Abstract
Natural killer (NK) cells are CD3− large granular lymphocytes (LGL) responsible for immunity against viral infections. A chronic lymphoproliferative disorder of NK cells has been described in which the expanded NK cells display a restricted phenotype and cytotoxic activity. These data raise the hypothesis that proliferating LGL in these patients result from discrete expansions of NK cells responding to an unknown, perhaps viral, antigen. Recently, it was found that mice transgenic for the tax gene of human T-cell leukemia/lymphoma virus (HTLV) develop NK leukemia. Therefore, we studied 15 patients with chronic NK lymphoproliferative disorder for evidence of HTLV infection. Sera were tested using an HTLV-I/II-enzyme linked immunosorbent assay and a modified Western blot assay containing recombinant env proteins. None of the sera met conventional criteria for HTLV seroreactivity. However, sera from 11 patients (73%) reacted with the recombinant HTLV env protein p21E. The anti-p21E reactivity of these sera was then mapped employing the recombinant proteins GD21 and BA21. No reactivity to the immunodominant HTLV epitope GD21 was observed, suggesting that prototypical HTLV infection is unlikely in these patients. This was confirmed by finding no evidence for HTLV nucleic acids by PCR analyses employing primers specific for conserved regions in the env, pol, and pX genes. In contrast, 10 of the 15 sera reacted with the epitope BA21, documenting for the first time an association between a unique seroreactivity and disease. The high incidence of BA21 seroreactivity in these patients suggests that exposure to a protein containing homology to BA21 may be important in the pathogenesis of this lymphoproliferative disorder.
PATIENTS WITH increased numbers of circulating large granular lymphocytes (LGL) have been described under a number of terms, including LGL leukemia1 and lymphoproliferative disease of granular lymphocytes (LDGL).2 Clonal T cell (CD3+) and natural killer (NK) cell (CD3−) LGL disorders have been designated as T- and NK-LGL leukemia, respectively.1 Manifestations of T-LGL leukemia include chronic neutropenia, recurrent bacterial infections, and rheumatoid arthritis.1 NK-LGL leukemia is an aggressive disease characterized by massive hepatosplenomegaly and B symptoms, such as fever, night sweats, or weight loss.3 LDGL has been defined as chronic (greater than 6 month) increases in CD3+ or CD3− LGL greater than 2,000/μL; assessment of clonality is not incorporated into this classification scheme.2 However, X-linked gene analyses in patients with CD3− LDGL have shown that the CD3− LGL are polyclonal.4 Most patients with CD3− LDGL have a chronic clinical course different than that described for NK-LGL leukemia.2 4
Four subsets of normal NK cells with defined antigen specificity can be identified using monoclonal antibodies directed to a novel family of NK specific triggering molecules.5 It has also been shown that recognition of virally-infected targets in vitro is mediated by such unique NK subsets.6 In normal individuals, all four NK cell subsets are represented. In contrast, most patients with CD3− LDGL have expansions of only one of the normal NK subsets,7 suggesting that these LGL have proliferated in vivo in response to some unknown, perhaps viral, antigen.8 Recent data of Martin et al9 would support an indirect role for retroviral infection in such an antigen-driven expansion. They found that HTLV-II had infected CD3+, CD8+ cells but not CD3− LGL in an HTLV-II seropositive patient with CD3− LDGL.9 Animal models also support a role for retroviral infection in pathogenesis of LGL disease, as mice transgenic for the tax gene of HTLV develop NK-LGL leukemia.10 Therefore, we studied whether HTLV-I/II infection might be a common mechanism involved in the pathogenesis of CD3− LDGL. Although there was no evidence of prototypical HTLV-I or HTLV-II infection in these patients, we found that sera from 12 of 15 patients reacted against recombinant HTLV-I env p21e protein. Epitope mapping showed that seroreactivity was directed at the BA21 epitope rather than the GD21 epitope of this HTLV transmembrane envelope protein.
PATIENTS AND METHODS
Patients.All patients met criteria for CD3− LDGL.2 LGL counts ranged from 2,100 to 12,300/μL (normal, 223 ± 99/μL).1 Thirteen of 15 patients had unique expansions of restricted NK subsets, as defined by two-color flow cytometry analyses with monoclonal antibodies EB6 and GL183, as reported previously.7 Seven of these patients were women and potentially informative for clonal analyses using X-linked polymorphisms. In 5 patients, X-linked gene analyses showed polyclonal proliferation of CD3− LGL; in one patient these clonal analyses were indeterminate.4 Such analyses were not performed in the remaining patient.
In 14 of 15 patients, there was no history of blood transfusions, intravenous drug use, or at risk sexual behavior that could lead to retroviral infection. Patient 14 did receive factor VIII concentrate for treatment of hemophilia A, as necessary. Six of these patients had serologic evidence for past exposure to hepatitis B. Three patients were infected with hepatitis C, as shown by detection of hepatitis C RNA by polymerase chain reaction (PCR) analyses. Epstein-Barr virus genome was detected in six patients by PCR analyses.8 None of the patients was on treatment for the LDGL disorder.
Antibody studies.Sera were tested in an HTLV-I/II enzyme-linked immunosorbent assay (ELISA; Cellular Products, Buffalo, NY). To discriminate HTLV-I from HTLV-II infection, a modified recombinant HTLV-I/II Western blot assay (HTLV blot 2.3; Diagnostic Biotechnology, Ltd, Singapore) was used according to the manufacturer's instructions.11 The HTLV blot 2.3 contains the gp21 HTLV recombinant protein p21E, which expresses amino acids 307-439 of the HTLV-I env glycoprotein, as well as type specific recombinant proteins and native viral proteins. Sera were tested independently in two laboratories with one of the tests being performed in a blinded fashion. Epitope mapping of p21E reactivity was performed employing the previously described recombinant proteins GD21 and BA21.12,13 GD21 expresses amino acids 361-404 and BA21 expresses amino acids 397-430 of the HTLV-I env glycoprotein. GD21-1 refers to GD21 expressing the HTLV-I specific sequence and GD21-2 refers to GD21 expressing the HTLV-II specific sequence. All three sequences are expressed in modified versions of the vector pGEX-2 (Pharmacia, Piscataway, NJ) in which the HTLV specific sequences are fused to the carboxy terminal of the glutathione-S-transferase (sj26) of Shistosomiasis japonicum.12 13
Western blot assays of sera with lysates from bacteria expressing recombinant proteins were performed as described.12,14 Recombinant GD21-1 and GD21-2 were purified as described.13 Nonrecombinant sj26 protein and BA21 were purified employing glutathione agarose. Western blot assays with purified GD21 and BA21 were performed as described12 except that: (1) The sera were incubated concurrently with strips containing sj26 blotted at a concentration of 2 μg/cm to control for antibody reactivity to the non-HTLV portion of the fusion proteins, and (2) sera were incubated with purified BA21 for 2 hours at a dilution of 1/100. This eliminated any low level background reactivity to purified sj26, but did not affect the number of sera reactive with BA21 or significantly alter the intensity of the observed reactivity to BA21.
Control sera consisted of sera from 47 normal blood donors, who tested negative at the Stanford Blood Center for the presence of transfusion-transmitted viruses (including HTLV and hepatitis B virus [HBV]). As an additional control, sera from 35 individuals with antibodies to HBV core protein were also evaluated. Positive controls were coded sera from 5 HTLV-I–infected and 5 HTLV-II–infected individuals, as determined by ELISA, Western blot, and PCR analyses, from diverse geographic locales including South America and Africa.
Competition assays.Ninety-six well plates (Nunclon; Nalge Nunc International, Rochester, NY) were coated with 100 μL of a 5 ng/μL solution of purified p21E protein15 in phosphate-buffered saline and incubated for 1 hour at 37°C. Wells were then washed one time with TBS (150 mmol/L NaCl, 20 mmol/L Tris pH 7.5) and blocked by the addition of 150 μL of BLOTTO (TBS plus 0.1% Tween-20, 2.5% normal goat sera, 2.5% nonfat dry milk). After 1 hour at room temperature, plates were washed one time with TBS followed by the addition of p21E reactive or control sera diluted 1/100 in 100 μL of BLOTTO containing 1 μg of purified BA21, 1 μg of purified sj26, or no additional protein. The wells were then incubated for 1.5 hours at room temperature with gentle rocking. The wells were then washed three times with TBS and 100 μL of antihuman IgG-horseradish peroxidase conjugate (Kirkegaard and Perry, Gaithersburg, MD), diluted 1/1000 in BLOTTO, was added. After 1 hour at room temperature, the wells were washed four times with TBS and bound conjugate detected by incubation with a 0.5 mg/mL solution of 2,2′-Azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS; Sigma, St Louis, MO) plus 0.3% hydrogen peroxide. Color development was allowed to proceed for 10 to 20 minutes and the absorbence of the wells was read at 405 nm using a multiwell plate reader (DuPont, Wilmington, DE).
DNA analyses.DNA from peripheral blood mononuclear cells from each patient was enzymatically amplified using PCR with the thermostable DNA polymerase Taq in a Perkin-Elmer Cetus Thermal Cycler (Norwalk, CT) for 60 cycles, as described.16,17 Amplified DNA was assayed using the Southern blot format. Primers and probes are as follows: primer pair 1 HTII pol (4735-4756)+/HTII pol (4920-4897)− and probes a HTII pol (4880-4899) + d and probe b HTI pol (4825-4850) + d; primer pair 2 HTIIpX (7248-7267)+/HTIIpX (7406-7386)− and probe c HTIIpX (7337-7376) + d. Of note, primer pairs 1 and 2 will amplify pol and pX gene sequences, respectively, which are common to HTLV-I and HTLV-II. Probe a will specifically detect the HTLV-II pol sequence, whereas probe b specifically detects the HTLV-I pol sequence. Probe c will detect either HTLV-I or HTLV-II pX sequences amplified with primer pair 2.16 Primer pair 3 HTII env (5620-5639) + and HTII env (5828-5846)− was designed to amplify the HTLV-II env gp46 protein used in the recombinant HTLV 2.3 Western blot assay,14 using probe d, HTII env (5670-5700) + d.
Degenerate primers were designed to amplify both HTLV-I and HTLV-II env p21e sequences encompassing the BA21 gene region. These primers utilized the HTLV-II MO-T sequence (HT env 6118-6135+/HTII env 6446-6465−), but incorporated degeneracies derived from 20 HTLV-I isolates and four other HTLV-II isolates. Probe e (HTI env 6390-6428 + d) will specifically detect the HTLV-I env sequence, whereas probe f (HTII env 6355-6394 + d) will specifically detect the HTLV-II env sequence. There was enough DNA remaining to test these degenerate primers in six CD3− LDGL patients who were BA21 seropositive.
RESULTS
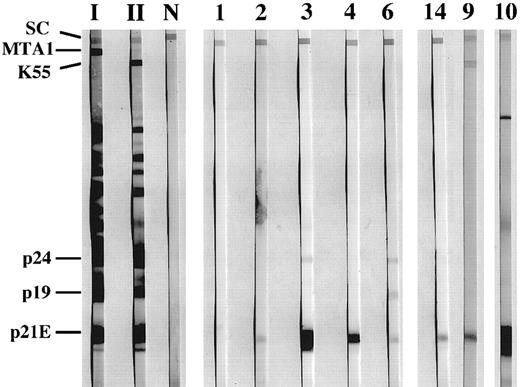
Antibody studies.Sera from all patients were negative in the HTLV-I ELISA. However, optical density readings from two of the sera were close to the positive cutoff value. Results of the modified Western blot assay (HTLV Blot 2.3) showed that none of the sera showed a positive pattern, ie, reactivity with gag p24 and env gp46 (Table 1). However, sera from 11 patients (73%) reacted against recombinant HTLV-I env p21e (Fig 1). One of these p21e reactive sera, which was close to the positive cutoff in the ELISA, also reacted against recombinant HTLV-II env gp46 (Fig 1). These results using HTLV Blot 2.3 were reproduced in an independent laboratory, when testing coded sera.
Summary of HTLV-I/II Antibody Reactivity in CD3-LDGL Patients*
| . | . | HTLV Blot 2.3 . | ||||||
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | |||
| SERA . | env . | gag . | p21e . | Epitope . | Mapping . | |||
| . | p21e . | gp46-I . | gp46-II . | p19 . | p24 . | GD21-1 . | GD21-2 . | BA-21 . |
| 1 | — | — | — | — | — | — | — | — |
| 2 | + | — | — | — | — | — | — | + |
| 3 | ++ | — | — | — | + | — | — | ++ |
| 4 | ++ | — | — | — | — | — | — | ++ |
| 5 | ± | — | — | — | — | — | — | + |
| 6 | + | — | — | + | + | — | — | + |
| 7 | — | — | — | — | — | — | — | — |
| 8 | + | — | — | — | — | — | — | + |
| 9 | + | — | + | ± | — | — | — | — |
| 10 | ++ | — | — | ± | — | — | — | ++ |
| 11 | — | — | — | ± | — | — | — | — |
| 12 | ++ | — | — | — | — | — | — | ++ |
| 13 | — | — | — | — | — | — | — | — |
| 14 | + | — | — | — | — | — | — | ++ |
| 15 | + | — | — | — | — | — | — | + |
| . | . | HTLV Blot 2.3 . | ||||||
|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | |||
| SERA . | env . | gag . | p21e . | Epitope . | Mapping . | |||
| . | p21e . | gp46-I . | gp46-II . | p19 . | p24 . | GD21-1 . | GD21-2 . | BA-21 . |
| 1 | — | — | — | — | — | — | — | — |
| 2 | + | — | — | — | — | — | — | + |
| 3 | ++ | — | — | — | + | — | — | ++ |
| 4 | ++ | — | — | — | — | — | — | ++ |
| 5 | ± | — | — | — | — | — | — | + |
| 6 | + | — | — | + | + | — | — | + |
| 7 | — | — | — | — | — | — | — | — |
| 8 | + | — | — | — | — | — | — | + |
| 9 | + | — | + | ± | — | — | — | — |
| 10 | ++ | — | — | ± | — | — | — | ++ |
| 11 | — | — | — | ± | — | — | — | — |
| 12 | ++ | — | — | — | — | — | — | ++ |
| 13 | — | — | — | — | — | — | — | — |
| 14 | + | — | — | — | — | — | — | ++ |
| 15 | + | — | — | — | — | — | — | + |
Intensity of antibody reactivity is indicated by the following scale: — , no reactivity; ±, weak reactivity.
+, Moderate reactivity; ++, strong reactivity.
Reactivity of sera from CD3-LGL patients to HTLV-I proteins. Sera were tested against HTLV blot 2.3 (Genelabs Diagnostics, Redwood City, CA) containing multiple HTLV recombinant env proteins. Assays were run in accordance with manufacturer's instructions, except that all sera were incubated with the strips overnight. The strips marked I, II, and N were incubated with sera from an HTLV-I, HTLV-II, and an uninfected individual, respectively. Strips marked 1 through 6, 9, 10, and 14 were incubated with sera from individuals with CD3− LDGL. The same numbers are used to identify individual sera in the other figures and Table 1. The migration of the viral proteins p19 and p24 as well as the positions of the recombinant proteins p21e, K55 (HTLV-II gp46), and MTA1 (HTLV-I gp46) are indicated at left. SC indicates position of the serum control band, which is reactive when human antibodies are present in the tested sample.
Reactivity of sera from CD3-LGL patients to HTLV-I proteins. Sera were tested against HTLV blot 2.3 (Genelabs Diagnostics, Redwood City, CA) containing multiple HTLV recombinant env proteins. Assays were run in accordance with manufacturer's instructions, except that all sera were incubated with the strips overnight. The strips marked I, II, and N were incubated with sera from an HTLV-I, HTLV-II, and an uninfected individual, respectively. Strips marked 1 through 6, 9, 10, and 14 were incubated with sera from individuals with CD3− LDGL. The same numbers are used to identify individual sera in the other figures and Table 1. The migration of the viral proteins p19 and p24 as well as the positions of the recombinant proteins p21e, K55 (HTLV-II gp46), and MTA1 (HTLV-I gp46) are indicated at left. SC indicates position of the serum control band, which is reactive when human antibodies are present in the tested sample.
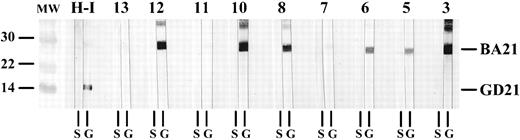
Epitope mapping studies of p21e reactivity were then performed. Serum from each of 5 HTLV-I–infected and 5 HTLV-II–infected individuals reacted to both GD21-1 and GD21-2. In contrast, none of the p21e reactive sera from the CD3− LDGL patients tested positive for GD21-1 or GD21-2. However, 10 of 11 p21e reactive sera from these patients tested positive against the BA21 epitope using whole cell lysates. This BA21 reactivity was confirmed by testing sera against purified recombinant BA21 protein (Fig 2). This high rate of BA21 seroreactivity (67%) was significantly different than that observed in normal blood donors, where 6 of 47 (13%) sera reacted against both p21e and BA21 (P < .001, Fisher's Exact T test). Although the CD3-LDGL patients did not belong to a high-risk group by history, six patients had serologic evidence for past exposure to HBV. To be certain that BA21 seroreactivity was not somehow artifactual due to HBV exposure, we tested 35 HBV core antibody positive sera for p21e and BA21 reactivity. We found that eight of 35 (23%) of these sera reacted against both p21e and BA21. Therefore, the rate of BA21 seroreactivity in the CD3− LDGL patients was also significantly different than that observed in these HBV positive sera (P = .004, Fisher's Exact T test). Three of 10 HTLV-infected sera (two HTLV-I, one HTLV-II) reacted against BA21.
Seroreactivity to purified GD21-1 and BA21 recombinant proteins. Representative Western blots displaying the reactivity of nine CD3− LGL patients (strips labeled 3, 5 through 8, 10 through 13) to HTLV-I gp21 recombinant proteins. All purified proteins were blotted at a concentration of 2 μg/cm nitrocellulose and sera were diluted and incubated as described.12 Strips containing BA21 (molecular weight ∼ 30 kD) coelectrophoresed with GD21-1 (molecular weight ∼ 15 kD) are indicated by the letter G at the bottom. Strips containing nonrecombinant sj26 are indicated by the letter S. The migration of BA21 and GD21-1 are indicated at right. Nonrecombinant sj26 has a molecular weight of 26.5. The strips labeled H-I were incubated with sera from an HTLV-I infected individual. The lanes marked MW are molecular weight standards whose size (in kD) are indicated at left.
Seroreactivity to purified GD21-1 and BA21 recombinant proteins. Representative Western blots displaying the reactivity of nine CD3− LGL patients (strips labeled 3, 5 through 8, 10 through 13) to HTLV-I gp21 recombinant proteins. All purified proteins were blotted at a concentration of 2 μg/cm nitrocellulose and sera were diluted and incubated as described.12 Strips containing BA21 (molecular weight ∼ 30 kD) coelectrophoresed with GD21-1 (molecular weight ∼ 15 kD) are indicated by the letter G at the bottom. Strips containing nonrecombinant sj26 are indicated by the letter S. The migration of BA21 and GD21-1 are indicated at right. Nonrecombinant sj26 has a molecular weight of 26.5. The strips labeled H-I were incubated with sera from an HTLV-I infected individual. The lanes marked MW are molecular weight standards whose size (in kD) are indicated at left.
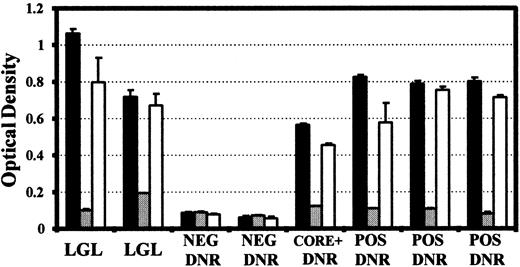
To ensure that the reactivity observed against the p21E and BA21 recombinant proteins was specific and directed at the same epitope, inhibition experiments were performed. Sera from both normal blood donors and CD3− LDGL patients that were reactive by Western blot to p21E and BA21 were also strongly reactive against purified p21E in an EIA format, with only background levels of reactivity observed from p21E negative blood donors. The reactivity of CD3− LDGL patient sera to p21E was substantially unaffected by the addition of excess nonrecombinant sj26 protein, but was completely inhibited in the presence of BA21 (Fig 3). Similar results were obtained with those control sera that were also p21E reactive (from both HBV core positive individuals and from normal donors). Titration experiments indicated that 100 ng of added BA21 was sufficient to result in 90% inhibition of the binding to p21E observed in sera from an LGL patient and a p21E reactive normal blood donor (not shown). Thus, the antibody reactivity detected against the p21E protein is directed primarily at an epitope contained within the HTLV env amino acid sequences (397-430) of the BA21 recombinant protein.
Inhibition of p21E reactivity by purified BA21. Reactivity towards purified p21E protein obtained in the absence (▪) or presence of 1 μg of added BA21 (▨) or 1 μg nonrecombinant sj26 protein (□). Sera tested were from BA21 reactive CD3-LGDL patients (LGL), BA21 negative blood donors (NEG DNR), BA21 reactive blood donors (POS DNR) and a BA21 reactive, HBV core antibody positive donor (CORE + DNR). All sera were diluted 1/100 in BLOTTO and the assay was performed as described in Materials and Methods. The bars represent the mean of two determinations and the error bar indicates the absorbence obtained from the higher replicate.
Inhibition of p21E reactivity by purified BA21. Reactivity towards purified p21E protein obtained in the absence (▪) or presence of 1 μg of added BA21 (▨) or 1 μg nonrecombinant sj26 protein (□). Sera tested were from BA21 reactive CD3-LGDL patients (LGL), BA21 negative blood donors (NEG DNR), BA21 reactive blood donors (POS DNR) and a BA21 reactive, HBV core antibody positive donor (CORE + DNR). All sera were diluted 1/100 in BLOTTO and the assay was performed as described in Materials and Methods. The bars represent the mean of two determinations and the error bar indicates the absorbence obtained from the higher replicate.
DNA analyses.Gene sequences shared by HTLV-I and HTLV-II (pol and pX) were not detected in any patient. We could not detect HTLV-II env gp46 sequences in the one patient with antibody reactivity against this env protein (not shown). We could also not detect sequences related to HTLV BA21 using degenerate primers designed to amplify either HTLV-I or HTLV-II BA21 gene regions (not shown).
DISCUSSION
We found that the great majority of sera from CD3-LDGL patients reacted against p21e, the recombinant transmembrane env protein of HTLV. The incorporation of p21e, expressing amino acids 306-439 of HTLV env gene, in modified Western blot assays has greatly increased the detection of env specific antibodies in HTLV-infected individuals.11,14 Sera with antibodies to HTLV-I or HTLV-II react equally to p21e.15 An immunodominant epitope, GD21, has been identified within the p21e protein. GD21-1 contains amino acids 361 to 404, and Western blot assays incorporating the recombinant GD 21-1 protein (HTLV Blot 2.4) have shown 100% sensitivity and 98% specificity in detecting HTLV-I or HTLV-II infection.13 None of the CD3-LDGL sera reacted against GD21-1 or against GD21-2, a recombinant protein containing the corresponding HTLV-II env amino acids. Likewise, we could not detect HTLV-I or HTLV-II gene sequences in any patient. Taken together, these data show that none of the CD3− LDGL patients was infected with prototypical HTLV-I or HTLV-II.
We found that the p21e immunoreactive sera from the CD3− LDGL patients were specifically reactive to the BA21 epitope, which contains amino acids 397-430 of HTLV-I env. Previously, it was reported that approximately 60% of sera from HTLV-infected individuals react to BA21.12 Here we found a lower rate (30%) of BA21 seroreactivity when studying a smaller number of HTLV-reactive sera collected from infected patients from more diverse geographic areas. Previous studies have also shown antibodies to p21e in 0.6% to 8.5% of serologically indeterminant normal donors who were negative for HTLV by PCR.11,18,19 Epitope mapping studies have shown that sera from these p21e positive, HTLV negative individuals react to BA21 and not to GD21-1.12 In this study we observed that sera from 13% of HTLV ELISA-negative blood donors had antibodies to both p21e and BA21. In contrast, we found a very high incidence of BA21 seroreactivity in patients with CD3− LDGL. Of interest, sera from about 50% of patients with T-LGL leukemia also react to both p21e and BA21 (manuscript submitted). One explanation for this antibody reactivity would be nonspecific interaction due to high levels of immune complexes or rheumatoid factor. However, reactivity to p21E in CD3− LDGL sera was demonstrated to be specifically inhibited by incubation with excess BA21. Additionally, all sera from patients with CD3− LDGL were negative when tested with control nonrecombinant sj26. These data argue against the supposition that general nonspecific reactivity was responsible for the observed results. Furthermore, in contrast to patients with T-LGL leukemia, sera from patients with CD3− LDGL do not contain rheumatoid factor or elevated levels of immune complexes.4
The high incidence of BA21 seroreactivity and lack of evidence for HTLV-I or HTLV-II infection suggests that exposure to a protein containing homology to BA21 may be important in the pathogenesis of this lymphoproliferative disorder of NK cells. Such antibody reactivity could result from autoantibodies directed against a cross-reactive cellular antigen. Alternatively, such seroreactivity could reflect antibodies reactive to the transmembrane glycoprotein of a novel endogenous or exogenous retrovirus. For example, antibodies reactive with proteins derived from endogenous retroviruses have been described in autoimmune diseases.20 However, little or no conservation of the amino acid sequence of BA21 could be discerned amongst the known endogenous retroviruses. Indeed, a search of available protein and nucleic acid databases employing the amino acid sequence of HTLV-I BA21 did not reveal any protein sequences with significant homology to BA21, other than those of HTLV-I and related primate retroviruses. Identification of cross-reactivity to the 34 amino acid sequence of BA21 may facilitate research aimed at understanding the etiology of this lymphoproliferative disorder of NK cells.
Supported by the Veterans Administration Grant CA54552 awarded by the National Cancer Institute; Grant HB67021 awarded by the National Heart, Lung and Blood Institute; Public Health Service grants DA60596 and HL33811 by the Italian Association for Cancer Research (Milan); and by the National Council for Research, Oncology Project (Rome).
Address reprint requests to Thomas P. Loughran, Jr, MD, H. Lee Moffitt Cancer Center and Research Institute, 12902 Magnolia Dr, Tampa, FL 33612.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal