To the Editor:
Hereditary hemochromatosis (HH) is an autosomal-recessive iron storage disease associated with widespread tissue injury including cirrhosis, endocrine failure, joint inflammation, and cardiac disorders. HLA-H has recently been reported to be a candidate gene for HH on the short arm of chromosome 6, as two separate mutations have been found in HH patients. Eighty-seven percent of HH patients have a G to A transition at nucleotide 845 of the open reading frame of HLA-H that results in a cysteine to tyrosine substitution at amino acid 282 (C282Y).1 Subsequently, other groups have confirmed the finding and demonstrated this missense mutation in 90% and 100% of HH patients, respectively.2,3 The implications of the second missense variant H63D caused by a C to G transversion at 187 nucleotides (nt) of the open reading frame, however, were uncertain as it occurred at a much lower frequency than the C282Y mutation in HH patients and occurred at the same frequency in the general population. The C282Y mutation appears to be closely related to HH but the H63D variant is most likely a polymorphism.2 3
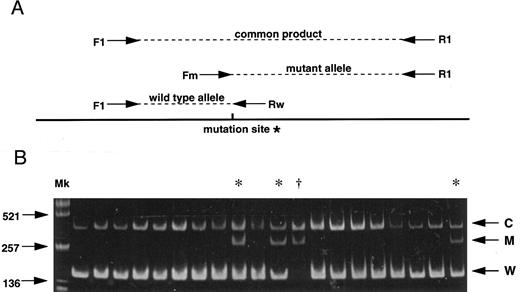
(A) Strategy for the AS-PCR. Each allele is distinguished by its product size. A common product is amplified from both alleles by the primers F1 and R1. The primer Fm recognizes only the mutant allele sequence while the primer Rw recognizes only the wild-type sequence. *G to A mutation (C282Y) creates a new Rsa I site. (B) Results of 20 subjects screening demonstrating 16 normal subjects, 3 carriers (*) and 1 homozygote (†). C, common product; M, mutant allele; W, wild-type allele; Mk, size marker (Alu I–digested pUC19).
(A) Strategy for the AS-PCR. Each allele is distinguished by its product size. A common product is amplified from both alleles by the primers F1 and R1. The primer Fm recognizes only the mutant allele sequence while the primer Rw recognizes only the wild-type sequence. *G to A mutation (C282Y) creates a new Rsa I site. (B) Results of 20 subjects screening demonstrating 16 normal subjects, 3 carriers (*) and 1 homozygote (†). C, common product; M, mutant allele; W, wild-type allele; Mk, size marker (Alu I–digested pUC19).
HH is a prevalent disorder and is often diagnosed only after significant organ damage has occurred. Treatment with regular phlebotomy will normally provide effective control of excessive iron stores but damage to organs such as liver and pancreas cannot be reversed by this treatment. Effective prevention of organ damage from HH will require screening of large population groups to identify heterozygotes and homozygotes at risk of the disorder and will only be feasible if reliable and cost-effective methods are available.
As both single base substitutions of the HLA-H gene create or abolish restriction endonuclease recognition sites, polymerase chain reaction (PCR) products spanning the regions of interest were digested with appropriate restriction enzymes for investigating the base substitutions in all previous reports.2-4 6-8 Here we report a more rapid and simple method for large scale screening of the C282Y mutation by allele specific PCR (AS-PCR) combined with facilitated genomic DNA extraction from whole blood. AS-PCR is a multiplex PCR method based on the fact that a successful PCR amplification requires the sequence of 3′ oligonucleotide ends to be absolutely complementary to the DNA template. Figure 1A shows a schematic of the AS-PCR strategy which includes four different oligonucleotide primers consisting of two non–allele-specific primers (F1: AAGCAGCCAATGGATGCCAAG and R1: CCACTGATGACTCCAATGACTA) and two allele-specific primers (Rw: CCTGGGTGCTCCACCTGGC, Fm: GGGAAGAGCAGAGATATACGTA). Genomic DNA was prepared by heating 5 μL whole blood in 20 μL of 0.05 N NaOH at 95°C for 20 minutes, followed by neutralization in 20 μL of 0.15 mol/L Tris-HCl (pH 6.5) and 3 μL of the sample was then directly used for the AS-PCR. PCR reaction mixtures consisted of 3 μL of the sample DNA as template, 200 μmol/L of each dNTPs, 200 nmol/L Rw, 1,000 nmol/L Fm, 1,000 nmol/L F1, 1,000 nmol/L R1, and 0.7 U Amplitaq Gold (Perkin-Elmer, Branchburg, NJ) for the hot start PCR in 20 μL total volume. Conditions for amplification were an initial denaturation at 95°C for 11 minutes followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 64°C for 30 seconds, and extension at 72°C for 30 seconds, with a final extension at 72°C for 10 minutes.
We screened 1,032 unrelated whites in the greater Dunedin area of New Zealand (ethical approval was obtained). Figure 1B shows results obtained from 20 subjects among whom 16 are normal, demonstrating the wild-type allele (W) and common products (C), while 3 are carriers with the wild-type allele, the C282Y mutant allele (M) and common products, and 1 is a homozygote with the mutant allele and common products. One hundred ninety-two heterozygotes and 10 homozygotes for the mutant allele were observed among a toatl of 1,032 subjects. Estimated frequencies for normal and mutant alleles were 0.897 and 0.103, respectively. The mutant allele frequency was one of the highest as yet reported.1 3-8
Evaluation of costs has shown that disposable reagents and supplies cost of the order of $1.37 (US) per sample. The costing was based on testing 90 samples in a batch 96-well process that included use of a single gel for the entire batch. Operation time was estimated at less than 8 hours for testing, interpretation, and reporting per batch (approximately 5 minutes per sample tested). Scope exists for further reduction in operation time through automation. The AS-PCR combined with the simple DNA extraction method offers more rapid, simple, economical, and reproducible means for detection of the HLA-H gene mutation than the conventional enzyme digestion method. Therefore, it is ideally suited to presymptomatic large scale population screening.
ACKNOWLEDGMENT
Supported by the Cancer Society of New Zealand and the Lottery Health Research.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal