Abstract
Responses of cells to cytokines typically involve the activation of a family of latent DNA binding proteins, referred to as signal transducers and activators of transcription (STAT) proteins, which are critical for the expression of early response genes. Of the seven known STAT proteins, STAT5 (originally called mammary gland factor) has been shown to be activated by several cytokines, such as granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3), and IL-5, which are known to play important roles in growth and differentiation of hematopoietic precursors. In this report we have used mice that are deficient in STAT5A (one of two homologues of STAT5) to study the role of STAT5A in GM-CSF stimulation of cells. When bone marrow–derived macrophages were generated by differentiation with macrophage-CSF (M-CSF), exposure of cells from wild-type mice to GM-CSF resulted in a typical pattern of assembly of DNA binding proteins specific for the gamma activation sequence (GAS) element within the β-casein promoter. However, in cells from the STAT5A null mouse one of the shifted bands was absent. Immunoblotting analysis in the null mice showed that lack of STAT5A protein resulted in no alteration in activation of STAT5B by tyrosine phosphorylation. Proliferation experiments revealed that, when exposed to increasing concentrations of GM-CSF, cells derived from the null mice grew considerably more slowly than cells derived from the wild-type mice. Moreover, expression of GM-CSF–dependent genes, CIS and A1, was markedly inhibited in cells derived from null mice as compared with those of wild-type mice. The decreased expression observed with A1, a bcl-2 like gene, may account in part for the suppression of growth in cells from the null mice. These data suggest that the presence of STAT5A during the GM-CSF–induced assembly of STAT5 dimers is critical for the formation of competent transcription factors that are required for both gene expression and cell proliferation.
CYTOKINES, such as interferons (IFNs), regulate the growth, differentiation, and functional status of cells within the immune system by activation of a family of transcription factors known as signal transducers and activators of transcription (STATs), which bind to specific enhancers and induce expression of early response genes that are thought to be critical for these events.1 These proteins are latent transcription factors that are activated by tyrosine phosphorylation following binding of specific ligands to cognate receptors, leading to the dimerization and subsequent translocation of the STAT proteins to the nucleus.2 Although initially thought to be a unique characteristic of IFN-mediated signaling, most cytokines have been shown to activate STATs, which in most cases play important roles in mediating the specific actions of these cytokines.3 We previously have defined a STAT-like protein, unrelated to those STAT proteins activated by IFNs, that was activated by granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3), and IL-5, all of which bind to receptors that share a common β chain (βc ).4 This protein, initially characterized by others as a growth factor for mammary tissue (termed mammary gland factor),5 has subsequently been renamed STAT5 and demonstrated to be activated by several groups of cytokines.6-8
Once it became apparent that activation of STAT5 occurred in response to these cytokines, the role of STAT5 in cell proliferation and differentiation was assessed by transfecting dominant negative constructs of STAT5 into cell lines and evaluating their effects on proliferation and differentiation. In one study,9 a dominant negative STAT5 suppressed IL-3–induced growth. However, in another study10 there was no effect on the rate at which cells lost viability after removal of IL-3 from the cultures. Other studies have commented on the ability of either IL-3 or GM-CSF to induce c-fos and the role of AP-1 on downstream proliferative effects. Other cytokines that activate STAT5 have also been evaluated with respect to the effects of STAT5 activation on proliferation, yielding disparate results.11
During studies of STAT5 activation, it became evident that there were two separate STAT5 molecules, referred to as STAT5A and STAT5B.12-14 STAT5A and STAT5B have molecular weights of about 94 and 92 kD, respectively, varying primarily by 12 amino acids at the carboxyl terminal end. These proteins are encoded by separate but closely linked genes. Moreover, these proteins can also be expressed as lower molecular weight forms of about 80 kD.8,13 Indeed, it was this 80-kD form that was isolated by our laboratory initially as a STAT-like protein activated by GM-CSF.4 Subsequently, we have shown that both the full-length proteins and the 80-kD protein can be isolated from human monocytes, and that in these cells the 80-kD protein was entirely STAT5A derived.8 Interestingly, no 80-kD STAT5A proteins were observed in human T cells. In murine cells, both STAT5A and STAT5B have been shown to have an 80-kD form, which was presumed to be a carboxyl terminal truncation that may function as a dominant negative transcription factor.9,10 15
The role of STAT5 in the cytokine-mediated control of cell growth and differentiation remains clouded by conflicting data; inhibition of STAT5 activation suppressed proliferative responses in some systems but not in others. Clearly, the use of mice deficient in one or both STAT5 homologues would allow for a better determination of the effects of STAT5 on gene expression and cell proliferation. STAT5A-deficient mice have now been generated using gene targeting.16 These mice have been shown to have marked defects in mammary gland development and lactogenesis. We have used these mice to study the effects of the lack of STAT5A on the activation of myeloid cells by GM-CSF.
MATERIALS AND METHODS
Mice.The generation of STAT5A-deficient mice by targeted disruption of the STAT5A gene has been previously described.16 Heterozygous mice were intercrossed to generate wild-type, heterozygous, and STAT5A null mice. In some experiments, STAT5A null mice with a 129SvEv inbred background were used instead of the original null mice, which had been established in a NIH Black Swiss/129SvEv hybrid background.
Bone marrow (BM) culture-derived macrophages.Femurs and tibias were aseptically removed, and the marrow was washed out by syringe using a 25-gauge needle and Dulbecco's modified Eagle medium (DMEM). After drawing the cells through the syringe several times to break up the clumps, the cells were cultured in 100-mm Petri dishes (Falcon; 1029; Becton Dickinson Labware, Lincoln Park, NJ) in DMEM containing 10% fetal bovine serum (FBS), 50 μg/mL gentamycin, and 100 ng/mL recombinant human macrophage colony-stimulating factor (M-CSF) (Chiron, Emeryville, CA). Spent culture fluid was exchanged with fresh medium every 3 to 4 days. Under these conditions, confluent monolayers of macrophages were obtained within 7 to 10 days.17 BM-derived macrophages were obtained by gently scraping with a plastic policeman after incubating the monolayers with phosphate-buffered saline (PBS) containing 5 mmol/L EDTA for 5 to 10 minutes. Cells were washed free of M-CSF and cultured in DMEM containing FBS for 24 hours before use. Cell viability was determined by trypan blue exclusion and verified to be greater than 90% before performing an experiment. Cells were treated with various concentrations of recombinant murine GM-CSF (Pharmingen, San Diego, CA) for various times.
Electrophoretic mobility shift assays (EMSA).Cells were washed with cold PBS and scraped into an ice-cold whole cell extraction buffer containing 0.1% Triton X-100 (Pierce, Rockford, IL), 10 mmol/L HEPES (pH 7.3), 400 mmol/L KCl, l mmol/L dithiothreitol, 2 mmol/L EDTA, 1 mmol/L EGTA, 1 mmol/L sodium orthovanadate, 10% glycerol, 1 mmol/L phenylmethylsulfonyl fluoride (PMSF), and 5 μg/mL each of aprotinin, leupeptin, and pepstatin. The extracts were vortexed for 10 seconds, incubated at 4°C for 10 minutes and centrifuged at 12,000g for 10 minutes to remove cell debris. Protein concentrations for each extract were determined, and equal amounts of protein were assayed for DNA binding proteins by EMSA as previously described.18 Briefly, 32P-labeled double-stranded oligonucleotide probes, either the 21-bp GAS-like element from the promoter of the bovine β-casein gene (5′ AGATTTCTAGGAATTCAAATC 3′)6 or the 39-bp IFN-γ response region (GRR) from the promoter of the human fcgrl gene (5′ AGCATGTTTCAAGGATTTGAGATGTATTTCCCAGAAAAG 3′)19 were incubated with the extracts (5 μg of protein) in binding buffer. The samples were then electrophoresed on a 6% nondissociating polyacrylamide gel to separate free probe from probe bound to protein. Supershift experiments were performed by incubating extracts with either normal rabbit serum or anti-STAT antibodies at a final concentration of 1:50 for 1 hour at 4°C before the addition of radiolabeled probe.
Immunoprecipitations and immunoblot analysis.After treatment, cells (10 × 106/treatment group) were washed with cold PBS and solubilized in 1% Triton X-100 in a buffer consisting of 250 mmol/L NaCl, 50 mmol/L Tris-HCl, pH 7.4, 50 mmol/L NaF, 0.5 mmol/L Na pyrophosphate, 1 mmol/L sodium orthovanadate, 1 mmol/L PMSF, and 5 μg/mL each of aprotinin, leupeptin, and pepstatin. The postnuclear lysate was then precleared with a suspension of protein G agarose (Pharmacia, Piscataway, NJ) for 1 hour at 4°C. After separation of the protein G agarose from the lysate by centrifugation, the lysate was subdivided and incubated with either normal rabbit serum or rabbit anti-STAT5B14 for 2 to 18 hours at 4°C. Protein G agarose was then added for 1 hour at 4°C to isolate the immune complexes. The protein G conjugates were washed in lysis buffer and boiled in sodium dodecyl sulfate (SDS) sample buffer.
The samples were analyzed by 8% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by electrophoretic transfer to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore, Bedford, MA). The membranes were blocked using 1% ovalbumin (Sigma Chemical Co, St Louis, MO) in 150 mmol/L NaCl, 10 mmol/L Tris-HCI, pH 8.0 with 0.05% Tween 20 for 1 hour. Membranes were then incubated with the biotin-conjugated antiphosphotyrosine monoclonal antibodies (MoAbs) PY20 (Leinco, Ballwin, MO) and 4G10 (UBI, Lake Placid, NY) for at least 2 hours. After washing, the membranes were incubated with peroxidase-conjugated avidin (Southern Biotechnology, Birmingham, AL) and developed using enhanced chemiluminescence (Amersham, Arlington Heights, IL). The immunoblots were then reprobed with the immunoprecipitating anti-STAT5B antibody to show that equal amounts of protein were loaded onto each lane of the gel. These blots were developed using alkaline phosphatase–conjugated goat-antirabbit IgG (Southern Biotechnology) and nitroblue tetrazolium chemistry. In other experiments, whole cell extracts were analyzed by SDS-PAGE followed by immunblotting with anti-STAT5A, anti-STAT5B, or anti-STAT1. After washing, the membranes were probed with peroxidase-conjugated goat-antirabbit IgG and developed using enhanced chemiluminescence.20
Northern blot analysis.Total cellular RNA isolated by the guanidine isothiocyanate procedure (Ambion, Inc, Austin, TX) was electrophoresed on 1% agarose gels containing 0.66% formaldehyde, transferred to nylon membranes (Duralon-UV; Stratagene, La Jolla, CA), and crosslinked to the membrane by ultraviolet light (UV Stratalinker; Stratagene). Membranes were prehybridized in a formamide-containing buffer (FastPair; Digene, Silver Spring, MD) at 42°C. The membrane-bound RNA was hybridized with radiolabeled probes under the same conditions. For probes, a 0.9-kb fragment of the cDNA for the cytokine-inducible SH2-containing protein, CIS, was isolated by Sph I/Xho I digestion of the plasmid pME18S.21 The bcl-2 family member cDNA clone referred to as A1 was kindly provided by Drs Michael Prystowsky and Amos Orlofsky (Albert Einstein College of Medicine, Bronx, NY), and the 0.5-kb insert was isolated from the Bluescript SK(−) vector by digestion with Pst I/Xba I.22 The cDNA for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), pRGAPD-13, has been previously described23; the insert was isolated by PstI digestion. The inserts were random primer labeled with 32P-dCTP to specific activities of 2 to 3 × 109 cpm/mg with the Klenow fragment of DNA polymerase I using random hexamers as primers.24 The membranes were washed twice in 2X saline-sodium citrate (SSC), 0.1% SDS at room temperature, and twice in 0.1X SSC, 0.1% SDS for 15 minutes each at 63°C and then exposed to Kodak XAR film (Eastman Kodak, Rochester, NY) at −70°C for different time intervals using intensifying screens. Filters were stripped by washing with boiling 0.05X SSC, 0.1% SDS. All signals were quantitated by phosphorimager analysis (Molecular Dynamics, Sunnyvale, CA) and normalized to GAPDH expression.
Cell proliferation analysis.Cell proliferation was analyzed according to Mosmann.25 Briefly, confluent plates of BM-derived macrophages that had been starved of M-CSF for 24 hours were washed three times with PBS, and harvested in PBS containing 5 mmol/L EDTA by scraping. Optimal cell numbers and time in culture were determined before performing the proliferation analysis. Cells were plated in 96-well tissue culture dishes at 5 × 104 cells per well in 200 μL of DMEM containing 10% FBS and various concentrations of recombinant murine GM-CSF. After 5 days in culture, 25 μL of a 5 mg/mL solution of MTT (dimethyl thiazole diphenyl tetrazolium bromide; Sigma) in PBS was added to all wells and incubated for 2 hours at 37°C. After inversion of plates to remove liquid from the wells, 125 μL isopropanol/HCl (0.04 N HCl in 2-propanol) was added, and absorbance was read at 570 nm.
RESULTS
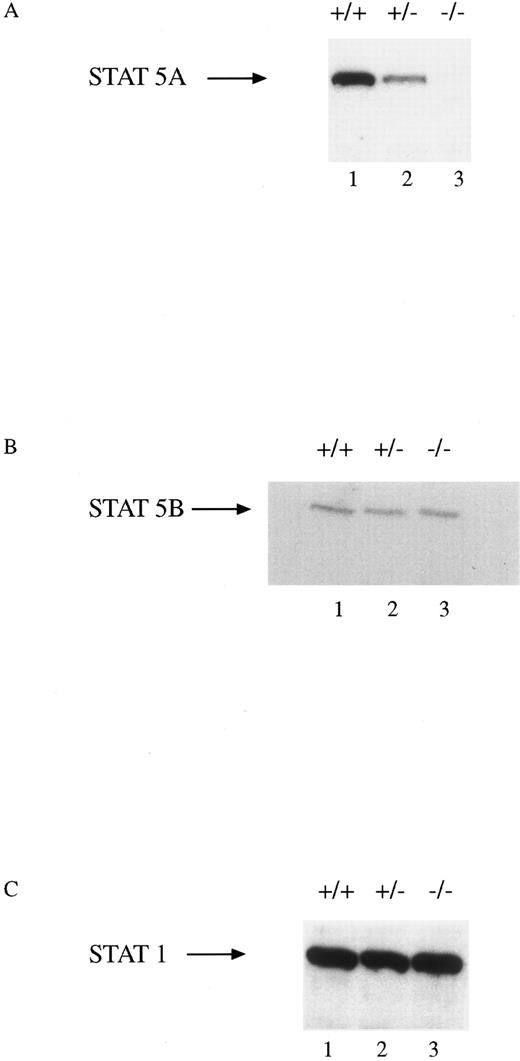
All mice appeared phenotypically normal at the time of birth and were raised in micro-isolator, filter-top cages without any apparent increase in the incidence of infectious diseases. The mice have been followed for up to 8 months without any apparent hematologic abnormalities. Total number of nucleated cells from the marrows of all three genotypes were indistinguishable. BM culture-derived macrophages were obtained from progenitor cells from the femurs and tibias of wild-type (WT, +/+), heterozygous (HZ, +/−), and STAT5A null mice (−/−). Extracts were obtained from these cells and assayed for the presence of STAT5A, STAT5B, and STAT1 by immunoblotting (Fig 1). There was a gene dosage effect in the cells from the three genotypes such that no STAT5A protein was present in the null mice, intermediate levels were observed in the HZ mouse, and normal levels were expressed in the WT mouse (Fig 1A). There was no difference in the level of expression of the STAT5B protein in the cells from the three groups (Fig 1B). Moreover, the expression of another STAT protein, STAT1, was unaffected by the absence of STAT5A (Fig 1C).
STAT5A protein expression in BM-derived macrophages from WT, HZ, and STAT5A null mice. Extracts (30 μg of protein per lane) of BM-derived macrophages from WT (+/+; lane 1), HZ (+/−; lane 2), and null (−/−; lane 3) mice were analyzed by SDS-PAGE followed by immunoblotting with (A) anti-STAT5A, (B) anti-STAT5B, or (C) anti-STAT1 as described in Materials and Methods. The membranes were developed by enhanced chemiluminescence.
STAT5A protein expression in BM-derived macrophages from WT, HZ, and STAT5A null mice. Extracts (30 μg of protein per lane) of BM-derived macrophages from WT (+/+; lane 1), HZ (+/−; lane 2), and null (−/−; lane 3) mice were analyzed by SDS-PAGE followed by immunoblotting with (A) anti-STAT5A, (B) anti-STAT5B, or (C) anti-STAT1 as described in Materials and Methods. The membranes were developed by enhanced chemiluminescence.
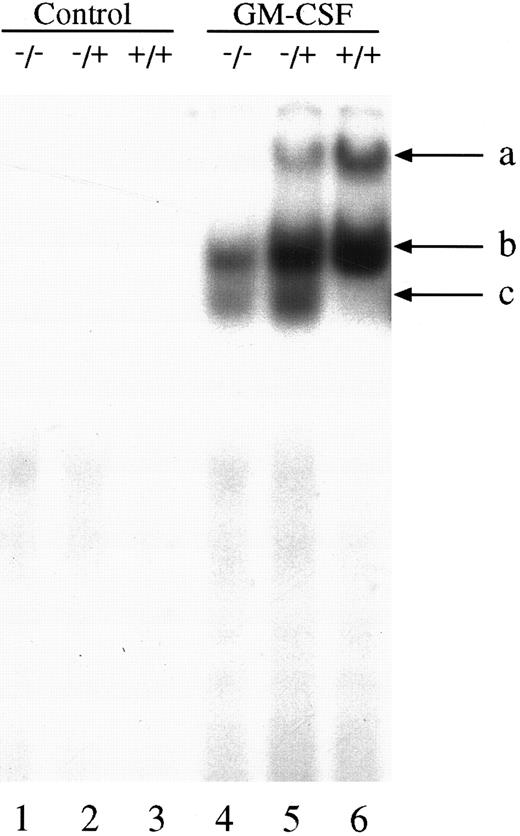
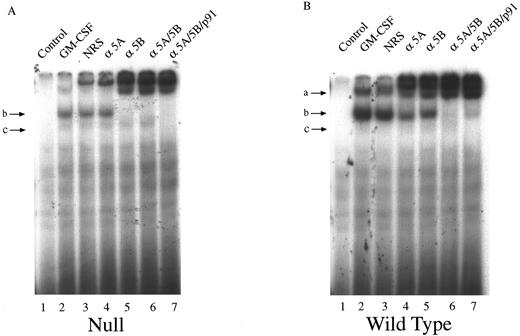
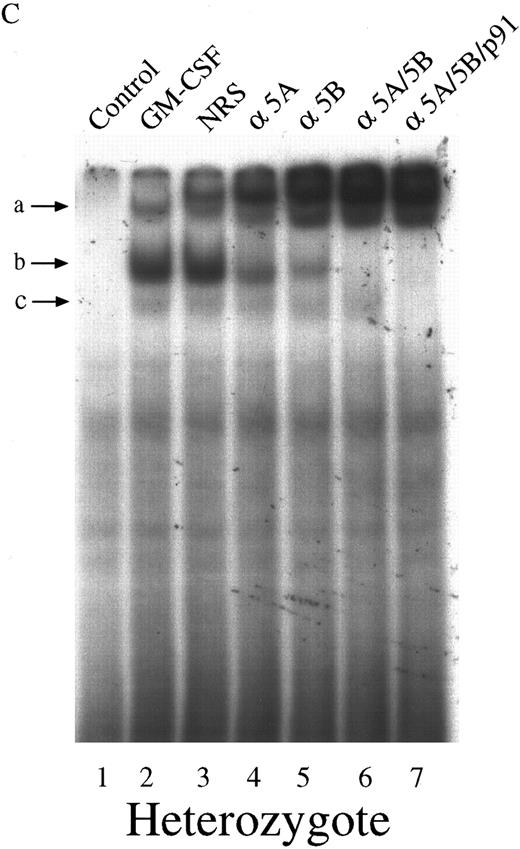
We next investigated the ability of extracts from these cells to bind to the GAS element from the promoter of the β-casein gene. Double-stranded, 32P-labeled β-casein probe was incubated with extracts before separation of bound and free probe by nondissociating PAGE (Fig 2). In control, untreated cells there was no activation of complexes capable of binding to the β-casein promoter (Fig 2, lanes 1 through 3). However, in cells treated with GM-CSF, there were three patterns of binding. Only a lowermost (c) and middle band (b) were observed in the null mouse (Fig 2, lane 4), whereas the lowermost (c), middle (b), and an uppermost band (a) were observed in the HZ mouse (Fig 2, lane 5). The WT mouse also showed bands (a) and (b), and, although in some experiments (Fig 2, lane 6) band (c) was difficult to observe, in other experiments (Fig 3) its presence was more evident. Furthermore, there was an increase in the intensity of band (a) in the WT compared with that observed in the HZ mouse (Fig 2, lane 6). Whether STAT5A or STAT5B were present in these shifted complexes was determined by “supershift” experiments. Extracts were incubated with anti-STAT antibody, and the loss of a band or a shift in the band was determined. Although incubation of extracts with normal rabbit serum (NRS) resulted in a minor nonspecific “supershift” band (Fig 3A-C, lane 3), there was essentially no loss of band intensity for any of the GM-CSF–induced shifted bands. For extracts from null mice, the predominant band (b) was only shifted by anti-STAT5B (Fig 3A, lane 5), and there was no shift with anti-STAT5A (Fig 3A, lane 4). For the WT mice, band (b) was partially shifted by anti-STAT5A (Fig 3B, lane 4) or anti-STAT5B (Fig 3B, lane 5) and totally shifted by addition of both antibodies (Fig 3B, lane 6). The content of the STATs in band (a) was difficult to assess because the lower bands were displaced into this band upon supershifts with antibodies such as the anti-STAT5B (Fig 3A, lane 5). The pattern of displacement of shifted bands in the HZ mice was similar to that of the WT mice (Fig 3C). For extracts derived from null, HZ, and WT mice, the addition of an anti-STAT1 antibody totally eliminated band (c) (Fig 3A-C, lane 7). Because responses to GM-CSF were observed in all three genotypes, changes in receptor structure seems to be an unlikely cause for these observations. EMSA and supershift experiments using the GRR probe yielded similar results (data not shown).
GM-CSF induction of GAS-binding complexes in BM-derived macrophages from STAT5A null, HZ, and WT mice. BM-derived macrophages from null (−/−; lanes 1 and 4), HZ (+/−; lanes 2 and 5), and WT (+/+; lanes 3 and 6) mice were incubated with either medium (lanes 1-3) or 10 ng/mL of GM-CSF (lanes 4-6) for 15 minutes at 37°C, and whole cell extracts (5 μg of protein per lane) were analyzed by EMSA using the 32P-labeled β-casein probe as described in Materials and Methods. Arrows labeled a, b, and c indicate the uppermost, middle, and lowermost bands containing GAS-binding complexes, respectively.
GM-CSF induction of GAS-binding complexes in BM-derived macrophages from STAT5A null, HZ, and WT mice. BM-derived macrophages from null (−/−; lanes 1 and 4), HZ (+/−; lanes 2 and 5), and WT (+/+; lanes 3 and 6) mice were incubated with either medium (lanes 1-3) or 10 ng/mL of GM-CSF (lanes 4-6) for 15 minutes at 37°C, and whole cell extracts (5 μg of protein per lane) were analyzed by EMSA using the 32P-labeled β-casein probe as described in Materials and Methods. Arrows labeled a, b, and c indicate the uppermost, middle, and lowermost bands containing GAS-binding complexes, respectively.
Characterization of the GAS-binding STAT protein complexes in GM-CSF–stimulated BM-derived macrophages from STAT5A null, WT, and HZ mice. BM-derived macrophages from null (A), WT (B), and HZ (C) mice were incubated with either medium (lane 1) or 10 ng/mL of GM-CSF (lanes 2-7) for 15 minutes at 37°C, and whole-cell extracts (5 μg of protein per sample) were analyzed by EMSA using the 32P-labeled β-casein probe as described in Materials and Methods. Extracts were preincubated with NRS (which resulted in a nonspecifically displaced band near the top of the gel) or the indicated anti-STAT antibodies before the addition of the probe. Arrows labeled a, b, and c indicate the uppermost, middle, and lowermost bands containing GAS-binding complexes, respectively. α5A, anti-STAT5A; α5B, anti-STAT5B; αp91, anti-STAT1.
Characterization of the GAS-binding STAT protein complexes in GM-CSF–stimulated BM-derived macrophages from STAT5A null, WT, and HZ mice. BM-derived macrophages from null (A), WT (B), and HZ (C) mice were incubated with either medium (lane 1) or 10 ng/mL of GM-CSF (lanes 2-7) for 15 minutes at 37°C, and whole-cell extracts (5 μg of protein per sample) were analyzed by EMSA using the 32P-labeled β-casein probe as described in Materials and Methods. Extracts were preincubated with NRS (which resulted in a nonspecifically displaced band near the top of the gel) or the indicated anti-STAT antibodies before the addition of the probe. Arrows labeled a, b, and c indicate the uppermost, middle, and lowermost bands containing GAS-binding complexes, respectively. α5A, anti-STAT5A; α5B, anti-STAT5B; αp91, anti-STAT1.
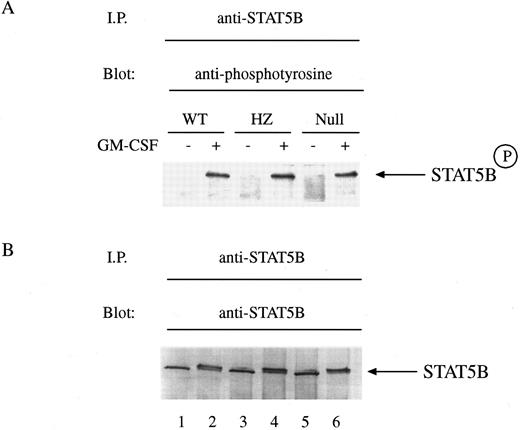
The role of STAT5A in the control of activation of STAT5B in hematopoietic cells is unexplored. Exposure of mammary tissue to prolactin during pregnancy results in the activation of both STAT5A and STAT5B equally well as determined by tyrosine phosphorylation.26 However, in STAT5A null mice there was noted to be a marked decrease in the ability of STAT5B to be tyrosine phosphorylated during pregnancy.16 To explore whether there was a similar requirement of STAT5A for GM-CSF–induced tyrosine phosphorylation of STAT5B, we exposed BM-derived macrophages to GM-CSF and measured tyrosine phosphorylation of STAT5B by immunoblotting with anti-phosphotyrosine antibodies. Equal numbers of cells were either left untreated or treated with GM-CSF for 15 minutes and then solubilized before immunoprecipitation with anti-STAT5B. Immunoblotting showed that GM-CSF could still induce the tyrosine phosphorylation of STAT5B in cells from null mice (Fig 4A, lanes 1-6). Reprobing of the immunoblot revealed that equal quantities of protein were loaded within each lane (Fig 4B, lanes 1-6).
GM-CSF–induced tyrosine phosphorylation of STAT5B in BM-derived macrophages from WT, HZ, and STAT5A null mice. (A) BM-derived macrophages from WT (lanes 1 and 2), HZ (lanes 3 and 4), and null (lanes 5 and 6) mice were incubated with either medium (lanes 1, 3, and 5) or 10 ng/mL of GM-CSF (lanes 2, 4, and 6) for 15 minutes at 37°C and then solubilized in a buffer containing 1% Triton X-100. STAT5B was immunoprecipitated from the lysates with anti-STAT5B, and the immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting as described in Materials and Methods. The membrane was probed with the antiphosphotyrosine MoAbs PY20 and 4G10, and developed by enhanced chemiluminescence. STAT5BP denotes tyrosine phosphorylated STAT5B. (B) The amount of protein loaded within each lane of the gel was determined by reprobing the immunoblot with the immunoprecipitating anti-STAT5B antibody and developing the blot with nitroblue tetrazolium chemistry.
GM-CSF–induced tyrosine phosphorylation of STAT5B in BM-derived macrophages from WT, HZ, and STAT5A null mice. (A) BM-derived macrophages from WT (lanes 1 and 2), HZ (lanes 3 and 4), and null (lanes 5 and 6) mice were incubated with either medium (lanes 1, 3, and 5) or 10 ng/mL of GM-CSF (lanes 2, 4, and 6) for 15 minutes at 37°C and then solubilized in a buffer containing 1% Triton X-100. STAT5B was immunoprecipitated from the lysates with anti-STAT5B, and the immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting as described in Materials and Methods. The membrane was probed with the antiphosphotyrosine MoAbs PY20 and 4G10, and developed by enhanced chemiluminescence. STAT5BP denotes tyrosine phosphorylated STAT5B. (B) The amount of protein loaded within each lane of the gel was determined by reprobing the immunoblot with the immunoprecipitating anti-STAT5B antibody and developing the blot with nitroblue tetrazolium chemistry.
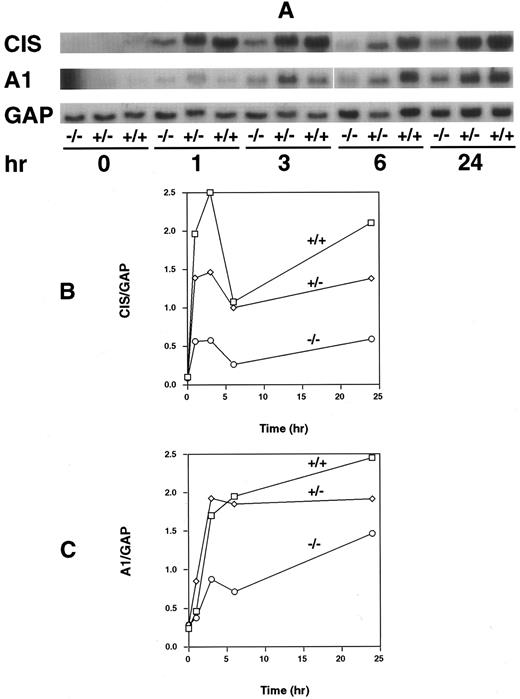
To assess whether the absence of STAT5A would affect expression of GM-CSF–induced genes including one (A1) which may influence proliferation, we measured steady-state levels of two genes known to be activated by GM-CSF: the CIS and A1 genes. Although both are known to be induced as early response genes following treatment with GM-CSF, only the CIS gene has been shown to have STAT5 binding elements within the promoter region. Cells were removed from M-CSF for 24 hours before stimulation with GM-CSF for various times. Total cellular RNA was isolated, and Northern analysis was performed. For the CIS gene, there was a gene dosage-dependent decrease in the ability of GM-CSF to induce CIS expression, with cells from WT, HZ, and null mice having the highest, intermediate, and lowest levels of expression, respectively (Fig 5A and B). Results for the GM-CSF–dependent gene, A1, were not dissimilar from that observed for the CIS gene (Fig 5A and C). Expression of A1 was also reduced (especially at the early time points) in the STAT5A null mice compared with that observed in cells from the WT and HZ mice.
Northern analysis of CIS and A1 mRNA induction in BM-derived macrophages. RNA was isolated from BM-derived macrophages from WT (+/+), HZ (+/−), and null (−/−) mice that had been treated for the indicated time periods with GM-CSF (10 ng/mL), and Northern analysis performed as described in Materials and Methods. The membrane was sequentially probed with CIS, A1, and GAPDH (GAP) probes (A). CIS (B) and A1 (C) mRNA levels normalized to levels of GAPDH (GAP) mRNA were plotted as a function of duration of GM-CSF exposure.
Northern analysis of CIS and A1 mRNA induction in BM-derived macrophages. RNA was isolated from BM-derived macrophages from WT (+/+), HZ (+/−), and null (−/−) mice that had been treated for the indicated time periods with GM-CSF (10 ng/mL), and Northern analysis performed as described in Materials and Methods. The membrane was sequentially probed with CIS, A1, and GAPDH (GAP) probes (A). CIS (B) and A1 (C) mRNA levels normalized to levels of GAPDH (GAP) mRNA were plotted as a function of duration of GM-CSF exposure.
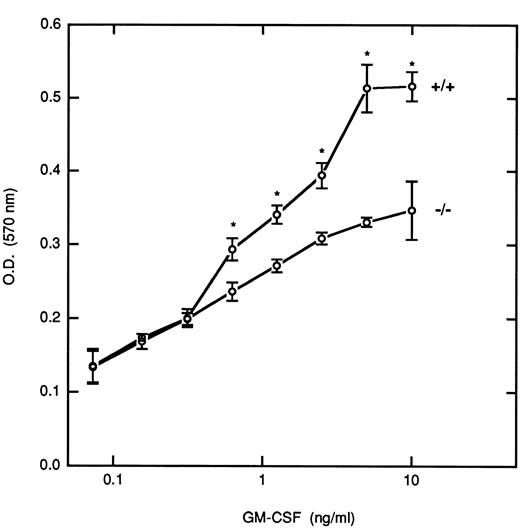
Although it has been reported that STAT5 is important for the growth of myeloid cells, the relative roles that the STAT5 isoforms play in controlling this growth have not been defined. To investigate this issue, we removed BM-derived macrophages from M-CSF for 24 hours before incubation with increasing concentrations of GM-CSF for 5 days, and the proliferative responses were measured by an MTT assay (Fig 6). At the lower doses of GM-CSF (<0.3 ng/mL) there was no differences in the proliferative responses between cells derived from WT and null mice. However, at higher doses of GM-CSF (>0.5 ng/mL) there were significant differences in the growth rate of the STAT5A null cells as compared with the WT cells (P < .05); at the highest dose studied there was a 33% inhibition of growth in the null cells, which was consistently observed in several experiments. At the concentrations of GM-CSF that resulted in logarithmic proliferation (between 0.5 and 5 ng/mL), cells from null mice required 5- to 10-fold more GM-CSF to obtain the same level of growth as that observed in the WT mice. Growth curves for the HZ mice were intermediate between the null and WT mice (data not shown).
Proliferative response of BM-derived macrophages to GM-CSF. BM-derived macrophages pooled from multiple inbred WT(+/+) and null (−/−) mice (5 × 104/well) were incubated with the indicated concentrations of GM-CSF for 5 days, and the proliferative response was assayed by the MTT assay as described in Materials and Methods. Results are presented as the mean ± the standard deviation of triplicate determinations. This experiment is representative of five independent experiments. Asterisks denote a significant difference (P < .05) in proliferation between cells from WT and null mice at that dose (Student's t-test).
Proliferative response of BM-derived macrophages to GM-CSF. BM-derived macrophages pooled from multiple inbred WT(+/+) and null (−/−) mice (5 × 104/well) were incubated with the indicated concentrations of GM-CSF for 5 days, and the proliferative response was assayed by the MTT assay as described in Materials and Methods. Results are presented as the mean ± the standard deviation of triplicate determinations. This experiment is representative of five independent experiments. Asterisks denote a significant difference (P < .05) in proliferation between cells from WT and null mice at that dose (Student's t-test).
DISCUSSION
Induction of early response genes and subsequent phenotypic changes within cells after exposure to cytokines are frequently the result of activation of latent transcription factors referred to as STATs. For members of the family of cytokines that activate cells through receptors that share a β subunit (IL-3, IL-5, and GM-CSF), the STAT protein that is most frequently activated is STAT5.27 STAT5 is both tyrosine and serine phosphorylated, translocates to the nucleus very rapidly, and is required for expression of several genes known to have STAT5 binding elements within their promoter.28 It has recently been appreciated that STAT5 consists of two homologues: STAT5A and STAT5B.12,14 26 Both proteins are usually present in cells, and both are activated by cytokines to form homodimers and heterodimers. The presence of a mouse model with the STAT5A gene disrupted by gene targeting allowed us to determine the role of STAT5A in the activation of cells by GM-CSF. We show that the absence of STAT5A results in clear differences in the ability of cells to assemble DNA-protein complexes in response to GM-CSF. This defect apparently results in diminished proliferation in response to GM-CSF and decreased expression of GM-CSF–dependent genes.
The GAS element in the promoter of the β-casein gene binds GM-CSF–activated STATs such that three distinct bands become shifted during an EMSA (see Fig 2). Although we did not observe the activation of STAT1 by GM-CSF in human peripheral blood monocytes,4 band (c) derived from the GM-CSF–stimulated BM-derived macrophages is clearly supershifted by an antibody specific for STAT1. Therefore, it appears that these cells can activate STAT1 in response to GM-CSF. Moreover, for reasons that are unclear, it appears that more STAT1 is activated in cells from the HZ mice than from null or WT mice. The most obvious difference between the null mice and the other two genotypes is the lack of band (a) (Fig 2, lane 4). Therefore, this band must represent either STAT5A homodimers or STAT5A-STAT5B heterodimers. It is also apparent that homodimers of STAT5B can migrate within band (b). This band could also be composed of STAT5A and STAT5B homodimers. The lack of STAT5A in band (a), which is also presumably the greatest in molecular weight, may imply that this band represents dimer-dimer formations between STAT5A and STAT5B. It has recently been shown for STAT1 and STAT4 that the ability to form tetramers consisting of two dimers on adjacent elements in the promoters of responsive genes may be critical for initiating cytokine-mediated gene induction.29,30 Indeed, it is clear (see Fig 2) that the amount of band (a) increases from nothing in the null mice, to intermediate levels in the HZ mice, and then to maximal levels in the WT mice. Such slower migrating bands on EMSA have been previously reported to be associated with STAT tetramer formation.30
Studies on a transgenic mouse strain with impaired mammary gland development have suggested that in mammary tissues the inability to express the whey acidic milk protein correlates with the lack of assembly of STAT5A/STAT5B heterodimers.26 This has been confirmed in the STAT5A-deficient mouse in that females, who do not lactate, cannot form STAT5A/STAT5B heterodimers and do not express the WAP milk protein.16 These results are highly suggestive of the possibility that the formation of STAT5A/STAT5B heterodimers may be critical for the induced expression of GM-CSF–dependent genes. This was also alluded to through the use of a STAT5A mutant construct that reduced the expression of STAT5-dependent genes such as CIS but not genes such as c-fos and myc.10 We have similarly shown that cells from STAT5A-deficient mice show a dramatically reduced ability to express a gene that is clearly dependent on STAT5, namely the CIS gene. Moreover, we were able to show that this inhibition occurs in a gene dosage-dependent manner. Although not as marked as the results with the CIS gene, the reduction in expression of the A1 gene is clearly evident in the null mice. Whether the differences in the results for CIS and A1 expression are due to differences in STAT5 binding sites within the promoters will require further study because the A1 promoter has only been partially sequenced.31 The ability of WT and HZ cells to respond to GM-CSF with enhanced A1 expression may, in part, explain their ability to demonstrate greater proliferation. If A1 functions as a bcl-2–like gene,22 enhanced expression should protect these cells from apoptosis.
BM-derived macrophages are sensitive to both M-CSF and GM-CSF with respect to the ability of these cytokines to induce proliferation and promote survival. BM precursor cells were expanded originally in M-CSF because this cytokine activates essentially no STAT5 in our system. Once starved of M-CSF for 24 hours, cells were incubated with increasing concentrations of GM-CSF and the proliferative response was measured. The curves generated from the WT mice start to separate from those of the null mice at a concentration of about 0.3 to 0.8 ng/mL in most of the experiments performed. At this dose of GM-CSF only a portion of the receptors (about 20% to 60%; D.S.F., unpublished observations) are presumably occupied. For the HZ mice (data not shown), the growth curve is in between that of the null and WT mice. These curves tends to plateau at concentrations of GM-CSF greater than 5 ng/mL, a dose that is known to result in occupancy of virtually all GM-CSF receptors (D.S.F., unpublished observations). In general, at concentrations of GM-CSF that occupy a small portion of the receptors, the ability of the three genotypes to be stratified by virtue of their proliferative response to GM-CSF is lost. As the dose of GM-CSF increases, normal tyrosine phosphorylation of STAT5A and STAT5B occurs, and proliferation reaches maximal levels in WT cells. Maximal differences are observed between cells of the null and WT mice during the logarithmic phase of the growth curve. Although it is known that STAT5 can bind to the β chain of the GM-CSF/IL-3/IL-5 receptor family,32,33 the detailed mechanisms by which tyrosine phosphorylation of STAT5 and subsequent heterodimer formation occur are not known. It is conceivable that some cells may not respond to GM-CSF until certain levels of receptor occupancy occur, and this may be occurring at doses that result in occupancy of virtually all the receptors. Moreover, it has recently been shown that the direct interaction of STAT5 with the JH2 domain of Jaks may also be a mechanism for STAT binding and activation.34 Therefore, the degree of activation of a Jak and STAT5 at various concentrations of ligand may need to be re-explored. The availability of mice deficient in STAT5A and STAT5B will enable us to investigate more fully the precise mechanism and role of heterodimer formation in both gene expression and cell proliferation.
ACKNOWLEDGMENT
The authors acknowledge Lisa Garrett and Theresa Hernandez for their contributions in animal husbandry.
L.A.R. was supported by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the Food and Drug Administration.
Address reprint requests to David S. Finbloom, MD, Division of Cytokine Biology, Center for Biologics Evaluation and Research, Food and Drug Administration, 29 Lincoln Dr MSC 4555, Bethesda, MD 20892-4555.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal