Abstract
We investigated the molecular genetic and biosynthetic basis of Bernard-Soulier syndrome in a severely affected white woman. Flow cytometric analysis showed a severe deficiency of glycoprotein (GP) Ib, GP IX, and GP V on the surface of her platelets. Similarly, GP Ibα was undetectable by immunoblot analysis of platelet lysates. Surprisingly, a large quantity of a 70-kD protein (which probably represents a GP Ibα degradation product) was found in the patient's plasma in much greater quantities than in the plasma of an unaffected individual. To analyze the molecular lesion responsible for the disorder, we amplified and sequenced gene segments corresponding to the entire coding regions of the GP Ibα, GP Ibβ, and GP IX genes. The patient was homozygous for a specific GP Ibα allele that contained two tandem VNTR repeats in the region encoding the macroglycopeptide (C variant) and three differences from the published GP Ibα gene sequence. Two mutations were unlikely to be involved in the disorder: the substitution of a single base (T → C) in the second nucleotide of exon 2, which is in the 5′ untranslated region of the GP Ibα transcript, and a silent mutation in the third base of the codon for Arg342 (A → G) that does not change the amino acid sequence. The third mutation was a deletion of the last two bases of the codon for Tyr492 (TAT). This mutation causes a frameshift that alters the GP Ibα amino acid sequence, beginning within its transmembrane region. The mutant polypeptide contains 81 novel amino acids and is 38 amino acids shorter than its wild-type counterpart. The new sequence changes the hydrophobic nature of the transmembrane domain and greatly decreases the net positive charge of what had been the cytoplasmic domain. The deletion mutation was introduced into the GP Ibα cDNA, alone and in combination with the 5′ mutation, and expressed in Chinese hamster ovary (CHO) cells. The deletion alone severely reduced GP Ibα expression on the cell surface. Expression was not decreased further by addition of the 5′ mutation, confirming that the deletion was the cause of the Bernard-Soulier phenotype. Stable cell lines expressing the mutant polypeptide secreted large amounts of the polypeptide into the medium, suggesting that the mutant anchors poorly in the plasma membrane. Nevertheless, a fraction of the mutant was able to associate with GP Ibβ, as demonstrated by their coimmunoprecipitation with a GP Ibβ antibody.
BERNARD-SOULIER SYNDROME is a rare hereditary bleeding abnormality, usually inherited as an autosomal recessive trait.1 Its clinical manifestations include purpura, epistaxis, gingival bleeding, and menorrhagia; the diagnosis is based on the presence of a prolonged skin bleeding time, thrombocytopenia, giant platelets on a peripheral blood smear, and defective platelet agglutination to ristocetin.
The disorder is due to defective adhesion of platelets to the vessel wall caused by quantitative or functional abnormalities of the platelet receptor for von Willebrand factor (vWf), the glycoprotein (GP) Ib-IX-V complex.2-5 This receptor complex contains four polypeptides: GP Ibα, GP Ibβ, GP IX, and GP V. GP Ibα is a heavily O-glycosylated surface mucin with a molecular mass of about 135 kD that contains the binding sites for vWf and thrombin.6 This polypeptide is disulfide-linked to GP Ibβ (24 kD); GP IX (22 kD) and GP V (83 kD) associate noncovalently with the other two subunits.7 All of the polypeptides of the complex are synthesized from distinct genes, and all are members of the leucine-rich motif protein family.7 These genes have several features in common: they are small and simple in structure, with only GP IX containing more than one intron.8-12 Except in the GP Ibβ gene, all of the introns are in the 5′ untranslated region, leaving the coding sequence uninterrupted.
Previous studies from our laboratory have shown that efficient expression of GP Ibα on the cell surface requires the presence of GP Ibβ and GP IX,13,14 whereas GP V plays only a minor role, if any, in surface expression of the complex.15 These data suggested that Bernard-Soulier syndrome could potentially arise from mutations affecting either GP Ibα, GP Ibβ, or GP IX. Indeed, mutations of the genes for these three polypeptides have now been identified in patients with Bernard-Soulier syndrome. Most of these mutations are in the GP Ibα gene and decrease expression of GP Ibα on the platelet surface,16-22 although mutations that affect the function more than the expression of GP Ibα have also been identified.23,24 Mutations of GP IX and GP Ibβ generally decrease expression of the entire complex on the platelet surface.25-28
In this report, we characterize the molecular genetic and biosynthetic defects underlying Bernard-Soulier syndrome in a patient whose platelets are severely deficient in the GP Ib-IX-V complex. The causative mutation is a deletion of two bases in the GP Ibα gene that shifts the translation reading frame, producing a GP Ibα polypeptide with novel transmembrane and cytoplasmic domains. This mutant polypeptide is poorly anchored in the plasma membrane, and much of it is secreted into the plasma or proteolyzed from the cell surface. The mutation occurs on the background of a unique haplotype that includes two other single-base substitutions that probably represent nucleotide polymorphisms in the population from which the patient derives.
SUBJECTS AND METHODS
The patient.The patient is a 36-year-old white female whose parents are third cousins. Her ancestors emigrated to Texas from Germany in the late 1800s. She has had a life-long history of excessive bleeding, starting with an episode of spontaneous gastrointestinal hemorrhage as an infant. She suffered from multiple episodes of prolonged epistaxis and has experienced menorrhagia since menarche. She was diagnosed with Bernard-Soulier syndrome at age 17, and has since then been treated with platelet transfusions for major bleeding episodes.
She has two siblings, an older sister who is unaffected and a younger brother who was diagnosed with Bernard-Soulier syndrome. The brother was unavailable for study.
Flow cytometric analysis of the patient's platelets.Flow cytometry analysis of GP Ib-IX-V polypeptides was performed on fixed whole blood.29 30 Venous blood was drawn slowly into a syringe containing acid-citrate-dextrose anticoagulant to a final concentration of 20% (vol/vol). Blood from a normal donor was drawn in parallel and treated identically. Anticoagulated whole blood was fixed by adding 900 μL 2% paraformaldehyde (vol/vol in phosphate-buffered saline [PBS]) for each 100 μL blood. The samples were incubated with two monoclonal antibodies, a phycoerythrin (PE)-conjugated monoclonal antibody against platelet GP IIb-IIIa (clone 5B12; Dako, Carpinteria, CA) and an antibody against one of three GP Ib-IX-V polypeptides, which was either directly conjugated to fluorescein isothiocyanate (FITC) or detected with a FITC-conjugated rabbit anti-mouse secondary antibody. The GP Ibα monoclonal antibody AN51 (Dako) was directly conjugated, whereas the GP IX (SZ1; AMAC Inc, Westbrook, ME) and GP V (SW16; Accurate Chemical and Scientific Corp, Westbury, NY) antibodies were unlabeled. Platelets were identified by labeling with PE and then analyzed for expression of GP Ib-IX-V polypeptides. Two techniques were used, depending on the antibody. To detect GP Ibα, 100 μL fixed blood was incubated with 10 μL PE-conjugated 5B12 and 10 μL FITC-conjugated AN51 for 1 hour at room temperature. At the end of incubation, the sample was diluted with 400 μL 2% paraformaldehyde and directly analyzed. To study GP IX and GP V, 100 μL fixed blood was first incubated with the primary monoclonal antibody (final concentration, 4 μg/mL) for 1 hour at room temperature. To remove unbound antibody, the sample was diluted with 1 mL PBS and centrifuged at 10,000g, and the pellet was resuspended in 100 μL 2% paraformaldehyde. The sample was then incubated with the FITC-conjugated secondary antibody (1:1,000 dilution) for 30 minutes at room temperature. After washing the sample with PBS, 10 μL PE-conjugated 5B12 was added and incubated for 1 hour. The sample was then diluted with 400 μL 2% paraformaldehyde and analyzed. All flow cytometric analyses were performed on a FACStar flow cytometer (Becton Dickinson, San Jose, CA). The platelets were first gated using the PE window (excitation at 488 nm and emission at 580 nm) to obtain cells expressing GP IIb-IIIa. Ten thousand cells from the GP IIb-IIIa–positive gate were analyzed for FITC fluorescence (excitation at 488 nm and emission at 520 nm). Fluorescence was compared in the patient's platelets versus platelets from a normal donor.
Immunoblotting of platelets and plasma.Platelet-rich plasma (PRP) was isolated as the upper clear layer from anticoagulated whole blood centrifuged at 100g for 15 minutes at room temperature. Platelets were isolated by centrifuging the PRP at 10,000g for 5 minutes. After removing the supernatant plasma (platelet-poor plasma), the platelet pellet was resuspended in RIPA lysis buffer (100 mmol/L Tris, 50 mmol/L NaCl, 0.5% sodium dodecyl sulfate [SDS], and 1% Triton X-100) that contained a protease inhibitor cocktail (1 mg/mL leupeptin, 1.6 mg/mL benzamidine, 0.1 mg/mL soybean trypsin inhibitor, and 1 mmol/L phenylmethylsulfonyl fluoride), 5 mmol/L EDTA, and 100 μg/mL DNase I. The platelet lysate and platelet-poor plasma were both mixed with equal volumes of 2× SDS sample buffer (4% SDS) containing 4% β-mercaptoethanol, boiled for 10 minutes, and then electrophoresed on a 7.5% SDS-polyacrylamide gel. Proteins were electrophoretically transferred to a nitrocellulose membrane. The membrane was incubated for 1 hour at room temperature in a solution containing 5% nonfat milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T) to block nonspecific binding. The membrane was then incubated for 1 hour with 4 μg/mL monoclonal antibody, either WM-23 to detect GP Ibα (kindly provided by Dr Michael C. Berndt, Baker Medical Research Institute, Prahran, Victoria, Australia) or G1.9 to detect platelet GP IIb (a kind gift from Dr Perumal Thiagarajan, University of Texas, Houston, TX). The membrane was washed twice with TBS-T and then incubated for 45 minutes with horseradish peroxidase–conjugated anti-mouse antibody (1:10,000 dilution; Amersham, Arlington Heights, IL) and washed as before. The bound antibody was then detected using a chemiluminescence detection kit (Amersham kit no. 2106)
DNA analysis.Genomic DNA was extracted from whole blood using the QIAamp blood kit (Qiagen Inc, Chatsworth, CA). Oligonucleotides for polymerase chain reaction (PCR) and DNA sequencing were based on the published sequences of the GP Ibα, GP Ibβ, and GP IX genes.31
PCR was performed in a DNA thermocycler (Perkin Elmer, Norwalk, CT). For each primer pair, conditions were optimized to produce maximum specificity and yield. Each reaction contained 100 to 300 ng genomic DNA, 250 ng of each primer, each dNTP at a final concentration of 200 μmol/L, 2.5 U Pfu DNA polymerase, and 5 μL Pfu buffer provided by the manufacturer (Stratagene, La Jolla, CA). The volume in each reaction tube was brought to 50 μL with water. Regions of the GP Ibα and GP IX genes were amplified as follows: the samples were heated to 95°C for 10 minutes and then subjected to 35 cycles of 1 minute at 95°C, 1 minute at 60°C, and 2 minutes at 72°C. At the end of 35 cycles, the mixture was incubated at 72°C for 10 minutes to increase the yield of amplification. Similar conditions were used to amplify the GP Ibβ gene, except that the annealing temperature was 61°C and the reaction mixture contained 5% dimethyl sulfoxide.
Both strands of each PCR fragment were directly sequenced using either the original PCR primers or specific internal primers. Dye-terminator cycle-sequencing reactions were performed with AmpliTaq DNA polymerase according to the protocol provided by the manufacturer (Perkin Elmer). The reaction products were analyzed using an ABI 373 DNA sequencer (Perkin Elmer).
Site-directed mutagenesis.Mutations found in the GP Ibα gene in the patient were introduced into the GP Ibα cDNA by site-directed mutagenesis. For each mutation, two complementary primers containing the mutant sequence were used to amplify the plasmid containing wild-type GP Ibα cDNA using Pfu DNA polymerase. The amplification reaction consisted of 16 cycles with the following steps: 30 seconds at 95°C, 60 seconds at 55°C, and 14 minutes at 68°C. The final products were digested with the restriction endonuclease Dpn I, which only recognizes methylated restriction sites present on the template DNA and does not digest the reaction products. The mixture was then used to transform DH5α Escherichia coli, which were plated on Luria broth plates containing 50 μg/mL ampicillin. For each mutagenesis reaction, plasmid DNA from several colonies was isolated and sequenced. cDNA inserts from mutant plasmids were sequenced completely to confirm the presence of the desired mutation and to exclude unwanted mutations.
Expression of mutant GP Ibα polypeptides.We analyzed expression of the mutant GP Ibα polypeptides in both transiently and stably transfected cell lines. Chinese hamster ovary (CHO) cells that stably express GP Ibβ and GP IX (CHO βIX cells14 ) were used for transient and stable expression of mutant cDNAs. The cells were transfected by liposome-mediated delivery of the plasmid DNA using a commercially available kit (LIPOFECTAMINE; GIBCO-BRL, Grand Island, NY). To compare expression of mutant and wild-type polypeptides, cells were analyzed 48 hours after the transfection either for surface expression of GP Ibα by flow cytometry or for total cellular expression of GP Ibα by immunoblotting (description follows).
Stable cell lines expressing wild-type or mutant GP Ibα were established by cotransfecting into CHO βIX cells the plasmid containing either wild-type or mutant cDNA (deletion mutation) and pREP4 (Invitrogen, Carlsbad, CA), a plasmid that carries a hygromycin resistance gene. Hygromycin at a final concentration of 400 μg/mL was used to select clones expressing the GP Ib-IX complex.
Analysis of the recombinant proteins.Plasma membrane expression of wild-type and mutant polypeptides in transiently transfected cells was compared by flow cytometry. Forty-eight hours after transfection, the cells were detached from the tissue culture plates with 0.54 mmol/L EDTA, washed twice with PBS, and incubated for 30 minutes with 4 μg/mL WM-23. The cells were then washed twice again and incubated for 30 minutes with FITC-conjugated rabbit anti-mouse IgG (1:1,000 dilution; Zymed, South San Francisco, CA). The cells were then washed twice more to remove unbound antibody. At least 10,000 cells from each transfection were analyzed by exciting the FITC label at 480 nm using an argon ion laser and evaluating emission at 520 nm. The analyses were performed on a Becton Dickinson FACStar flow cytometer.
Total cellular expression of the mutant polypeptides was also compared by immunoblotting versus that of the wild-type polypeptide in transiently transfected cells. Forty-eight hours after transfection, an equal number of cells expressing wild-type or mutant GP Ibα were lysed with RIPA lysis buffer as described for the platelets, and the lysates were electrophoresed on 7.5% SDS-polyacrylamide gels. The proteins were blotted onto nitrocellulose membranes, and the blots were probed with WM-23 as described for the platelets.
To analyze the possible association of the mutant GP Ibα polypeptide with GP Ibβ, GP Ibβ was immunoprecipitated from CHO βIX cells stably expressing the mutant (CHO αmutβIX cells), and the precipitate was analyzed for coprecipitation of GP Ibα. Aliquots of 2 × 106 cells were lysed in digitonin lysis buffer (50 mmol/L Tris, pH 7.4, 150 mmol/L NaCl, and 1% digitonin) containing the protease inhibitor cocktail, EDTA, and DNase I as described for the platelets. The lysate was centrifuged at 10,000g for 5 minutes to remove debris and then incubated overnight at 4°C with fixed Staphylococcus aureus cells (Pansorbin beads; Calbiochem Corp, La Jolla, CA) to remove proteins that bind the beads nonspecifically. The beads were then removed by centrifugation, and the lysate was incubated with 10 μg/mL monoclonal GP Ibβ antibody Gi27 (kindly provided by Dr Sentot Santoso, Giessen, Germany) for 1 hour at 4°C. Rabbit anti-mouse serum (1:1,000 dilution; Dako) was then added to the lysate and incubated for 1 hour at 4°C, after which 50 μL 10% slurry of Pansorbin beads was added. The beads were incubated with the lysate for 1 hour at 4°C, pelleted by centrifugation at 10,000g for 5 minutes, and resuspended in lysis buffer without digitonin. Bound proteins were eluted from the beads by adding an equal volume of 2× SDS buffer containing 4% β-mercaptoethanol and boiling for 10 minutes. The protein samples were applied to 7.5% SDS-polyacrylamide gels and transferred electrophoretically to nitrocellulose membranes, where the GP Ibα was detected with WM-23.
Statistical analysis.Data were analyzed by Student's two-tailed t-test for separate variances.
RESULTS
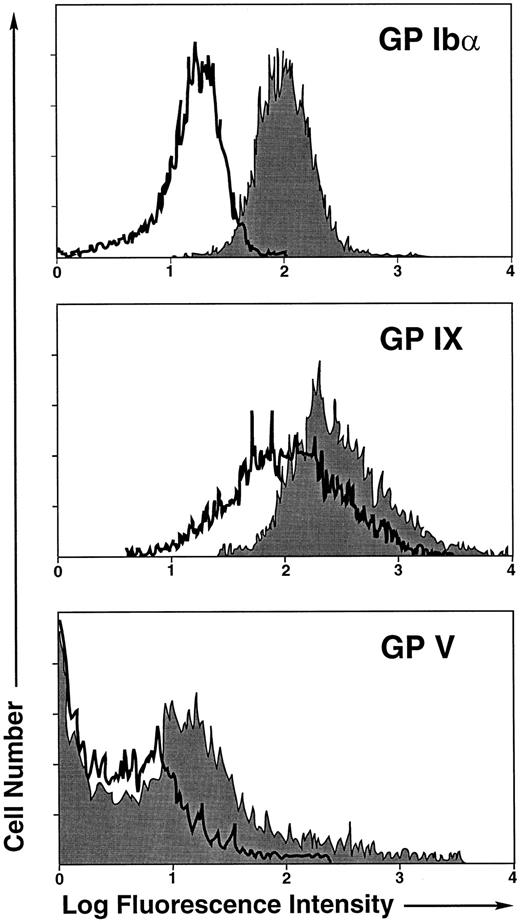
We sought to define the molecular basis of Bernard-Soulier syndrome in a 36-year-old white woman with the disorder. She was diagnosed with the syndrome at age 17, and since then her platelets have been studied extensively.32,33 The platelets in this patient have been demonstrated to have a severe defect in vWf binding and ristocetin agglutination,32 and to be incapable of shear-induced aggregation,33 among other defects. Despite these investigations, the molecular defect causing her illness had not been defined. To obtain clues as to the mutant gene causing the disorder, we first evaluated by flow cytometric analysis the surface expression of the GP Ib-IX-V complex on her platelets. The whole blood was labeled simultaneously with an antibody against platelet GP IIb-IIIa and an antibody against either GP Ib (AN51), GP IX (SZ1), or GP V (SW16). Platelets were first identified by GP IIb-IIIa positivity, and the positive population was evaluated for expression of the GP Ib-IX-V complex. This analysis revealed a severe deficiency of all three polypeptides on the patient's platelets, with GP Ibα being affected most severely (Fig 1).
Flow cytometric analysis of patient platelets. Analysis was performed on fixed whole blood with antibodies against GP Ibα (AN51), GP IX (SZ1), and GP V (SW16). (□) Patient platelets; (░) control platelets.
Flow cytometric analysis of patient platelets. Analysis was performed on fixed whole blood with antibodies against GP Ibα (AN51), GP IX (SZ1), and GP V (SW16). (□) Patient platelets; (░) control platelets.
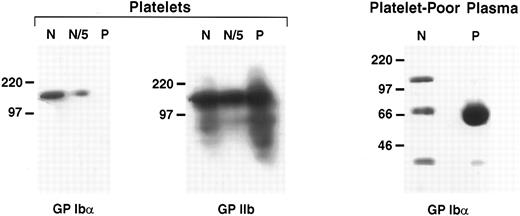
We also evaluated GP Ibα expression in the patient's platelets by immunoblotting with the monoclonal antibody WM-23. Platelets from the patient and a normal subject were prepared from an equivalent volume of PRP and analyzed. Here again, the patient's platelets were severely deficient in GP Ibα (Fig 2). This polypeptide was undetectable from a quantity of platelet lysate that contained approximately twice the amount of GP IIb-IIIa as that present in the control platelets (Fig 2, middle panel). GP Ibα was detected easily in the control platelets, even when diluted fivefold (Fig 2, left panel).
Western blot analysis of GP Ibα in platelet lysates and plasma. GP Ibα or its degradation products in the patient's platelets and platelet-poor plasma were analyzed by immunoblotting with the GP Ibα antibody WM-23. Platelets from the patient (P) and from an unaffected control subject (N) were prepared from an equal volume of PRP. Platelet lysates were compared for GP Ibα content (left panel) and GP IIb content (middle panel, antibody G1.9). N/5 represents a fivefold dilution of the normal plasma. The right panel is an immunoblot of 20 μL normal plasma and patient plasma.
Western blot analysis of GP Ibα in platelet lysates and plasma. GP Ibα or its degradation products in the patient's platelets and platelet-poor plasma were analyzed by immunoblotting with the GP Ibα antibody WM-23. Platelets from the patient (P) and from an unaffected control subject (N) were prepared from an equal volume of PRP. Platelet lysates were compared for GP Ibα content (left panel) and GP IIb content (middle panel, antibody G1.9). N/5 represents a fivefold dilution of the normal plasma. The right panel is an immunoblot of 20 μL normal plasma and patient plasma.
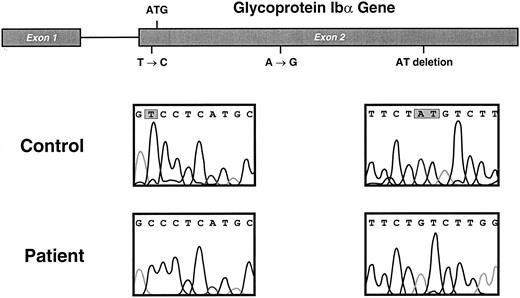
GP Ibα gene and mutations in the Bernard-Soulier patient. Sequencing results from the patient and from a control subject are compared in the bottom 2 panels, with the mutated nucleotides shown enclosed in gray boxes. The position of the mutations in the GP Ibα gene are depicted above.
GP Ibα gene and mutations in the Bernard-Soulier patient. Sequencing results from the patient and from a control subject are compared in the bottom 2 panels, with the mutated nucleotides shown enclosed in gray boxes. The position of the mutations in the GP Ibα gene are depicted above.
In contrast to the patient's platelets, the patient's plasma contained copious amounts of a 70-kD protein detected by the GP Ibα antibody, which presumably represents a GP Ibα degradation product.
The patient is homozygous for a mutant GP Ibα allele that contains three mutations.The genes encoding GP Ibα, GP Ibβ, and GP IX were each amplified by PCR, and the amplified fragments were directly sequenced. No mutations were found in GP Ibβ or GP IX genes, but the GP Ibα gene contained three differences from the published sequence, for which the patient was homozygous (Fig 3). The mutations were as follows: a single-base substitution (T → C) of the second nucleotide of exon 2, which is at position −5 from the ATG start codon; a silent A → G substitution of the third nucleotide of the codon for Arg342; and a deletion of the last two bases of the codon for Tyr492. Additional evidence that the patient was homozygous for this particular GP Ibα allele was provided by the fact that she was also homozygous for the polymorphism of a variable number of tandem repeats in the mucin-like region of the polypeptide (C variant, two repeats),34 and by the observation that each of her parents was heterozygous for the mutant allele (data not shown).
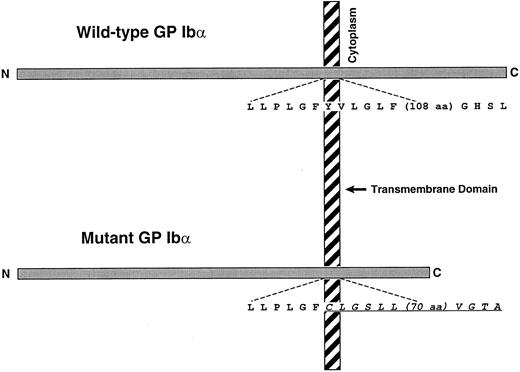
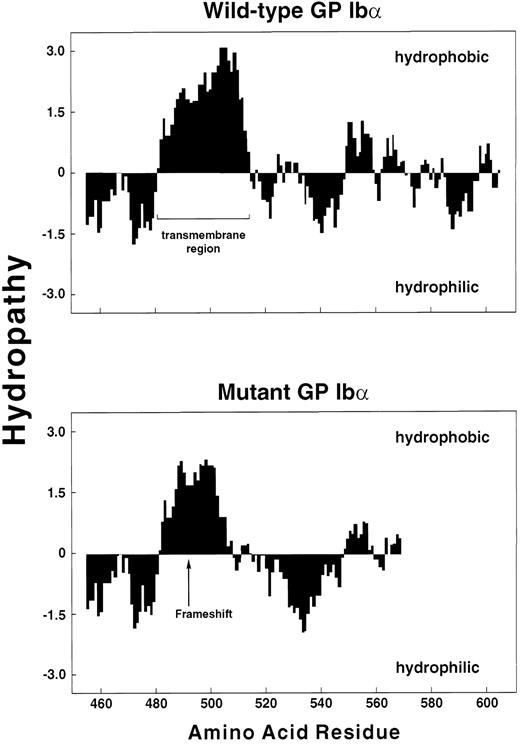
The mutation most likely responsible for the patient's phenotype is the two-base deletion, which leads to a reading frameshift, predicting a new stretch of amino acids that starts within the GP Ibα transmembrane domain and extends 81 amino acid residues downstream. The resultant full-length polypeptide is predicted to be 38 amino acids shorter than wild-type GP Ibα (Fig 4).
Schematic of wild-type GP Ibα and the mutant produced by the dinucleotide deletion of the second 2 bases of the codon for Tyr492 (TAT). The mutation shifts the translation reading frame beginning approximately 7 amino acids into the GP Ibα transmembrane domain. The new polypeptide sequence is indicated by the underlined italic portion. The full-length mutant is 38 amino acids shorter than wild-type GP Ibα.
Schematic of wild-type GP Ibα and the mutant produced by the dinucleotide deletion of the second 2 bases of the codon for Tyr492 (TAT). The mutation shifts the translation reading frame beginning approximately 7 amino acids into the GP Ibα transmembrane domain. The new polypeptide sequence is indicated by the underlined italic portion. The full-length mutant is 38 amino acids shorter than wild-type GP Ibα.
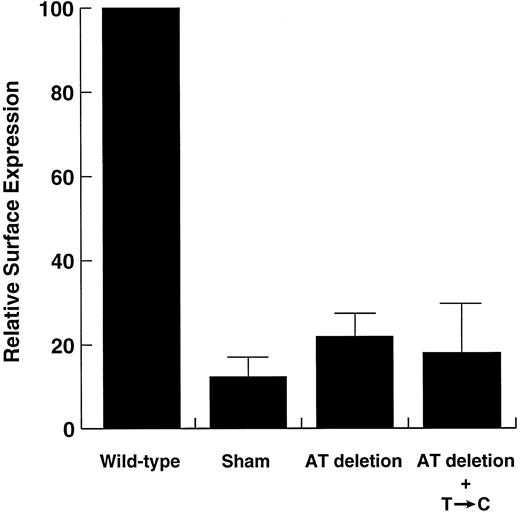
The dinucleotide deletion is responsible for the Bernard-Soulier phenotype.To determine the effects of the patient's genotype on her phenotype, we constructed mutant plasmids with different combinations of two mutations: the mutation in the 5′ untranslated region (T → C) and the dinucleotide deletion. The mutant polypeptides were transiently expressed in CHO βIX cells, and expression of the polypeptide on the cell surface was evaluated with WM-23. Flow cytometric analysis indicated that the dinucleotide mutation alone decreases surface expression of GP Ibα by 80%, with addition of the T → C mutation causing no further decrease in expression (Fig 5). Although it was low, binding of the antibody to the surface of cells transfected with mutant plasmids was significantly greater than binding to sham-transfected cells.
Flow cytometric analysis of mutant GP Ibα surface expression in transiently transfected CHO cells. CHO βIX cells were transiently transfected with plasmids containing a wild-type GP Ibα cDNA or cDNAs containing the AT deletion, alone and in combination with the 5′ mutation (see Fig 4). Both mutants produced lower surface levels of GP Ibα than did the wild-type plasmid (Student's t-test, n = 5, P < .0001), but were not significantly different from each other (P = .53). Surface fluorescence in sham-transfected cells was significantly lower than in cells transfected with the AT-deletion mutant (P < .05).
Flow cytometric analysis of mutant GP Ibα surface expression in transiently transfected CHO cells. CHO βIX cells were transiently transfected with plasmids containing a wild-type GP Ibα cDNA or cDNAs containing the AT deletion, alone and in combination with the 5′ mutation (see Fig 4). Both mutants produced lower surface levels of GP Ibα than did the wild-type plasmid (Student's t-test, n = 5, P < .0001), but were not significantly different from each other (P = .53). Surface fluorescence in sham-transfected cells was significantly lower than in cells transfected with the AT-deletion mutant (P < .05).
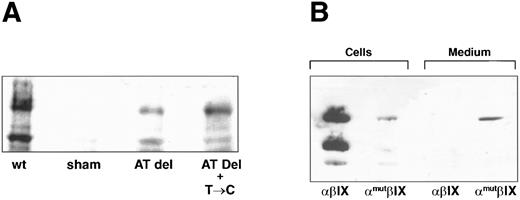
We also evaluated expression by immunoblot analysis of lysates of the transiently transfected cells (Fig 6A). Lysates from cells transfected with wild-type or mutant GP Ibα cDNA contain the respective polypeptides at different stages of maturation or degradation. The upper band (∼130 kD) represents the mature polypeptide, and the lower band (∼70 kD) represents either nonglycosylated or partially proteolyzed GP Ibα. As predicted, the dinucleotide deletion greatly decreased GP Ibα expression. However, in contrast to the patient's platelets, the mutant polypeptide was readily detected in the transfected cells.
Western blot analysis of recombinant polypeptides. (A) Cells transiently transfected with plasmids containing wild-type GP Ibα (wt), no insert (sham), the AT deletion alone (AT del), or the AT deletion in combination with the T → C mutation (AT Del + T → C) were analyzed for expression of GP Ibα with the monoclonal antibody WM-23. (B) Stable cell lines expressing the wild-type GP Ib-IX complex (αβIX cells) or a complex containing the AT-deletion mutant (αmutβIX) were analyzed for GP Ibα expression in the cells and in the cell supernatant (medium).
Western blot analysis of recombinant polypeptides. (A) Cells transiently transfected with plasmids containing wild-type GP Ibα (wt), no insert (sham), the AT deletion alone (AT del), or the AT deletion in combination with the T → C mutation (AT Del + T → C) were analyzed for expression of GP Ibα with the monoclonal antibody WM-23. (B) Stable cell lines expressing the wild-type GP Ib-IX complex (αβIX cells) or a complex containing the AT-deletion mutant (αmutβIX) were analyzed for GP Ibα expression in the cells and in the cell supernatant (medium).
To determine if the recombinant is secreted into the cell medium, as indicated from the patient's plasma, we compared the amount of GP Ibα in the cells and in the supernatant medium from the cells stably expressing the deletion mutant (Fig 6B). From this analysis, it appears that most of the polypeptide synthesized is secreted into the medium. The polypeptide in the medium is of a higher molecular weight than the polypeptide found in the patient's plasma, probably owing to the greater quantity of proteases in the plasma than in the cell medium. In the experiment shown in Fig 6B, the cell lysate represents one third of the cells from a confluent 75-cm2 cell-culture flask (about 2 × 106 cells), whereas the immunoblot of the supernatant required only 20 μL from a total media volume of 10 mL. This small amount of medium contained at least as much polypeptide as present in the cell lysate (Fig 6B, lane 2 v lane 4). Thus, it appears that in the cell lysate, as in the patient, the majority of mutant polypeptide is secreted or hydrolyzed from the cells. This finding was in marked contrast to the results from cells expressing wild-type GP Ibα. Although the cell lysate contained abundant GP Ibα (Fig 6B, lane 1), the polypeptide was not detected in the medium (Fig 6B, lane 3).
A fraction of mutant GP Ibα remains associated with GP Ibβ.One possible mechanism by which the frameshift mutation might decrease levels of GP Ibα on the platelet surface is by precluding the association of GP Ibα with GP Ibβ, which we have previously demonstrated to be important for surface expression of GP Ibα.13 To determine if this was the case, we evaluated the association of these two polypeptides by immunoprecipitating GP Ibβ and examining whether GP Ibα coprecipitated. As clearly indicated by the data in Fig 7, at least a fraction of the mutant GP Ibα polypeptide in CHO cells is in a complex with GP Ibβ. In this immunoblot, which is from a gel electrophoresed for a longer time than those in Fig 6, the difference in molecular weight between wild-type and mutant polypeptides (∼4 kD) is clearly discernible. Examination of the association by immunoblotting GP Ibα from reduced and nonreduced SDS-polyacrylamide gels indicates that the association is covalent (not shown).
Association of mutant GP Ibα with GP Ibβ. GP Ibβ was immunoprecipitated from cell lysates of stably transfected cells (see Fig 6). The samples were reduced, electrophoresed on a 7.5% SDS-polyacrylamide gel, and transferred to nitrocellulose membrane. The membrane was analyzed for GP Ibα content with WM-23. βIX, untransfected CHO βIX cells; other labeling as in Fig 6.
Association of mutant GP Ibα with GP Ibβ. GP Ibβ was immunoprecipitated from cell lysates of stably transfected cells (see Fig 6). The samples were reduced, electrophoresed on a 7.5% SDS-polyacrylamide gel, and transferred to nitrocellulose membrane. The membrane was analyzed for GP Ibα content with WM-23. βIX, untransfected CHO βIX cells; other labeling as in Fig 6.
DISCUSSION
In the present study, we have characterized the biochemical and genetic basis of a unique case of Bernard-Soulier syndrome. The platelets of the affected patient have been studied extensively in the past because of their almost complete lack of a functional receptor for vWf,32 33 but the nature of the genetic lesion was unknown until now. By flow cytometry and Western blot analysis, we identified a severe deficiency of the GP Ib-IX-V complex on the patient's platelets, with GP Ibα being the polypeptide affected most severely.
To determine the genetic basis of this abnormality, we sequenced the GP Ibα, GP Ibβ, and GP IX genes and found three mutations in the GP Ibα gene, for which the patient was homozygous. Both of her parents had one copy of the mutant allele. Among the changes in the mutant allele, we found a deletion in the last two bases of the codon for Tyr492 (TAT), which causes a frameshift that changes the amino acid sequence of GP Ibα beginning seven residues into the predicted transmembrane domain.6 This change predicts a mutant polypeptide that is 38 amino acids shorter than the wild-type polypeptide and contains a novel sequence of 81 amino acids. Because of this drastic change in the structure of the protein, we hypothesized that this mutation was the probable cause of Bernard-Soulier syndrome in this patient. We did not expect the other mutations of the mutant haplotype, a silent mutation in Arg342 and substitution of the second base of exon 2 (in the 5′ untranslated region of the transcript), to significantly influence the patient's phenotype. Both of these mutations have been reported as polymorphisms in a Finnish population, with the 5′ mutation being more frequent than the third-base change at codon 342.35 Clearly, neither of these mutations causes Bernard-Soulier syndrome by itself, but we considered it possible that the 5′ mutation might contribute to the low GP Ibα expression because of a potential influence on the efficiency of mRNA translation (it lies immediately upstream of the ATG start codon). This was not the case. The polypeptide produced by cells expressing the deletion alone was expressed poorly on the plasma membranes of transfected cells. Addition of the substitution in the 5′ untranslated region did not further decrease expression of the mutant polypeptide, indicating that the deletion is both necessary and sufficient to produce the clinical phenotype in this patient.
Hydropathy analysis of wild-type and mutant polypeptides. The regions from amino acid residue 450 to the end of each polypeptide were analyzed by the method of Kyte and Doolittle,39 using a window of 10 residues, with GenePro software (Riverside Scientific, Seattle, WA).
Hydropathy analysis of wild-type and mutant polypeptides. The regions from amino acid residue 450 to the end of each polypeptide were analyzed by the method of Kyte and Doolittle,39 using a window of 10 residues, with GenePro software (Riverside Scientific, Seattle, WA).
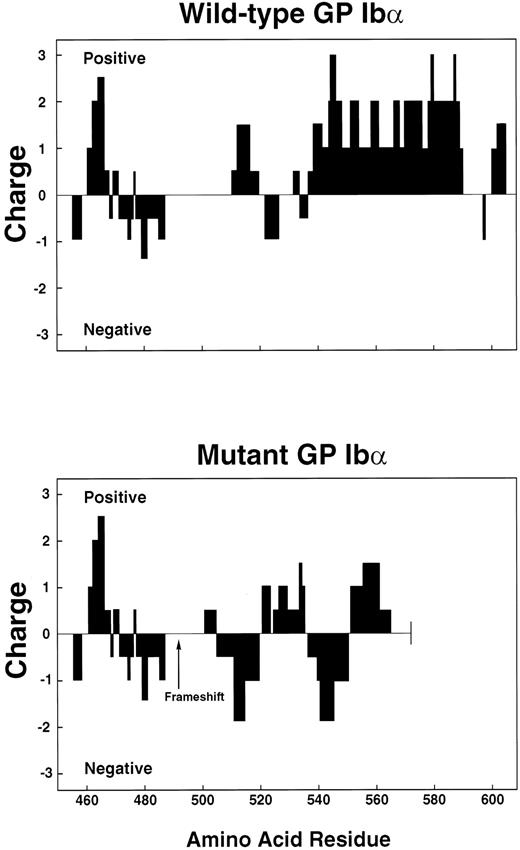
Charge profile of wild-type and mutant polypeptides. Charge profiles of the 2 polypeptides were analyzed by assigning values of +10 to Arg and Lys, +5 to His, −10 to Asp and Glu, and 0 to the other amino acid residues, using GenePro software. The scan window was set at 10.
Charge profile of wild-type and mutant polypeptides. Charge profiles of the 2 polypeptides were analyzed by assigning values of +10 to Arg and Lys, +5 to His, −10 to Asp and Glu, and 0 to the other amino acid residues, using GenePro software. The scan window was set at 10.
However, these results did not completely explain the platelet phenotype. GP Ibα was undetectable in the patient's platelets, but was readily detectable in the transfected CHO cells, albeit at greatly reduced amounts. This discrepancy has two probable causes. First, unlike CHO cells, platelets lack a nucleus, and their limited capability to synthesize proteins depends on a small pool of preformed mRNA. In contrast, CHO cells are able to continually synthesize proteins, and even a small amount of expression on the cell surface might be sustained by replacement of lost or cleaved proteins by newly synthesized proteins. Second, the mutation may render the polypeptide more susceptible to proteolytic cleavage, and platelets in the circulation are in a much more protease-rich environment than cells in culture media.
The mutant polypeptide contains several structural features that may explain its poor expression on the cell surface. The sequence following the frameshift, while still hydrophobic, is less so than the corresponding sequence of wild-type GP Ibα (Fig 8). In addition, the positively charged amino acids that follow the transmembrane domain of the wild-type polypeptide are replaced in the mutant by a region that tends to be negatively charged. In fact, the mutation dramatically reduces the positive charge of the entire cytoplasmic region (Fig 9). Positively charged regions on the cytoplasmic faces of transmembrane segments seem to be a general feature of transmembrane proteins, and may serve to orient the protein or to help anchor it within the membrane.36 These two features may prevent the polypeptide from anchoring in the membrane of the endoplasmic reticulum as it is being synthesized, although the residual hydrophobicity of the transmembrane segment may slow the transit of the polypeptide during translocation. Because of its decreased time of residence in the membrane, the mutant GP Ibα may have insufficient time to efficiently associate with GP Ibβ. This scenario is consistent with our observations that a large percentage of the mutant polypeptide or its proteolytic fragments can be found in the patient's plasma and in the culture media of cells expressing the mutant, while in the cells at least a fraction of the remaining GP Ibα is associated with GP Ibβ.
Another consequence of the mutation is that the sequences through which GP Ibα associates with actin-binding protein37 and the 14.3.3 protein38 are both missing. This may also affect stability of the protein.
Interestingly, we learned during the course of this study that another patient with Bernard-Soulier syndrome who was characterized independently has the same two-base deletion, also on the background of the haplotype that contains the other two mutations found in our patient. As best as we could ascertain from their family histories, the two patients are unrelated. However, they both are of German ancestry, with both families having emigrated to the United States sometime in the last century. These findings suggest either that the mutant allele was brought to the United States by one family of Northern European immigrants, representing a founder effect, or that the allele may be similarly prevalent in the German population in Europe. Those investigating Bernard-Soulier syndrome in Northern European countries should be alert to this possibility.
In summary, we have characterized the molecular and synthetic basis of Bernard-Soulier syndrome in a patient homozygous for a GP Ibα mutation that apparently arose on the background of a distinct haplotype from a German population. The Bernard-Soulier phenotype is caused by deletion of the second two bases of the codon for Tyr492, which produces a frameshift and yields a novel polypeptide segment of 81 amino acids that is markedly different from wild-type GP Ibα in both hydropathy and charge. The mutant polypeptide is poorly anchored in the plasma membranes of both platelets and transfected cells and appears in increased quantities in the plasma and cell supernatant.
ACKNOWLEDGMENT
We thank Drs Jing-fei Dong, Joel Moake, Konstantinos Konstantopoulos, and Brett Casey, and David Smith for helpful suggestions.
Supported by National Institutes of Health Grants No. HL02463 and HL46416, American Heart Association Grants No. 96012670 and 96002750, and an Established Investigator Award from the American Heart Association (to J.A.L.).
Address reprint requests to José A. López, MD, Veterans Affairs Medical Center, Hematology/Oncology (111H), 2002 Holcombe Blvd, Houston, TX 77030.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal