNA protein complexes, as shown in Fig 1A. In addition to the two major complexes observed with extracts from uninduced K562 cells, nuclear extracts from cells induced along the megakaryocytic pathway produced a selective increase in formation of the slower migrating complex that was reproducibly observed with multiple preparations of nuclear extract. Shorter exposure time showed that the enhancement was largely due to the formation of a doublet in the induced cells. These findings suggest that nuclear proteins bind to the core promoter in the uninduced K562 cells. Activation of the α2 gene during megakaryocytic differentiation is accompanied by enhanced binding of nuclear proteins to the core promoter region.
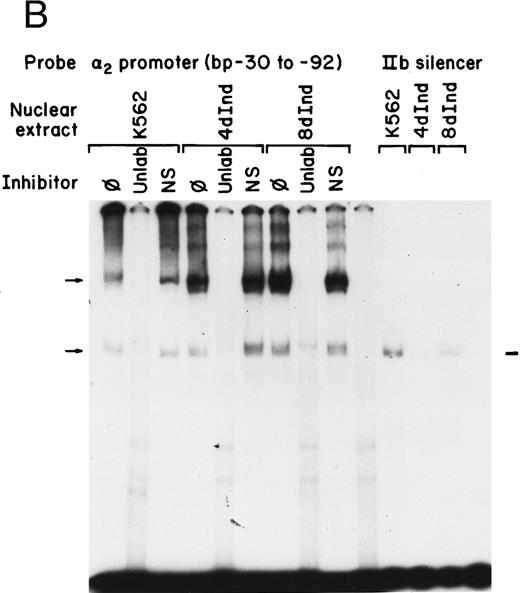
Gel mobility shift analysis of the α2 integrin core promoter. (A) Gel mobility shift analysis was performed with the 32P-end–labeled double-stranded DNA fragment containing the core promoter region (bp −30 to −92) and 1 μg of nuclear protein from uninduced or K562 cells (K562) induced for 4 days (4d Ind) or 8 days (8d Ind) with phorbol dibutyrate. The two major DNA-protein complexes are indicated with arrows. (B) Binding of nuclear proteins from uninduced and induced K562 cells to the 62-bp core fragment is specific. DNA-proteins are formed in the absence of competitive inhibitor (⊘) or in the presence of unlabeled specific inhibitor (Unlab) or a nonspecific (NS) inhibitor consisting of 119-bp double-stranded DNA fragments from the coding region of the α2 cDNA. Gel mobility shift analysis was performed as described in Materials and Methods. The DNA-protein interaction formed with the αIIb integrin silencer domain is indicated with a dash. Gel mobility shift analysis using the αIIb silencer domain served as a positive control for nuclear extracts.
Gel mobility shift analysis of the α2 integrin core promoter. (A) Gel mobility shift analysis was performed with the 32P-end–labeled double-stranded DNA fragment containing the core promoter region (bp −30 to −92) and 1 μg of nuclear protein from uninduced or K562 cells (K562) induced for 4 days (4d Ind) or 8 days (8d Ind) with phorbol dibutyrate. The two major DNA-protein complexes are indicated with arrows. (B) Binding of nuclear proteins from uninduced and induced K562 cells to the 62-bp core fragment is specific. DNA-proteins are formed in the absence of competitive inhibitor (⊘) or in the presence of unlabeled specific inhibitor (Unlab) or a nonspecific (NS) inhibitor consisting of 119-bp double-stranded DNA fragments from the coding region of the α2 cDNA. Gel mobility shift analysis was performed as described in Materials and Methods. The DNA-protein interaction formed with the αIIb integrin silencer domain is indicated with a dash. Gel mobility shift analysis using the αIIb silencer domain served as a positive control for nuclear extracts.
Complex formation was specifically inhibited by unlabeled core promoter fragment, but not by a similar concentration of an irrelevant 119-bp double-stranded DNA fragment (Fig 1B). An oligonucleotide containing the silencer domain of the integrin αIIb gene served as a positive control for our nuclear extracts. Based on recently published data from Fong and Santoro,24 nuclear extracts from uninduced K562 cells bind to this element, thereby silencing αIIb gene activity. However, nuclear extracts from induced K562 cells fail to bind.
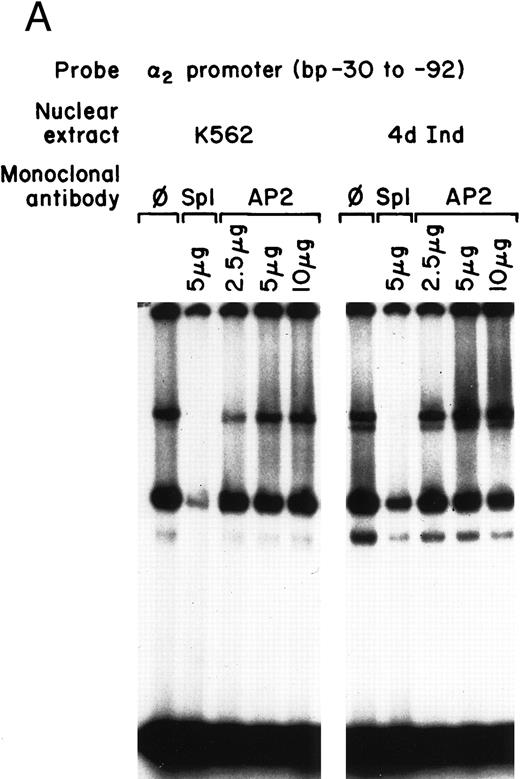
Identification of two potential consensus binding sites for Sp1 and one site for AP2 suggested that one, both, or all three of these sites may be important for core promoter activity. To determine the role of these transcription factors in core promoter activity, inhibitory monoclonal antibodies (MoAbs) to Sp1 and AP2 were used in gel mobility shift analysis. The inhibitory anti-Sp1 MoAb completely inhibited formation of the slower migrating DNA-protein complexes and partially inhibited formation of the faster migrating complex when nuclear extracts from either uninduced or induced K562 cells were used (Fig 2A). The monoclonal anti-Sp1 antibody used in these experiments has been previously shown to inhibit complex formation.31 In contrast to the ability of the anti-Sp1 antibody to inhibit complex formation, an MoAb against AP2, known to effectively inhibit AP2-DNA complex formation, had no effect on DNA-protein interactions in our assay (Fig 2A). These findings suggest that Sp1 is a major component of the DNA-protein complexes identified by the gel mobility shift assay of the 62-bp core promoter and support a role for Sp1, rather than AP2, in the function of the core promoter of the α2 integrin gene.
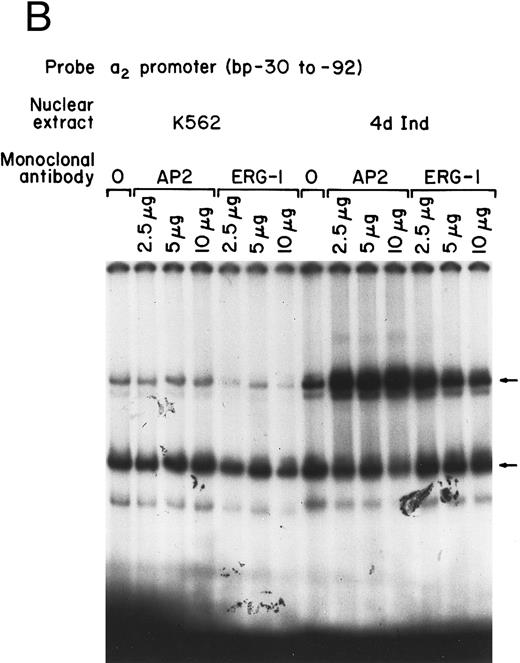
Core promoter-nuclear protein complex formation is inhibited by anti-Sp1 but not anti-AP2 or anti–Egr-1 MoAbs or oligonucleotides. Gel mobility shift experiments were performed with 32P-end–labeled 62-bp core promoter fragment (bp −30 to −92) and 1 μg of nuclear extract from uninduced and phorbol ester-induced K562 cells. MoAbs against AP2 (A) or Egr-1 (B) (at the indicated concentration) were preincubated with nuclear extract for 18 hours at 4°C before the addition of 32P-labeled probe. Unlabeled oligonucleotides containing the Egr or Sp1 consensus sequence (at the indicated molar excess) were incubated with nuclear extracts before the addition of the 32P-labeled 62-bp probe (C).
Core promoter-nuclear protein complex formation is inhibited by anti-Sp1 but not anti-AP2 or anti–Egr-1 MoAbs or oligonucleotides. Gel mobility shift experiments were performed with 32P-end–labeled 62-bp core promoter fragment (bp −30 to −92) and 1 μg of nuclear extract from uninduced and phorbol ester-induced K562 cells. MoAbs against AP2 (A) or Egr-1 (B) (at the indicated concentration) were preincubated with nuclear extract for 18 hours at 4°C before the addition of 32P-labeled probe. Unlabeled oligonucleotides containing the Egr or Sp1 consensus sequence (at the indicated molar excess) were incubated with nuclear extracts before the addition of the 32P-labeled 62-bp probe (C).
The possible role of early growth response (EGR) family of transcription factors in binding to and activating the core promoter was also considered. Egr-1 is involved in the phorbol ester-induced megakaryocyte differentiation of K562 cells. The inhibitory activity of an anti-Egr-1 MoAb shown to either supershift or inhibit EGR-binding was compared with the inhibition by anti-Sp1 and anti-AP2 antibodies.32,33 At high concentrations, the monoclonal anti–Egr-1 antibody slightly inhibited DNA-protein complex formation using either induced or uninduced K562 cells (Fig 2B). The significance of the slight inhibition is uncertain because a classic consensus binding site for Egr-1 is not present in the 62-bp core promoter. Swirnoff and Milbrandt34 determined the binding site preferences for Egr family members by PCR-mediated random site selection. The combined consensus sequence T-G-C-G-T/g-G/A-G-G-C/a/t-G-G/T is not present in the 62-bp core fragment.34 In addition, molar excess of oligonucleotide containing the Egr-1 consensus sequence failed to inhibit complex formation between the 62-bp core and nuclear extracts from induced or uninduced K562 cells. The same molar excess of oligonucleotide containing the Sp1 consensus sequence almost completely eliminated DNA-protein complex formation (Fig 2C). In addition, recombinant Egr-1 protein failed to bind the 62-bp core promoter (data not shown).
To determine the role of Sp1 protein in the two separate DNA-protein complexes formed with the nuclear extracts, gel mobility shift analysis was also performed with recombinant Sp1 protein and the 62-bp core promoter fragment. Nuclear extracts from uninduced and induced K562 cells served as controls. As shown in Fig 3, lower concentrations of recombinant Sp1 protein bound the 62-bp core to form a complex of the same size as the more rapidly migrating complex. At higher concentrations of Sp1 protein, the additional slower migrating complex was also formed. The presence of the slower migrating DNA-protein complex only observed at high concentrations of Sp1 protein in a system reconstituted of only recombinant Sp1 protein and the 62 bp DNA fragment suggests that two Sp1 sites, one of higher and one of lower affinity, are present in the 62-bp DNA fragment. The two complexes formed between the 62-bp core and recombinant Sp1 protein are similar in electrophoretic mobility to those formed with crude nuclear extracts.
Recombinant Sp1 protein bound the core promoter to form the same DNA-protein complexes as nuclear extracts from K562 cells or from K562 cells induced with phorbol dibutyrate. Gel mobility shift experiments were performed using 32P-end–labeled core promoter (bp −30 to −92) and either recombinant Sp1 protein (at the indicated footprinting units [fu]) or nuclear extracts from uninduced and induced K562 cells. A footprinting unit is defined as the amount of protein required to yield a complete footprint on a fragment of the SV40 early promoter. Two DNA-protein complexes are indicated by arrows.
Recombinant Sp1 protein bound the core promoter to form the same DNA-protein complexes as nuclear extracts from K562 cells or from K562 cells induced with phorbol dibutyrate. Gel mobility shift experiments were performed using 32P-end–labeled core promoter (bp −30 to −92) and either recombinant Sp1 protein (at the indicated footprinting units [fu]) or nuclear extracts from uninduced and induced K562 cells. A footprinting unit is defined as the amount of protein required to yield a complete footprint on a fragment of the SV40 early promoter. Two DNA-protein complexes are indicated by arrows.
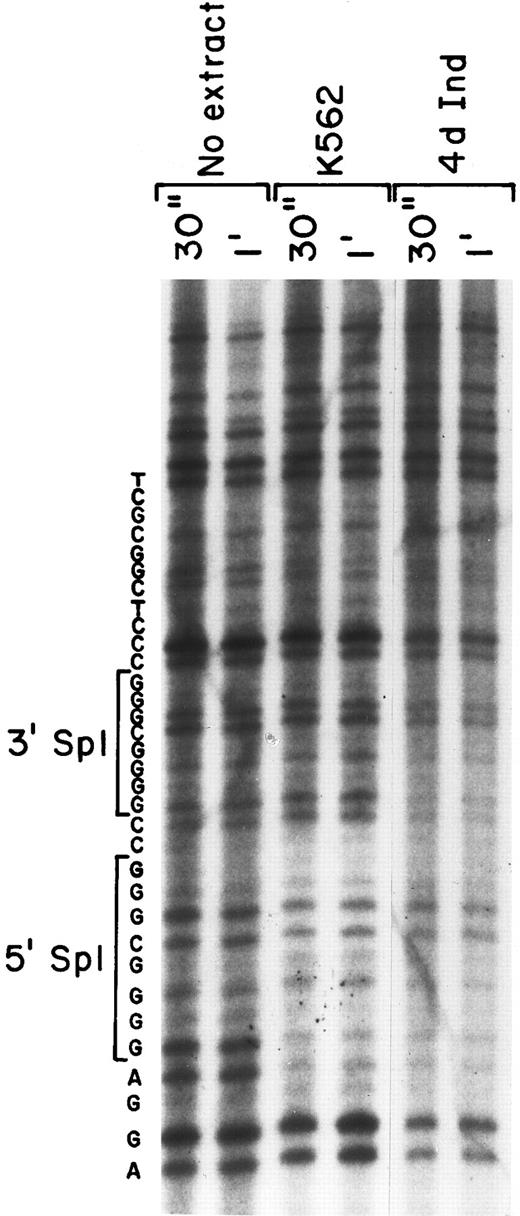
Nuclear proteins interact with both Sp1 sites in DNA footprinting.To verify the binding of nuclear proteins to the DNA motif containing Sp1 consensus sites, we performed in vitro DNA footprinting analysis (Fig 4). Nuclear extracts from both uninduced K562 cells and K562 cells induced for 4 days with phorbol dibutyrate were allowed to bind to the 5′-end–labeled 201 bp fragment (bp −92 to +109) containing the two tandem Sp1 binding sites. After DNase I digestion, the digested products were analyzed on a denaturing SDS-polyacrylamide gel. A sequencing gel of the identical region was run in parallel to allow exact identification of the protected region. The region containing the 5′ Sp1 site was protected from DNase I digestion when incubated with extracts derived from either uninduced or induced K562 cells (Fig 4). In addition, the 3′ Sp1 site was protected with nuclear extracts from induced K562 cells, but was not protected with extracts derived from uninduced K562 cells, as shown in Fig 4. The intensity of the slower migrating bands beyond the protected region at the 3′ end of the probe is similar, documenting approximately equal loading. The nucleotide sequence of the protected region is shown to identify the location of the two Sp1 binding sites.
Two protected regions corresponding to the Sp1 sites were detected by DNA-footprinting analysis. The ability of nuclear proteins to bind and protect the core promoter fragment of the α2 integrin core promoter was assessed. DNase 1 digestion for 30 seconds and 1 minute of the 32P-end–labeled DNA fragment (bp −92 to +109) was performed with no nuclear extract and in the presence of nuclear extracts from uninduced and induced K562 cells (at 70 μg) for 30 seconds and 1 minute. The digested products were analyzed on a denaturing SDS-polyacrylamide gel. Sequence reaction of the DNA region was run in parallel and the sequence of the region is shown.
Two protected regions corresponding to the Sp1 sites were detected by DNA-footprinting analysis. The ability of nuclear proteins to bind and protect the core promoter fragment of the α2 integrin core promoter was assessed. DNase 1 digestion for 30 seconds and 1 minute of the 32P-end–labeled DNA fragment (bp −92 to +109) was performed with no nuclear extract and in the presence of nuclear extracts from uninduced and induced K562 cells (at 70 μg) for 30 seconds and 1 minute. The digested products were analyzed on a denaturing SDS-polyacrylamide gel. Sequence reaction of the DNA region was run in parallel and the sequence of the region is shown.
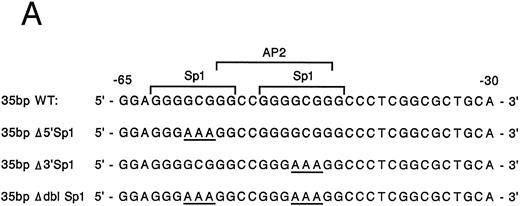
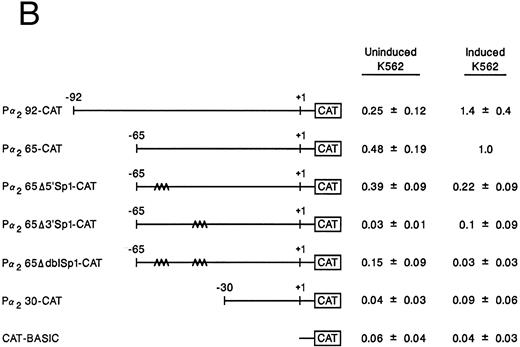
Mutation of one or both Sp1 binding sites diminishes promoter activity.Gel mobility shift analysis suggested that one or probably both of the Sp1 consensus binding sites were important for promoter activity. We therefore focussed on the role of the two Sp1 consensus binding sites in the function of the core promoter. A shorter double-stranded DNA fragment extending from bp −30 to −65, which included the two tandem Sp1 sites as well as the AP2 site but eliminated the more 5′ 27-bp region (bp −65 to −92), was created by PCR and placed upstream of CAT structural gene sequences to form the pα265-CAT construct. Next, mutations in either the 5′ Sp1 site (pα2Δ5′ Sp1-CAT), the 3′ Sp1 site (pα2Δ3′ Sp1-CAT), or both sites (pα2ΔdblSp1-CAT) were made by site-directed mutagenesis of the pα265-CAT construct (Fig 5A). Promoter activity of the mutant constructs pα2Δ5′ Sp1-CAT, pα2Δ3′ Sp1-CAT, and pα2Δdbl Sp1-CAT in uninduced and induced K562 cells were compared with the activity of pα265-CAT construct in induced K562 cells, which was assigned a value of 1.0. Results are recorded as the mean and standard deviation of at least four separate electroporations. The shorter pα265-CAT construct retained much of the activity of the longer pα292-CAT construct in K562 cells induced for 6 days with phorbol dibutyrate. In contrast, the pα265 Δ5′ Sp1-CAT construct was only 22% as active, suggesting that significant but not complete loss of function resulted from mutation of the 5′ Sp1 site. Mutation of the 3′ Sp1 site markedly diminished the ability of the construct to direct CAT reporter gene activity (10% compared with the pα265-CAT construct). Mutation of both Sp1 sites completely eliminated reporter gene activity to the background of pCAT-Basic construct.
Site-directed mutagenesis of the Sp1 binding site within the α2 integrin core promoter altered activity. (A) The diagram shows the sequence of the 35-bp region of the α2 integrin core promoter extending from bp −30 to −65 containing the intact Sp1 and AP2 binding sites and the sequence of the three mutant constructs prepared by PCR containing either a mutated 5′ Sp1 site, a mutated 3′ Sp1 site, or mutations of both Sp1 recognition sites. The shorter double-stranded DNA fragment extending from bp −30 to −65 that included the two tandem Sp1 sites as well as the AP2 site, but lacked 27 bp at the 5′ end (bp −65 to −92), was created by PCR and placed upstream of CAT structural sequences to form the p α265-CAT construct (shown in B). Mutations of the 5′ Sp1 site, the 3′ Sp1 site, or both sites were made by site-directed mutagenesis of the p α265-CAT construct. (B) The activity of the intact or mutant α2 integrin core promoter. The promoter activity of the original construct pα292-CAT; the mutant constructs pα2Δ 5′ Sp1-CAT, pα2Δ 3′ Sp1-CAT, and pα2Δ dbl Sp1-CAT; and pCAT Basic was compared with the activity of the pα2 65-CAT in induced K562 cells after transient transfection of the constructs into either uninduced or induced K562 cells. Cotransfection with RSV luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined by thin-layer chromatography and differential extraction. The mean and standard deviation of CAT activity of the mutant constructs from four separation electroporations was determined relative to the pα265-CAT construct in induced K562 cells that was assigned a value of 1.0.
Site-directed mutagenesis of the Sp1 binding site within the α2 integrin core promoter altered activity. (A) The diagram shows the sequence of the 35-bp region of the α2 integrin core promoter extending from bp −30 to −65 containing the intact Sp1 and AP2 binding sites and the sequence of the three mutant constructs prepared by PCR containing either a mutated 5′ Sp1 site, a mutated 3′ Sp1 site, or mutations of both Sp1 recognition sites. The shorter double-stranded DNA fragment extending from bp −30 to −65 that included the two tandem Sp1 sites as well as the AP2 site, but lacked 27 bp at the 5′ end (bp −65 to −92), was created by PCR and placed upstream of CAT structural sequences to form the p α265-CAT construct (shown in B). Mutations of the 5′ Sp1 site, the 3′ Sp1 site, or both sites were made by site-directed mutagenesis of the p α265-CAT construct. (B) The activity of the intact or mutant α2 integrin core promoter. The promoter activity of the original construct pα292-CAT; the mutant constructs pα2Δ 5′ Sp1-CAT, pα2Δ 3′ Sp1-CAT, and pα2Δ dbl Sp1-CAT; and pCAT Basic was compared with the activity of the pα2 65-CAT in induced K562 cells after transient transfection of the constructs into either uninduced or induced K562 cells. Cotransfection with RSV luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined by thin-layer chromatography and differential extraction. The mean and standard deviation of CAT activity of the mutant constructs from four separation electroporations was determined relative to the pα265-CAT construct in induced K562 cells that was assigned a value of 1.0.
In contrast to the activity in induced K562 cells, CAT activity generated by the pα292-CAT or pα265-CAT construct in uninduced K562 cells is extremely low. The activity of the shorter pα265-CAT construct with two intact Sp1 binding sites was similar to the longer construct pα292-CAT. The mutations of Sp1 in the context of the pα265-CAT construct diminished (the 5′ Sp1 mutation) or eliminated (the 3′ Sp1 mutation) the activity in uninduced K562 cells. The very low levels of CAT activity generated by uninduced K562 cells creates difficulties in comparison. All experiments were conducted in parallel with the induced K562 cells that generate very high levels of CAT activity.
Mutations that inhibit function interfere with DNA-protein complex formation.To determine if the site-directed mutations of the Sp1 binding sites interfered with the DNA-protein complex formation, the gel mobility shift assays were repeated using the original 62-bp fragment extending from bp −30 to −92 as the radio-labeled probe and the 35-bp fragments (bp −30 to −65) containing either the intact or mutated Sp1 sites as unlabeled competitors, as shown in Fig 6A and B. The 35-bp double-stranded fragment effectively inhibited formation of both DNA-protein complexes with nuclear extracts prepared from uninduced K562 cells. The degree of inhibition was similar to that seen with the entire 62-bp fragment (compare Figs 1 and 6A and B). The fragment containing a mutation in the 5′ Sp1 site but an intact 3′ site retained the ability to effectively inhibit formation of the slower migrating complex and partially inhibit formation of the more rapidly migrating complex using extracts from uninduced K562 cells. Similar results were obtained with extracts from uninduced K562 cells. In contrast, the fragment containing a mutated 3′ Sp1 site but an intact 5′ site had little effect on the formation of either complex. The fragment in which both the 5′ and 3′ Sp1 sites were mutated failed completely to inhibit complex formation. These findings are consistent with the functional reporter gene data showing that both the 5′ and 3′ Sp1 sites are necessary for full activity.
Mutations in Sp1 sites interfere with DNA-protein complex formation. Gel mobility shift assays were performed using 32P-end–labeled core promoter fragment (bp −30 to −92) and 1 μg of nuclear extracts from uninduced (A) or 4-day induced (B) K562 cells. Two protein complexes were formed, as indicated by the arrows. Unlabeled double-stranded DNA fragments consisting of either the 35-bp fragment containing intact Sp1 binding sites (bp −30 to −65) or the 35-bp fragments containing the 5′ mutated Sp1 binding site (Δ 5′ Sp1), the 3′ mutated Sp1 binding site (Δ 3′ Sp1), or mutations of both Sp1 binding sites (Δ dbl Sp1) or an irrelevant 119-bp double-stranded DNA fragment were used as competitive inhibitors. The molar excess of the unlabeled competitor is indicated.
Mutations in Sp1 sites interfere with DNA-protein complex formation. Gel mobility shift assays were performed using 32P-end–labeled core promoter fragment (bp −30 to −92) and 1 μg of nuclear extracts from uninduced (A) or 4-day induced (B) K562 cells. Two protein complexes were formed, as indicated by the arrows. Unlabeled double-stranded DNA fragments consisting of either the 35-bp fragment containing intact Sp1 binding sites (bp −30 to −65) or the 35-bp fragments containing the 5′ mutated Sp1 binding site (Δ 5′ Sp1), the 3′ mutated Sp1 binding site (Δ 3′ Sp1), or mutations of both Sp1 binding sites (Δ dbl Sp1) or an irrelevant 119-bp double-stranded DNA fragment were used as competitive inhibitors. The molar excess of the unlabeled competitor is indicated.
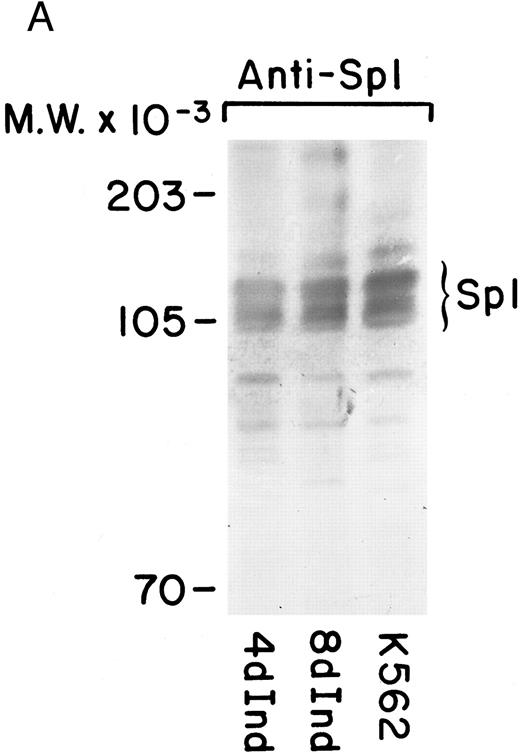
Increased Sp1 binding to the α2 promoter.Because DNA protein interactions were found in both uninduced and induced K562 cells, we were interested in determining the differences in Sp1 activity in uninduced and induced K562 cells. Western blot analysis showed no marked differences in the amount of Sp1 species detectable by immunoblot technique from the induced versus uninduced protein lysate. Furthermore, there was no appreciable difference in the pattern of molecular weight forms in the two samples (Fig 7A). Sp1 existed as a triplet of bands in both uninduced and induced K562 cells, suggesting that Sp1 is phosphorylated in both uninduced and induced K562 cells.
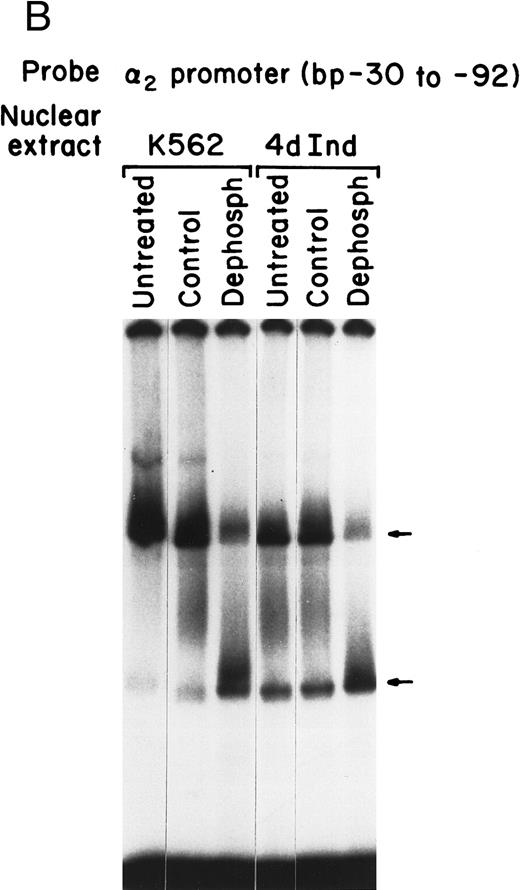
Sp1 binding to the α2 integrin promoter requires phosphorylation. (A) Immunoblot analysis was performed by SDS-PAGE on 100 μg of total protein from cell lysates of uninduced K562 cells and K562 cells induced for 4 days with phorbol dibutyrate using a polyclonal antiserum specific for the Sp1 protein. A triplet of Sp1 bands was shown. (B) Gel mobility shift analysis was performed using 32P-end–labeled α2 integrin core promoter (−32 to −92 bp) and 2 μg of nuclear extracts from uninduced or phorbol ester-induced K562 cells (untreated), nuclear extracts dephosphorylated by calf intestinal phosphatase (dephosph), or nuclear extracts treated with 2 μg of calf intestinal alkaline phosphatase in the presence of phosphatase inhibitors (control). The two specific DNA-protein complexes are indicated with arrows.
Sp1 binding to the α2 integrin promoter requires phosphorylation. (A) Immunoblot analysis was performed by SDS-PAGE on 100 μg of total protein from cell lysates of uninduced K562 cells and K562 cells induced for 4 days with phorbol dibutyrate using a polyclonal antiserum specific for the Sp1 protein. A triplet of Sp1 bands was shown. (B) Gel mobility shift analysis was performed using 32P-end–labeled α2 integrin core promoter (−32 to −92 bp) and 2 μg of nuclear extracts from uninduced or phorbol ester-induced K562 cells (untreated), nuclear extracts dephosphorylated by calf intestinal phosphatase (dephosph), or nuclear extracts treated with 2 μg of calf intestinal alkaline phosphatase in the presence of phosphatase inhibitors (control). The two specific DNA-protein complexes are indicated with arrows.
A role for phosphorylation in posttranslational modification of Sp1 and in regulating DNA-binding activity of Sp1 during differentiation has recently been described by a number of investigators.26 35 To further assess the role of phosphorylation in binding of Sp1 to the α2 integrin core promoter, dephosphorylation of nuclear extracts with calf intestinal alkaline phosphatase or treatment with calf intestinal alkaline phosphatase in the presence of phosphatase inhibitors as a control was performed before gel mobility shift analysis. Using the radiolabeled 62-bp fragment (bp −30 to −92) of the α2 core promoter, Sp1-DNA complex formation in both uninduced and induced K562 nuclear extracts was dependent on phosphorylation. Dephosphorylation of either nuclear extract resulted in enhanced formation of the faster migrating complex and diminished formation of the slower migrating complex, as shown in Fig 7B. The inclusion of phosphatase inhibitors in the dephosphorylation reaction efficiently blocked the phosphatase activity and the consequent inhibition of complex formation. Although there was no difference in the effect of dephosphorylation on complex formation with uninduced or induced K562 nuclear extracts, dephosphorylation diminished binding to the Sp1 site that mediates formation of the slower migrating DNA-protein complex.
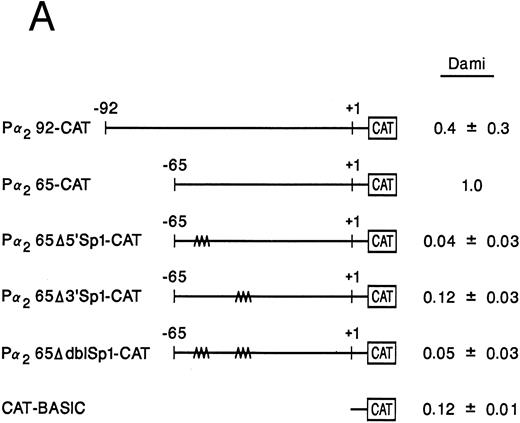
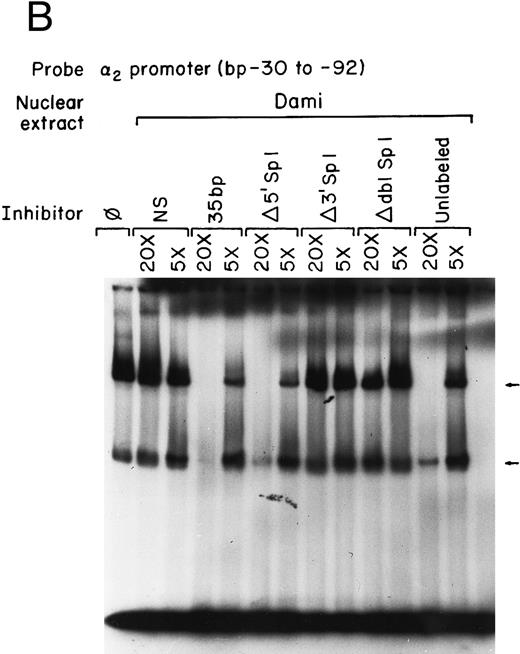
Sp1 binding sites in the core promoter are important for α2 gene expression in other models of megakaryocytic differentiation.To determine the role of Sp1-mediated promoter activity in another model of megakaryocytic differentiation, we determined the functional importance of the two Sp1 sites in the megakaryoblastic leukemia cell line, Dami. The core promoter is active in Dami cells in a manner similar to induced K562 cells. Mutation of the 3′ Sp1 site, the 5′ Sp1 site, or both sites either partially diminished or eliminated activity in a manner similar to that seen in K562 cells (Fig 8A). Nuclear extracts from Dami cells form similar DNA-protein complexes as recombinant Sp1 protein and as nuclear extracts from uninduced and induced K562 cells (Fig 8B). The mobility shift assays support a role for Sp1 in regulating the α2 integrin gene in other models of megakaryocytic differentiation.
The two Sp1 binding sites are required for α2 integrin gene regulation in other models of megakaryocytic differentiation. (A) The relative activities of the intact α2 integrin core promoter construct and the Sp1 mutant constructs were determined in the megakaryoblastic leukemia cell, Dami. The promoter activity of the original construct pα292-CAT, the mutant construct pα2Δ5′ Sp1-CAT, pα2Δ3′ Sp1-CAT, pα2Δdbl Sp1-CAT, and pCAT-Basic was compared with the pα2 65-CAT construct after transient transfection into Dami cells. Cotransfection with pRSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined by thin-layer chromatography and differential extraction. The average and standard deviation of the CAT activity of the mutant constructs relative to the pα265-CAT construct was determined. (B) Nuclear extracts from Dami cells bind the α2 integrin core promoter. The 62-bp fragment of the α2 integrin promoter (extending from bp −30 to −92) specifically bound nuclear proteins from Dami cells producing two DNA-protein complexes. Gel mobility shift analysis was performed with the 32P-end–labeled double-stranded DNA fragment containing bp −30 to −92 and 1 μg of nuclear protein from Dami cell lines. Two DNA-protein complexes are indicated with arrows. Unlabeled double-stranded DNA fragments consisting of either the 35-bp fragment containing intact Sp1 binding sites (bp −30 to −65) or the 35-bp fragment containing the 5′ mutated Sp1 binding site (Δ5′ Sp1), the 3′ mutated Sp1 binding site (Δ3′ Sp1), or mutations of both Sp1 binding sites (Δdbl Sp1) or an irrelevant 119-bp double-stranded DNA fragment were used as competitive inhibitors. The molar excess of the unlabeled competitor is indicated.
The two Sp1 binding sites are required for α2 integrin gene regulation in other models of megakaryocytic differentiation. (A) The relative activities of the intact α2 integrin core promoter construct and the Sp1 mutant constructs were determined in the megakaryoblastic leukemia cell, Dami. The promoter activity of the original construct pα292-CAT, the mutant construct pα2Δ5′ Sp1-CAT, pα2Δ3′ Sp1-CAT, pα2Δdbl Sp1-CAT, and pCAT-Basic was compared with the pα2 65-CAT construct after transient transfection into Dami cells. Cotransfection with pRSV-luciferase was used to control for transfection efficiency. After 48 hours of incubation, cell extracts were assayed. After normalization for transfection efficiency, CAT activity of the constructs was determined by thin-layer chromatography and differential extraction. The average and standard deviation of the CAT activity of the mutant constructs relative to the pα265-CAT construct was determined. (B) Nuclear extracts from Dami cells bind the α2 integrin core promoter. The 62-bp fragment of the α2 integrin promoter (extending from bp −30 to −92) specifically bound nuclear proteins from Dami cells producing two DNA-protein complexes. Gel mobility shift analysis was performed with the 32P-end–labeled double-stranded DNA fragment containing bp −30 to −92 and 1 μg of nuclear protein from Dami cell lines. Two DNA-protein complexes are indicated with arrows. Unlabeled double-stranded DNA fragments consisting of either the 35-bp fragment containing intact Sp1 binding sites (bp −30 to −65) or the 35-bp fragment containing the 5′ mutated Sp1 binding site (Δ5′ Sp1), the 3′ mutated Sp1 binding site (Δ3′ Sp1), or mutations of both Sp1 binding sites (Δdbl Sp1) or an irrelevant 119-bp double-stranded DNA fragment were used as competitive inhibitors. The molar excess of the unlabeled competitor is indicated.
DISCUSSION
Regulation of the α2 integrin gene in cells of hematopoietic origin requires three regions of the 5′ flanking region, a core promoter, a silencer, and cell-type–specific enhancers in the distal 5′ flank.19 20 In this report, we describe a detailed analysis of the core promoter region.
Sequence analysis of the core promoter region showed two Sp1 sites overlapping a potential AP2 site between bp −30 and −92.19,20 The core promoter requires the two tandem Sp1 binding sites, but not the AP2 site, for high-level promoter activity in the hematopoietic cell line K562 induced to acquire megakaryocytic features or Dami cell line derived from a patient with megakaryoblastic leukemia. Because Sp1 is generally considered a ubiquitously expressed transcription factor involved in regulation of many housekeeping genes with TATA-less promoters, we initially considered the AP2 site to be a more likely candidate to mediate the induced expression of the α2 integrin gene.27,29,36 However, gel mobility shift assays, DNA footprinting analysis, and reporter gene assays all showed that the two tandem Sp1 binding sites rather than the AP2 site were responsible for the core promoter activity. In addition, gel mobility shift analysis and DNA footprinting showed increased Sp1 binding to the core promoter in cells induced to become megakaryocytic. Our data suggest that enhanced Sp1 binding to the core promoter mediates the greatly enhanced promoter activity in cells expressing the integrin. It seems likely that modification of the Sp1 protein or its association with other undetected proteins is responsible for enhanced binding to the core promoter and consequently α2 integrin promoter activity.35 37-39
Although Sp1 is ubiquitously expressed and was initially thought to regulate ubiquitously expressed genes, the role of Sp1 in transcriptional regulation of cell type and differentiation dependent genes is now established.40-43 Sp1 is involved in transcription of many cell-type–specific genes, including the myeloid specific integrin gene CD11b, the monocyte specific gene CD14, the liver specific α1 acid glycoprotein, and the neural specific acetylcholine receptor α3 gene.40,44-46 Although these genes all exhibit cell-type–specific and differentiation-specific patterns of expression, they are regulated through a TATA-less promoter interacting with Sp1. In such TATA-less promoters, interaction between Sp1 and transcription factor IID complexed to an initiator element can initiate transcription.47
Some cell-type–specific promoters require cooperative activity of the ubiquitous Sp1 protein with cell-type–restricted transcription factors, such as Egr-1 for thrombospondin and platelet-derived growth factor (PDGF ) B-chain, AP2 for the acetylcholine receptor α3 gene, or GATA 1 for the γ-globin gene.46 48-50 Based on our mutational analysis, no sites in the core promoter of the α2 integrin gene other than the two Sp1 binding sites are required for transcriptional activation. Our mutational analysis shows that the 3′ Sp1 site is more important than the 5′ site, but that both sites together are required for full transcriptional activation. Gel mobility shift analysis did not identify DNA-protein complexes other than those formed by Sp1. Gel mobility shift analysis using inhibitory MoAbs directed against AP2 further excluded a role for AP2 binding.
Members of the EGR family were also considered because Egr-1 binds a GC-rich target sequence that closely resembles the Sp1 binding site.51-53 In addition, Egr-1 is involved in the phorbol ester-induced megakaryocytic differentiation of K562 cells, and Egr-1 can function in association with Sp1.54,55 Overlapping Egr-1 and Sp1 sites have been identified in the promoters of the Egr1, the tumor necrosis factor α, the mouse adenosine deaminase, the mouse thrombospondin, the human interleukin-2, and the PDGF B-chain genes.48,50,55-58 Although the core promoter of the α2 integrin is highly GC-rich, the consensus binding site in the core promoter region is not identical to the consensus site for binding to Egr family members.34 The time course for Egr-1 mRNA expression by K562 cells induced to become megakaryocytic does not correspond to the time course of α2 integrin gene expression. Egr-1 mRNA is induced by 0.5 hours, peaks by 1 to 2 hours, and is completely gone by 24 hours.54 In contrast, our own results show that α2 mRNA is not detected until at least 24 hours and peaks at 4 to 6 days.18 Inhibitory MoAbs to Egr-1 and an oligonucleotide containing the Egr consensus binding site failed to alter DNA-complex formation in gel mobility shift analysis. In addition, recombinant Egr-1 failed to bind the core promoter. These experiments suggest that only Sp1 is required for DNA-binding to the core region and activation of the α2 integrin gene.
Transcriptional activation of the α2 integrin gene requires two tandem Sp1 binding sites. Enhanced DNA-Sp1 complex formation is associated with high-level activity of the α2 integrin gene promoter. One mechanism by which Sp1 may be modified to activate gene expression is phosphorylation.22,35 We showed that phosphorylation is required for Sp1 to bind to the lower affinity Sp1 site of the core promoter in both uninduced and induced K562 cells. The protein target(s) of phosphatase activity may be Sp1 itself, other proteins involved in coactivater complexes, or upstream mediators involved in the signaling pathways. These findings and those of our DNA footprint analysis suggest that enhanced binding of Sp1 by phosphorylation of Sp1 or an associated protein leads to greater DNA-protein complex formation after induction along the megakaryocytic pathway. The formation of Sp1-coactivator complexes that modify Sp1 binding affinity or activity is a potential mechanism for the increased promoter activity. Sp1 has been known to interact with other factors such as the retinoblastoma protein in vivo and to form protein-protein complexes not easily detected by routine binding studies.39 Enhanced DNA-protein complex formation appears to be required for high-level promoter activity of the α2 integrin gene.
ACKNOWLEDGMENT
The authors thank Samuel A. Santoro for encouragement and advice during the course of this work and for constructive criticism and review of the manuscript. The authors also thank Marian Bentz for excellent secretarial assistance.
Supported by National Institutes of Health Grant No. RO1 HL51450.
Address reprint requests to Mary M. Zutter, MD, Box 8118, Department of Pathology, Washington University School of Medicine, 660 S Euclid Ave, St Louis, MO 63110.

![Fig. 3. Recombinant Sp1 protein bound the core promoter to form the same DNA-protein complexes as nuclear extracts from K562 cells or from K562 cells induced with phorbol dibutyrate. Gel mobility shift experiments were performed using 32P-end–labeled core promoter (bp −30 to −92) and either recombinant Sp1 protein (at the indicated footprinting units [fu]) or nuclear extracts from uninduced and induced K562 cells. A footprinting unit is defined as the amount of protein required to yield a complete footprint on a fragment of the SV40 early promoter. Two DNA-protein complexes are indicated by arrows.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/2/10.1182_blood.v90.2.678/4/m_bl_0025f3.jpeg?Expires=1767958460&Signature=a3TvtPMyi~c29C2FwmaKwzgWRK239sPHBQuJEKjgflHkCmoeXoKWdwn48brFanTs890R6CVrMXZIy6q3pgQ1CTJ23it2d24efCROl31iWwybvsz5Bc6-B5aufZY9U7IeHJEb-xOKVhg0cxLyFAQ6xhTAMiOssrXPutL0tKqtdzfQDnQM6E3EBlJeFw9NPFnaNt59qFRxZSRKPHgq3upV4Mzdj3RFFdGAd6XnnBB48IfJUxmNTZsN9tlt7HTlQiALe4nVuGy4j7lzc6aldiSojUuVHuxGxLkLjT4ajRgqOUvxNSTo61fAbpDIejz7sbLl2INuB4ICSTQpmxvGGPKBLA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal