Abstract
Previous work showed that acute myelocytic leukemia blasts accumulate less long chain polyglutamates of methotrexate (MTX) than acute lymphocytic leukemia blasts when incubated with this radiolabeled antifolate. This difference likely explains the increased sensitivity of lymphoid leukemias to short-term exposure of MTX as compared with myeloid leukemias. In this study, we examined the basis for differences between long chain MTX polyglutamate accumulation between different leukemia cell types using both leukemia cell lines and blasts freshly isolated from blood of leukemic patients. The major difference found between leukemia cells that accumulate long chain polyglutamates and those that do not were differences in Km values for the enzyme folylpolyglutamate synthetase. Km values did not change with partial purification of this enzyme, indicating that interfering substances in crude lysates were not responsible for this difference. We postulate that there may be differences in the properties of this enzyme related to tissue specific expression. In contrast to MTX, both Tomudex (Zeneca Pharmaceuticals, Wilmington, DE) and 1843U89, potent inhibitors of thymidylate synthetase, have low Kms for folylpolyglutamate synthetase, and polyglutamate forms of these inhibitors are accumulated to the same degree in both myeloid and lymphoid acute leukemia cells, paralleling the equivalent cytotoxicity found between myeloid and lymphoid leukemia cell lines. Based on these results, we believe a clinical trial of Tomudex in patients with acute myeloid leukemia is warranted.
METHOTREXATE (MTX), a key drug in the curative regimen of children with acute lymphocytic leukemia (ALL), is less effective in the treatment of acute myeloid leukemia (AML).1,2 An important correlate of this difference in sensitivity to MTX was found to be the high levels of MTX acccumulation in pre-B ALL cells as compared with AML cells, due to polyglutamylation of this drug.1,3,4 However, as has been observed previously, there are some myeloid cell lines such as THP-1 that show adequate accumulation of intracellular MTX polyglutamates and as a result are sensitive to MTX,5 as are approximately 10% of AML patients.6 Like folates, intracellular MTX is extensively metabolized to polyglutamate derivatives, which are retained within the cell in the absence of extracellular drug, whereas, under the same conditions, the parental compound rapidly effluxes from the cell. The ability to remain within the cell is influenced by the chain length so that the long chain MTX polyglutamates (three to six glutamates) are retained much longer than MTX or MTX diglutamate.1,2 7 MTX polyglutamates result in a prolonged dihydrofolate reductase (DHFR) inhibition, and thymidylate synthetase (TS), glycinamide ribonucleotide (GAR), and aminoimidazole carboxamide (AICAR) transformylases are inhibited, as well. 8,9
The amount of long chain polyglutamates in cells represents a result of the balance between the activity of two different enzymes, folylpolyglutamate synthetase (FPGS), which adds up to 4 glutamyl groups in γ-peptide linkage to MTX,10-12 and γ-glutamyl hydrolase (GGH), a lysosomal exopeptidase that hydrolyses MTX to the monoglutamate form.13 In the present study, we investigated possible causes for the reduced accumulation of long chain MTX polyglutamates observed in AML blast cells as compared with ALL blasts. In addition, two new antifolate inhibitors of thymidylate synthetase (TS), tomudex (ZD1694)14,15 and 1843U89,16 were evaluated as substrates for FPGS in representative myeloid (K562, HL-60) and lymphoid cell lines (CCRF-CEM, MOLT-3), as well as blast cells from patients.
MATERIALS AND METHODS
Cell lines and materials.Cell lines CCRF-CEM, K562, HL-60, and MOLT-3 were obtained from American Tissue Type Culture Collection (Rockville, MD) and were grown at 37°C in a 5% CO2 humidified atmosphere in RPMI 1640 media containing 10% fetal bovine serum (FBS) and 1% glutamine. MTX was obtained from Lederle Laboratories (Pearl River, NJ). [3H] MTX (20 Ci/pmol) was purchased from Moravek Biomedical (Brea, CA). Tomudex and [3H]tomudex (19.1 Ci/pmol) were supplied by Zeneca Pharmaceuticals (Wilmington, DE). 1843U89 and [3H]1843U89 (31 Ci/pmol) were gifts from Dr Robert Ferone from Glaxo-Wellcome Laboratories (Research Triangle Park, NC). L-[2,3-3H]Glutamic acid NET 395(22 Ci/pmol) was purchased from Dupont NEN (Boston, MA). All other chemicals were obtained from Sigma Chemicals (St Louis, MO).
Cytotoxicity tests.A colorimetric assay using the tetrazolium salt, sodium 3′-{1 -[(phenylamino)-carbonyl]-3,4-tetrazolium}-bis (4-methoxy-6-nitro)benzene-sulfonic acid hydrate (XTT) was used to measure the cytotoxicity of MTX, Tomudex, and 1843U89, as previously described.17 Briefly, cells growing in midlog phase were exposed to the drugs for 24 hours, then washed twice with cold phosphate-buffered saline (PBS). Drug-free medium was added and growth was followed for an additional 96 hours. XTT/phenazine methosulfate (PMS) solution was added to the culture and the optical density of the wells was determined using a EL340 microplate reader from Biotek Instruments (Winooski, VT) at a test wavelength of 450 nm and a reference wavelength of 650 nm.
Polyglutamylation assay.Intracellular polyglutamylate accumulation was measured using a method previously described.18 Briefly, intracellular levels of MTX, Tomudex, and 1843U89 and their polyglutamates were measured by exposing cell lines to 1 μmol/L radiolabeled drug for 24 hours. The cells were then washed twice with PBS at 4°C and the pellets were suspended in 0.5 mL of boiling 50 mmol/L sodium phosphate (pH 5.5), boiled for an additional 5 minutes and centrifuged at 15,000g for 10 minutes at 4°C. Supernatants were stored at −20°C until high performance liquid chromatography (HPLC) analysis was performed. HPLC separations were done on an absorbosphere C18 column (5-μm particle size, 4.6 × 250 mm) equipped with a 1 cm guard cartridge column of the same material from Alltech Associates (Deerfield, IL) using a Spectra Physics 8800 HPLC gradient pump from Thermo Separation (Fremont, CA) with an appropriate gradient specific to each drug. Radioactivity in each 1 mL fraction was measured by a Beckman LS-5801 liquid scintillation counter from Beckman Instruments (Fullerton, CA).19
FPGS assay.FPGS activity in cell extracts was determined as previously described.10 20 Crude extracts were prepared from cell pellets by suspension in 5 vol of extraction buffer (50 mmol/L Tris-HCl, 20 mmol/L Kcl, 10 mmol/L MgCl2 , 5 mmol/L dithiotreitol, pH 7.6) at 4°C, sonication (5 seconds × 3 at intervals of 30 seconds) and centrifugation (4°C) at 20,000g for 20 minutes. The FPGS assay mix contained, in a final volume of 250 μL and at pH 8.4, 100 mmol/L Tris-HCl (pH 8.85), 10 mmol/L adenosine triphosphate (ATP), 20 mmol/L MgCl2 , 20 mmol/L KCl, 10 mmol/L dithiothreitol (DTT), 4 mmol/L L-[3H]glutamate, enzyme and substrate at different concentrations (either MTX, Tomudex, or 1843U89). The reactions were stopped after 2 hours by the addition of 1 mL of 5 mmol/L L-glutamate (pH 7.5) at 4°C. The unreacted [3H]glutamate was separated from the polyglutamates by a Sep-Pack C18 cartridge reversed phase chromatography from Waters (Milford, MA). Activity of FPGS was expressed in pmol [3H]glutamate incorporated/mg protein/hour at 37°C.
Partial purification of folylpolyglutamate synthetase.FPGS was partially purified based on previously described methods.10 12 Cell extracts were prepared as follows: 9 × 109 cells (CCRF-CEM) and 12 × 109 cells (K562) were suspended in 40 mL and 65 mL extraction buffer, sonicated as described and centrifuged at 20,000g for 20 minutes. Streptomycin sulfate at a final concentration of 1% was added to the supernatant solution and the mixture stirred for 1 hour. The precipitate was removed by centrifugation at 16,000g for 30 minutes and discarded. Solid ammonium sulfate was slowly added to the streptomycin sulfate supernatants with stirring at 4°C until 40% saturation was reached . The solution was stirred for 1 hour and then centrifuged for 30 minutes at 27,000g. The pellets were redissolved in 12 mL and 16 mL extraction buffer, respectively. After overnight dialysis against one change of 5 L of Tris-HCl 50 mmol/L, DTT 5 mmol/L, and benzamidine 10 mmol/L pH 7.5 buffer, the dialyzed fractions were applied to P11 phosphocellulose (Whatman Ltd, Maidstone, UK) columns that had been equilibrated with the same buffer. The columns were eluted with a linear gradient of KCl (0 to 200 mmol/L) in the same buffer. Enzyme activity was detected in one peak in both preparations.
Patient samples.Leukemia cells were obtained from heparinized bone marrow aspirates or leukophoresis collections after obtaining written informed consent. The samples contained greater than 90% blast cells. Mononuclear cells were isolated using a Ficoll Hypaque centrifugation gradient at 4°C and washed twice with PBS. The cells for the FPGS assay were pelleted and stored at −70°C until analysis. Cells were resuspended in RPMI 1640 with 10% dFBS and 1% glutamine, for the polyglutamylation assay.
Subcellular fractionation.For the subcellular fractionation of CCRF-CEM and K562 cells and the measurement of subcellular distribution of FPGS, we used a modification of the method described by Lin et al21 and the same procedures for the enzymatic determination of the mitochondrial, cytosolic, and lysosomal markers. Briefly, cells were pelletted by centrifugation at 300g for 5 minutes and resuspended in homogenization solution (HMS, 250 mmol/L sucrose,1 mmol/L EDTA, pH 6.9). The cells were disrupted in a nitrogen cavitation device after exposing them to a pressure of 360 p.s.i. for 30 minutes and then centrifuged at 900g for 6 minutes to allow sedimentation of nuclei and unbroken cells. The supernatant was transferred to another tube and stored on ice, the pellet (nuclear fraction) was washed and resuspended in HMS. The second supernatant was combined with the first and then centrifuged. The supernatant (total postnuclear supernatant, PNS) was centrifuged at 10,000g for 15 minutes and the pellet was stored on ice. The supernatant was recentrifuged at 10,000g for 15 minutes to give a final supernatant (cytosolic fraction). The two last pellets were combined and resuspended in HMS to obtain the mitochondrial fraction.
RESULTS
Four cell lines, two lymphoid (CCRF-CEM and MOLT-3) and two myeloid (K562 and HL-60) were studied. CCRF-CEM, MOLT-3, and HL-60 were found to be more sensitive to 24-hour exposure of MTX, as compared with K562 cells, varying from 4- to 15-fold, as measured by IC50 values (Table 1). In contrast, all four cell lines were equally and much more sensitive to Tomudex and to 1843U89 than MTX (Table 1). When polyglutamate levels were measured after incubation with radiolabeled MTX, Tomudex, and 1843U89, respectively, the lowest levels of MTX polyglutamates were found in K562 cells, while a high percentage of Tomudex and 1843U89 (80% or greater) were found as polyglutamates in all the cell lines. All four cell lines did not show any difference in MTX uptake as measured by the PT430 displacement assay.22 Differences in DHFR protein levels were not found by Western blot analysis (data not shown).
Inhibitory Effects, Accumulation of Polyglutamates and Kinetic Parameters of FPGS in ALL and AML Cell Lines With MTX, Tomudex, and 1843U89 as Substrates
| . | IC50 . | Long Chain Polyglutamates . | Long Chain Polyglutamates as % of Total Polyglutamates . | Km . | Vmax . |
|---|---|---|---|---|---|
| . | (nmol/L) . | (pmol/107 cells) . | . | (μmol/L) . | (pmol/mg/h) . |
| MTX | |||||
| CCRF-CEM | 171 ± 15 | 59.6 ± 2.6 | 85 | 44 ± 0.2 | 1,575 ± 152 |
| MOLT-3 | 50 ± 10 | 35.2 ± 3.3 | 80 | 23 ± 4.5 | 1,230 ± 41 |
| K562 | 740 ± 14 | 14.5 ± 2.3 | 28 | 92 ± 9 | 1,473 ± 13 |
| HL-60 | 103 ± 23 | 53.7 ± 8.6 | 73 | 89 ± 4.6 | 5,067 ± 1,167 |
| Tomudex | |||||
| CCRF-CEM | 15 ± 0.5 | 7.43 ± 2.2 | 90 | 0.5 ± 0.12 | 1,566 ± 350 |
| MOLT-3 | 12 ± 0.5 | 5.78 ± 0.4 | 83 | 1.2 ± 0.2 | 1,574 ± 89 |
| K562 | 11 ± 0.5 | 7.23 ± 1.2 | 88 | 0.6 ± 0.2 | 1,358 ± 365 |
| HL-60 | 8 ± 1 | 5.43 ± 0.4 | 85 | 1.6 ± 0.5 | 2,400 ± 575 |
| 1843U89 | |||||
| CCRF-CEM | 4 ± 1 | 102 ± 7 | 97 | 2.4 ± 0.9 | 3,274 ± 1,214 |
| MOLT-3 | 4 ± 0.6 | 41 ± 6 | 87 | 6.2 ± 0.3 | 3,320 ± 170 |
| K562 | 10 ± 4 | 59 ± 6 | 97 | 1.2 ± 0.3 | 1,423 ± 180 |
| HL-60 | 3 ± 1 | 56 ± 17 | 92 | 4.5 ± 0.7 | 4,200 ± 260 |
| . | IC50 . | Long Chain Polyglutamates . | Long Chain Polyglutamates as % of Total Polyglutamates . | Km . | Vmax . |
|---|---|---|---|---|---|
| . | (nmol/L) . | (pmol/107 cells) . | . | (μmol/L) . | (pmol/mg/h) . |
| MTX | |||||
| CCRF-CEM | 171 ± 15 | 59.6 ± 2.6 | 85 | 44 ± 0.2 | 1,575 ± 152 |
| MOLT-3 | 50 ± 10 | 35.2 ± 3.3 | 80 | 23 ± 4.5 | 1,230 ± 41 |
| K562 | 740 ± 14 | 14.5 ± 2.3 | 28 | 92 ± 9 | 1,473 ± 13 |
| HL-60 | 103 ± 23 | 53.7 ± 8.6 | 73 | 89 ± 4.6 | 5,067 ± 1,167 |
| Tomudex | |||||
| CCRF-CEM | 15 ± 0.5 | 7.43 ± 2.2 | 90 | 0.5 ± 0.12 | 1,566 ± 350 |
| MOLT-3 | 12 ± 0.5 | 5.78 ± 0.4 | 83 | 1.2 ± 0.2 | 1,574 ± 89 |
| K562 | 11 ± 0.5 | 7.23 ± 1.2 | 88 | 0.6 ± 0.2 | 1,358 ± 365 |
| HL-60 | 8 ± 1 | 5.43 ± 0.4 | 85 | 1.6 ± 0.5 | 2,400 ± 575 |
| 1843U89 | |||||
| CCRF-CEM | 4 ± 1 | 102 ± 7 | 97 | 2.4 ± 0.9 | 3,274 ± 1,214 |
| MOLT-3 | 4 ± 0.6 | 41 ± 6 | 87 | 6.2 ± 0.3 | 3,320 ± 170 |
| K562 | 10 ± 4 | 59 ± 6 | 97 | 1.2 ± 0.3 | 1,423 ± 180 |
| HL-60 | 3 ± 1 | 56 ± 17 | 92 | 4.5 ± 0.7 | 4,200 ± 260 |
Cell lines were exposed to different concentrations of these drugs and cell viability measured as described in Materials and Methods. Polyglutamates formation was measured after a 24-hour exposure to radiolabeled drugs as also described in Materials and Methods. Cells extracts were prepared as described in Materials and Methods. Substrates at different concentrations were used to measure enzyme activity and to determine Km and Vmax values. The results are the average of at least 3 or more experiments with standard deviations provided.
In CCRF-CEM, MOLT-3, and HL-60 cells, the predominant polyglutamate form of MTX was MTX glu(3) and MTX glu(4) , while the predominant form of MTX in K562 cells was MTX glu(1) . Tomudex polyglutamate forms in all four cell types were glu(3) and glu(4) , while 1843U89 diglutamates were found almost exclusively in the four leukemia cell types (data not shown).
Of the three drugs, 1843U89 is clearly the most potent and correlated with high levels of the diglutamate of this drug achieved in all four cell lines. As cytotoxicity of K562 cells to MTX was much less than the other three cell types and was correlated to low levels of MTX polyglutamates formed, we then attempted to understand the basis for these results. The activity of FPGS in crude extracts of K562 cells was similar to that in CCRF-CEM and MOLT-3 cells, (1,220 ± 30 v 1,380 ± 180 and 1,140 ± 40 pmol/mg/h, respectively), but lower than that in HL-60 cells (3,330 ± 880 pmol/mg/h), indicating that the level of this enzyme activity was not responsible for the low level of polyglutamates formed. When γ-glutamyl hydrolase activity was measured in extracts of the four cell lines, K562 cells were found to have a lower level of this activity when compared with CCRF-CEM cells (data not shown), therefore, the lack of accumulation of MTX polyglutamates in K562 cells could not be attributed to increased MTX polyglutamate breakdown.
MTX is transported into leukemia cells via the reduced folate transporter, a carrier-mediated saturable transport process. Therefore, the Km for various folate substrates for FPGS is important for conversion of folate antagonists to polyglutamate forms, as the intracellular concentration of drugs are limited by this transport process. As has been observed in other cell lines,19 the FPGS enzyme has a relatively high Km for MTX as compared with Tomudex and 1843U89 (Table 1). In particular, the Kms for MTX for the FPGS enzyme from K562 and HL-60 cell extracts were twofold greater than Kms measured for CCRF-CEM and MOLT-3 cells, respectively. Vmax values were approximately the same for the CCRF-CEM, MOLT-3, and K562 enzyme, while it was fourfold higher in HL-60 cells. In contrast, the low Km values for Tomudex (0.5 to 1.6 μmol/L) and 1843U89 (1.2 to 6.7 μmol/L) and comparable Vmax values resulted in a high level of intracellular conversion of these drugs to their polyglutamate forms (Table 1).
To investigate if the difference in Km values between lymphocytic and myelocytic leukemia cells was caused by interfering substances in the crude extracts, we partially purified FPGS from K562 and CCRF-CEM cell lines. After a 30-fold to 80-fold purification following chromatography on phosphocellulose (see Materials and Methods), the difference in the Km value between the two cell lines persisted, making it unlikely that the differences were due to inhibitors or stimulating substances present in the crude extracts (data not shown).
Kinetic Parameters of FPGS With MTX as Substrate and Accumulation of MTX Polyglutamates in ALL and AML Patients
| Diagnosis . | Km . | Vmax . | Long Chain MTX Polyglutamates . | Long Chain MTX Polyglutamates as % of Total Polyglutamates . |
|---|---|---|---|---|
| . | (μmol/L) . | (pmol/mg/h) . | (pmol/107 cells) . | . |
| ALL | 48 | 14,450 | 9 | 90 |
| ALL | 23 | 1,065 | 12.7 | 66 |
| ALL | 47 | 1,832 | 16.3 | 86 |
| Lymphoid BC/CML | 45 | 6,600 | 30.8 | 88 |
| AML | 75 | 1,490 | 3.36 | 24 |
| AML | 83 | 2,030 | ND | ND |
| AML | 100 | 1,000 | ND | ND |
| Diagnosis . | Km . | Vmax . | Long Chain MTX Polyglutamates . | Long Chain MTX Polyglutamates as % of Total Polyglutamates . |
|---|---|---|---|---|
| . | (μmol/L) . | (pmol/mg/h) . | (pmol/107 cells) . | . |
| ALL | 48 | 14,450 | 9 | 90 |
| ALL | 23 | 1,065 | 12.7 | 66 |
| ALL | 47 | 1,832 | 16.3 | 86 |
| Lymphoid BC/CML | 45 | 6,600 | 30.8 | 88 |
| AML | 75 | 1,490 | 3.36 | 24 |
| AML | 83 | 2,030 | ND | ND |
| AML | 100 | 1,000 | ND | ND |
Cells extracts were prepared as described in Materials and Methods. MTX at different concentrations was used to measure enzyme activity and to determine Km and Vmax values. Polyglutamates formation was measured after 24-hour exposure to radiolabeled MTX as described in Materials and Methods.
Abbreviation: ND, not done (insufficient sample).
The possibility that differing proportions of cytosolic and mitochondrial FPGS enzyme with different properties could explain the different kinetic parameters of the enzyme in the two cell lines was also investigated. After differential centrifugation of the crude extract of the two cell lines (see Materials and Methods), we detected activity only in the cytosolic fraction and no activity was found in the mitochondrial or the nuclear fractions. When we measured the kinetic parameters of FPGS with MTX as a substrate in the cytosolic fraction, Km values did not change in the two cell lines as compared with the values obtained with the crude extracts.
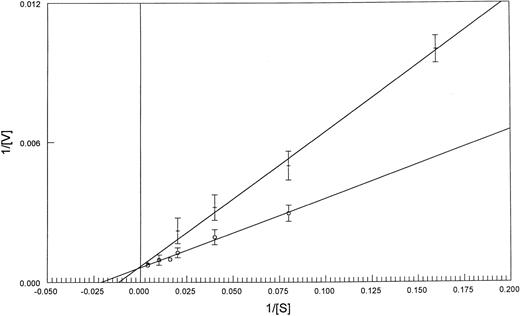
To investigate if this difference in FPGS Km values extended to fresh leukemia samples, we compared the Km and the Vmax of FPGS with MTX as a substrate in blasts from patients: three with AML, three with ALL, and from one patient with blastic crisis of chronic myeloid leukemia in which 90% of the blast cells expressed lymphoid markers (Table 2). Five of the seven patient samples were tested for polyglutamylation (Table 2). The PT430 competitive displacement flow cytometric assay performed on these blasts did not show impaired MTX transport (data not shown). PT430 accumulation has been used to measure DHFR levels, as well,23 and no evidence of increased DHFR binding was observed. The Km values obtained from extracts from blast cells from the four patients with lymphocytic leukemia were similar to those obtained for the CCRF-CEM cell line (40 ± 12 and 44 μmol/L, respectively), while the AML blast samples had values similar to those of K562 cell line (86 ± 12 and 92 ± 9 μmol/L, respectively) (Figs 1 and 2).
Lineweaver-Burk plots of FPGS activity as a function of MTX concentration in CCRF-CEM (o-o) and K562 (+-+) cell lines. Values are the mean of three experiments, with standard deviations indicated. See Materials and Methods for details.
Lineweaver-Burk plots of FPGS activity as a function of MTX concentration in CCRF-CEM (o-o) and K562 (+-+) cell lines. Values are the mean of three experiments, with standard deviations indicated. See Materials and Methods for details.
Lineweaver-Burk plots of FPGS activity as a function of MTX concentration in blast from two patients. AML (+-+), ALL (▵). See Materials and Methods for details.
Lineweaver-Burk plots of FPGS activity as a function of MTX concentration in blast from two patients. AML (+-+), ALL (▵). See Materials and Methods for details.
DISCUSSION
AML is considered intrinsically resistant to therapy with MTX.1-3,7 The reason for this resistance is believed to be the reduced formation of long chain MTX polyglutamates.4 When we studied MTX polyglutamylation in cell lines, we observed that the K562 cell line accumulated a lower amount of long chain polyglutamates than the other cell lines. The observation that FPGS activity was not low and the γ-glutamyl hydrolase activity in the K562 cells was not higher than the CCRF-CEM cell line prompted us to study the properties of FPGS in these cell lines and in fresh leukemia cells.
The consistent finding that emerged from our kinetic studies was the different Km values of FPGS with MTX as substrate between the AML and the ALL cell lines, which suggested that the enzyme may be different in the two forms of leukemia. The finding that the AML patient samples had a Km value similar to the AML cell line, whereas the ALL patients sample were similar to the ALL cell line, showed this did not occur solely in cell lines. Of interest is the lower Km also found from blast cells from the patient with blast crisis of chronic myelogenous leukemia, consistent with the diagnoses of a blast crisis of the lymphoid type.
The different Km values for FPGS between AML and ALL raised the possibility that there are two different enzymes or an alternative possibility examined was that an inhibitor of FPGS may be present in myelocytic leukemia. This latter hypothesis is made unlikely by the unchanged Km values we obtained after partial purification of the FPGS proteins from AML and ALL cells. It is also unlikely that difference in enzyme properties from mitochondria and the cytoplasm accounts for the results observed, as neither cell type was found to have appreciable mitochondrial enzyme activity.
Our findings of different FPGS activity in AML and ALL with different affinities for MTX may be explained by studies performed by Chen et al24 in which alternative splice variants of the FPGS gene were identified. These additional splice variants may be responsible for the tissue-specific encoding of different isoforms of FPGS, which may explain the differences observed by us in leukemias of different cellular origin. We are currently in the process of examining AML and ALL cells for alterations in RNA expression level of these different isoforms.
The finding of a low Km for FPGS when using Tomudex or 1843U89 as a substrate in both leukemia cells and efficient formation of polyglutamates of tomudex and 1843U89 provides an explanation for the potency of these TS inhibitors in both leukemia subtypes, as compared with MTX. These new TS inhibitors may therefore be effective in the treatment of AML, as well as in ALL, in contrast to what is observed with MTX. Based on these findings, a clinical trial of Tomudex in the treatment of AML is planned at our institution.
Supported by American Cancer Society Grant No. BC-561C, the Charles A. Dana Foundation, and Grant No. CA09512 from the National Cancer Institute, Bethesda, MD. G.S.A.L. is the recipient of an American-Italian Cancer Foundation Award (New York, NY) and a grant from Dompe' Biotec Italia. R.G. is the recipient of an ASCO Young Investigator Award.
Address reprint requests to Joseph R. Bertino, MD, Program of Molecular Pharmacology and Therapeutics, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, Box 78, New York, NY 10021.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal