Abstract
Human C/EBPε is a newly cloned gene coding for a CCAAT/enhancer binding protein that may be involved in the regulation of myeloid differentiation. Our studies showed that levels of C/EBPε mRNA were markedly increased in NB4 cells (promyelocytic leukemia line), because they were induced by 9-cis retinoic acid (9-cis RA) to differentiate towards granulocytes. Accumulation of C/EBPε mRNA occurred as early as 1 hour after exposure of NB4 cells to 9-cis RA (5 × 10−7 mol/L); and at 48 hours, levels were increased by 5.1-fold. Dose-response studies showed that 10−7 to 10−6 mol/L 9-cis RA (12 hours) resulted in peak levels of C/EBPε mRNA; but even 10−10 mol/L 9-cis RA increased levels of these transcripts. NB4 cells pulse-exposed (30 minutes) to all-trans retinoic acid (ATRA), washed, and cultured (3 days) with either dimethylsulfoxide (DMSO) or hexamethylene bisacetamide (HMBA) had a prominent increase in levels of C/EBPε mRNA and an increase in granulocytic differentiation, but exposure to either DMSO or HMBA alone had no effect on base levels of C/EBPε and did not induce differentiation. Macrophage-differentiation of NB4 reduced levels of C/EBPε mRNA. Nuclear run-off assays and half-life studies showed that accumulation of C/EBPε mRNA by 9-cis RA was due to enhanced transcription. Furthermore, this C/EBPε mRNA accumulation did not require synthesis of new protein factors because 9-cis RA induced C/EBPε mRNA accumulation in the absence of new protein synthesis. ATRA also induced expression of C/EBPε protein in NB4 cells, as shown by Western blotting. In contrast to the increase of C/EBPε in 9-cis RA–mediated granulocytic differentiation, the DMSO-induced differentiation of HL-60 cells down the granulocytic pathway was associated with an initial reduction of C/EBPε mRNA levels. In summary, we have discovered that expression of C/EBPε mRNA is markedly enhanced as the NB4 promyelocytes are induced by retinoids to differentiate towards granulocytes. This induction of C/EBPε mRNA expression is transcriptionally mediated and occurs in the absence of synthesis of additional protein factors. We suspect that the C/EBPε promoter/enhancer contains a retinoic acid-response element that is directly stimulated by retinoids.
HEMATOPOIETIC CELLS committed to the myeloid lineage progress through a complex series of differentiation steps before reaching the morphologically and functionally distinct end-stage of macrophages and neutrophils. While differentiating, these cells are continually reprogrammed to express stage-specific genes through the activation of regulatory transcription factors.1-4 Consequently, as studies on myeloid genes suggest, transcription factors play a critical role in myelopoiesis and untimely transcriptional activity may result in a failure to differentiate and promote leukemic growth.5-7 To understand the process of normal myeloid differentiation, identification and characterization of the important regulatory factors integral to the myeloid lineage are important.
Members of the C/EBP family of transcription factors have been implicated in a number of developmental and growth regulatory processes in several cell types.8-11 In the hematopoietic system, C/EBP-α, -β, and -δ showed a coordinated pattern of expression in differentiating myeloid cells and appeared to play decisive roles in myelopoiesis.12 These three C/EBP isoforms also collaborated with the Myb oncoprotein to induce the expression of myeloid-specific target genes, even in cells of heterologous tissues such as erythroblasts or fibroblasts.4 Expression of C/EBP-β was found to parallel the differentiation of myeloid cells to macrophages.12 Furthermore, mice containing a homozygous null mutation of either the C/EBP-β or the C/EBP-α were found to be defective in either macrophage functions or granulocyte development, respectively.13,14 Another C/EBP-like factor, C/EBP-γ, was found to be expressed ubiquitously15 and cooperated with Fos to bind to positive regulatory element-I (PRE-I), which is a strong enhancer element essential for the expression of the human interleukin-4–encoding gene.16
Recently, we and others have isolated another member of the C/EBP family of transcription factors, C/EBPε.17,18 This gene has two exons and encodes a nuclear phosphoprotein of 32 kD.18 The DNA sequence analysis of C/EBPε shows that the gene encodes a protein highly homologous to rat CRP1.17 Similar to other C/EBP isoforms, C/EBPε binds to a consensus C/EBP binding site.19,20 Remarkably, C/EBPε shows a restricted expression in cells of the myeloid lineage, especially during their differentiation toward granulocytes.17 18 Expression studies of this gene showed that it is expressed independently of other C/EBP proteins. Such a restricted pattern of expression suggests greatly that the gene may play an important role in the regulation of myelopoiesis. Therefore, in the present study, we examine the regulation of C/EBPε expression during differentiation of myeloid cells by culturing several myeloid cell lines with various differentiating agents. Our data suggest that the expression of C/EBPε is closely associated with the retinoid-mediated differentiation of early myeloid cells to granulocytes and that this expression is controlled by transcriptional mechanisms in the absence of new protein synthesis.
MATERIALS AND METHODS
Cells and culture conditions. HL-60 cells were obtained from the American Tissue Culture Collection (Rockville, MD). NB4 cells were generously provided by M. Lanotte (St Louis Hospital, Paris, France). Cells were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO/BRL, Gaithersburg, MD). Cell viability was determined in all experimental procedures using trypan blue exclusion.
Chemicals. All-trans retinoic acid (ATRA; Sigma, St Louis, MO) was dissolved in 95% (vol/vol) ethanol to a stock concentration of 10−2 mol/L and stored at −70°C. 9-cis retinoic acid (9-cis RA; kindly provided by M. Dawson, Stanford Research Institute, Palo Alto, CA) was dissolved in 95% ethanol to a stock concentration of 10−2 mol/L and stored at −70°C. The physiologically active vitamin D compound, 1,25 dihydroxyvitamin D3 [1,25(OH)2D3 , vit D3 ; generously provided by M. Uskokovic, Hoffmann La Roche, Nutley, NJ] was dissolved in 100% ethanol at a stock concentration of 10−3 mol/L and stored at −20°C. 12-O-tetradecanoylphorbol 12-acetate (TPA; Sigma) was dissolved in 100% acetone to a stock concentration of 10−4 mol/L and kept at −20°C. Dimethylsulfoxide (DMSO) and hexamethylene bisacetamide (HMBA) were purchased from Sigma. Actinomycin-D (Sigma) was dissolved in 20% (vol/vol) ethanol to a stock concentration of 1 mg/mL. Cycloheximide (CHX; Sigma) was dissolved in 1× phosphate-buffered saline (GIBCO/BRL) to a stock concentration of 2 mg/mL and stored at −20°C.
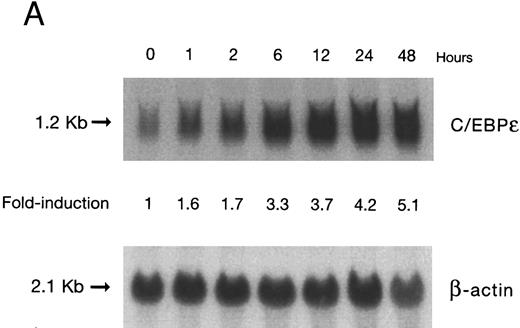
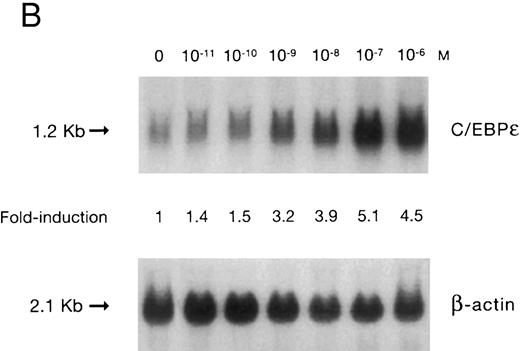
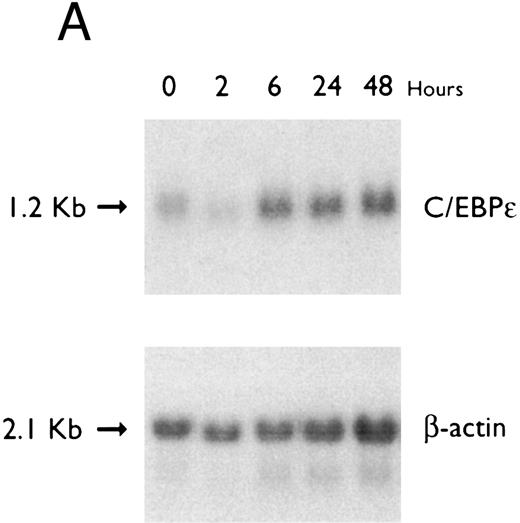
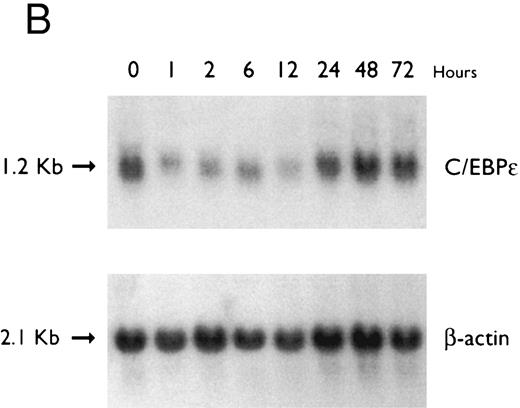
(A) Time-response: C/EBPε mRNA expression in NB4 cells cultured with 9-cis RA. Northern blot with total RNA (30 μg/lane) of NB4 exposed for various lengths of time to 9-cis RA (5 × 10−7 mol/L) and probed with full-length C/EBPε cDNA. The bottom panel shows ethidium bromide-staining of the 28S and 18S rRNA on the gel as assessment of RNA quantities in each lane. Fold-inductions were calculated by the ratio of densitometry readings of C/EBPε to β-actin. (B) Dose-response: C/EBPε mRNA levels in NB4 cells treated with 9-cis RA. Northern blot of total RNA (30 μg/lane) of NB4 cells exposed to various concentrations of ATRA for 12 hours and hybridized with full-length C/EBPε cDNA probe. The bottom panel shows hybridization with the β-actin probe as assessment of RNA quantities in each lane. Fold-inductions were calculated by the ratio of densitometry readings of C/EBPε to β-actin.
(A) Time-response: C/EBPε mRNA expression in NB4 cells cultured with 9-cis RA. Northern blot with total RNA (30 μg/lane) of NB4 exposed for various lengths of time to 9-cis RA (5 × 10−7 mol/L) and probed with full-length C/EBPε cDNA. The bottom panel shows ethidium bromide-staining of the 28S and 18S rRNA on the gel as assessment of RNA quantities in each lane. Fold-inductions were calculated by the ratio of densitometry readings of C/EBPε to β-actin. (B) Dose-response: C/EBPε mRNA levels in NB4 cells treated with 9-cis RA. Northern blot of total RNA (30 μg/lane) of NB4 cells exposed to various concentrations of ATRA for 12 hours and hybridized with full-length C/EBPε cDNA probe. The bottom panel shows hybridization with the β-actin probe as assessment of RNA quantities in each lane. Fold-inductions were calculated by the ratio of densitometry readings of C/EBPε to β-actin.
Induction of differentiation. ATRA-priming was performed by incubating the NB4 cells with 10−7 mol/L of ATRA for 30 minutes. The cells were then washed twice in culture medium without ATRA and reincubated with various differentiating agents for 3 days. To stop protein synthesis, NB4 cells were pretreated for 30 minutes with 10 μg/mL CHX and then induced with 5 × 10−7 mol/L 9-cis RA. For analysis of RNA stability, NB4 cells were treated with actinomycin D (10 μg/mL) with or without a previous 12 hours of exposure to 9-cis RA (5 × 10−7 mol/L). ATRA and 9-cis RA were used interchangeably because they had similar biologic activities. Differentiation of cell lines was assessed by the ability of the cells to produce superoxide as measured by reduction of nitroblue tetrazolium (NBT)21 and by analysis of the membrane-bound differentiation marker, CD11b, using one- and two-color immunofluorescence. The latter was performed as described.22 Briefly, cells were incubated at 4°C for 60 minutes in 10% human AB serum (Sigma) to block Fc receptors and then stained with fluorescein isothiocyanate (FITC)-conjugated murine monoclonal antibody (Becton Dickinson, Mountain View, CA). Control samples were incubated with FITC-conjugated mouse IgG1 isotype control (Becton Dickinson). Analysis of fluorescence was performed on a FACScan flow cytometry, using a LYSIS II program (Becton Dickinson).
RNA isolation and Northern blot analysis. Total RNA was extracted using Trizol (GIBCO/BRL) according to the manufacturer's instructions. Twenty to 30 μg of total RNA was electrophoresed on 1.1% agarose gel containing MOPS and formaldehyde and transferred onto nitrocellulose membranes (MSI, Westboro, MA) as described.23 Blots were hybridized for 16 to 24 hours at 42°C in 50% formamide, 5× SSC, 5× Denhardt's, 0.1% sodium dodecyl sulfate (SDS), and 100 μg/mL salmon sperm DNA (Sigma) plus 3 × 106 cpm/mL of the full-length C/EBPε cDNA probe after labeling with α-32P-dCTP by random primer DNA-labeling system (Stratagene, La Jolla, CA). Filters were washed to a stringency of 0.1× SSC at 65°C and exposed to Kodak Biomax film (Eastman Kodak, New Haven, CT). Radioactive counts were measured through Ambis Core Software, Version 4.0 according to the manufacturer's instructions (CSPI, Billerica, MA). Fold-inductions were calculated by the ratio of radioactive counts of C/EBPε to β-actin.
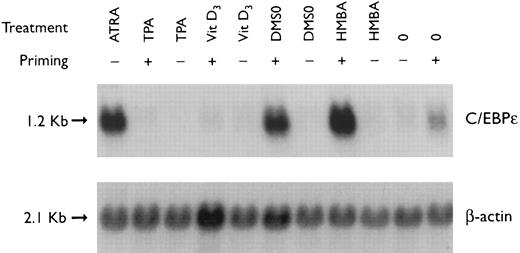
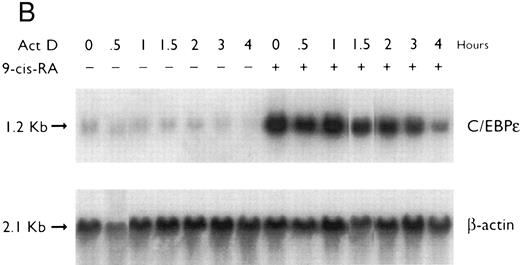
Regulation of C/EBPε expression in NB4 cells induced to differentiate after ATRA-priming. Northern blot of total RNA (30 μg/lane) from NB4 cells treated with various differentiating agents for 3 days with or without 30 minutes of priming with 1 × 10−7 mol/L ATRA. The top panel shows the hybridization of full-length C/EBPε cDNA probe. The bottom panel shows the hybridization with the β-actin probe as assessment of RNA quantities on each lane.
Regulation of C/EBPε expression in NB4 cells induced to differentiate after ATRA-priming. Northern blot of total RNA (30 μg/lane) from NB4 cells treated with various differentiating agents for 3 days with or without 30 minutes of priming with 1 × 10−7 mol/L ATRA. The top panel shows the hybridization of full-length C/EBPε cDNA probe. The bottom panel shows the hybridization with the β-actin probe as assessment of RNA quantities on each lane.
Nuclear run-off assays. For run-on experiments, nuclei were harvested from NB4 cells before and after treatment with 5 × 10−7 mol/L 9-cis RA for 12 hours. Nuclei were isolated by resuspending the cells in 1 mL RSB solution (10 mmol/L Tris-HCl [pH 7.5], 10 mmol/L KCl, 3 mmol/L MgCl2 ) and adding 100 μL of 3% NP-40 (Sigma) for 10 minutes. After pelleting at 500g for 10 minutes at 4°C, the nuclei were resuspended at 108 nuclei/mL in nuclei storage buffer (40% glycerol, 50 mmol/L Tris-HCl [pH 8.3], 5 mmol/L MgCl2 , 0.1 mmol/L EDTA) and kept on ice. For each reaction, 200 μL of nuclei was incubated at 30°C for 20 minutes in 100 μL of run-on assay buffer (15 mmol/L Tris-HCl [pH 8.0]; 7.5 mmol/L MgCl2 ; 450 mmol/L KCl; 0.75 mmol/L each of ATP, GTP, and CTP; 1 U/μL RNasin [Promega, Madison, WI]; and 250 μCi α-32P-UTP). RNase-free DNase I (3 U; Promega) was then added and the reaction was incubated for 5 minutes at 30°C. Digestion with 15 μg of proteinase K was performed at 37°C for 45 minutes in 1% SDS and 10 mmol/L EDTA. The RNA was then extracted, precipitated, and hydrolyzed as described.24 For the detection of run-on labeled transcripts on the dot blots, 10 μg of each cDNA plasmid (denatured by boiling in 0.3 mol/L NaOH and neutralized with 60 mmol/L Tris-HCl [pH 8.0] and 6× SSC) was applied to a nitrocellulose strip (MSI) in a dot blot apparatus. The immobilized plasmids were then washed with 500 μL of filtered 6× SSC. The filters were baked and prehybridized as for Northern blot analysis. For each reaction, equal counts of run-on labeled transcripts as purified by ProbeQuant G-50 micro-columns (Pharmacia, Piscataway, NJ) were hybridized at 106 cpm/mL for 36 hours at 42°C. Filters were washed to a stringency of 0.1× SSC at 55°C and exposed to Kodak Biomax film. Fold-inductions were calculated by the ratio of radioactive counts of C/EBPε to β-actin measured through Ambis Core Software, Version 4.0, according to the manufacturer's instructions (CSPI).
Western blot analysis. Samples of total cell lysate (30 μg) were denatured by the addition of 2× electrophoresis sample buffer (120 mmol/L Tris-HCl [pH 6.8], 4% SDS, 25% glycerol, 3% β-mercaptoethanol) and incubated for 10 minutes at 90°C. Samples were separated on a 15% polyacrylamide gel containing SDS.25 The proteins were electroblotted overnight onto Immobilon-P membrane as described by the manufacturer (Millipore, Bedford, MA). Purified rabbit polyclonal anti-C/EBPε antibody (18 μg/mL)18 was incubated with the previously blocked membrane for 1 hour at room temperature, and bound antibody was detected using antirabbit horseradish peroxidase antibody conjugate and the ECL substrate according to the manufacturer's instructions (Amersham, Arlington Heights, IL). COS cells transfected with either a human C/EBPε cDNA expression plasmid or empty vector were used as positive and negative controls, respectively.
RESULTS
Accumulation of C/EBPε mRNA during induction of differentiation of NB4 cells to granulocytes. The NB4 cells differentiated along the granulocytic pathway after exposure to 9-cis RA26,27; after exposure of these cells to 9-cis RA (10−7 mol/L for 6 days), 55% and 93% became NBT+ and CD11b+ as compared with less than 5% and less than 10%, respectively, in control NB4 cells (data not shown). Increased accumulation (1.6-fold) of C/EBPε mRNA occurred as early as 1 hour after exposure of the NB4 cells to 9-cis RA (5 × 10−7 mol/L). By 48 hours after induction, C/EBPε mRNA levels were 5.1-fold greater than the baseline (Fig 1A). Longer exposure of the retinoid did not result in increased levels of C/EBPε mRNA (data not shown). Dose-response studies with NB4 cells induced with 9-cis RA showed that 10−7 to 10−6 mol/L 9-cis RA (12 hours) induced peak levels (5-fold) of C/EBPε mRNA, but even 10−10 mol/L 9-cis RA increased levels of the transcripts by 1.4-fold (Fig 1B).
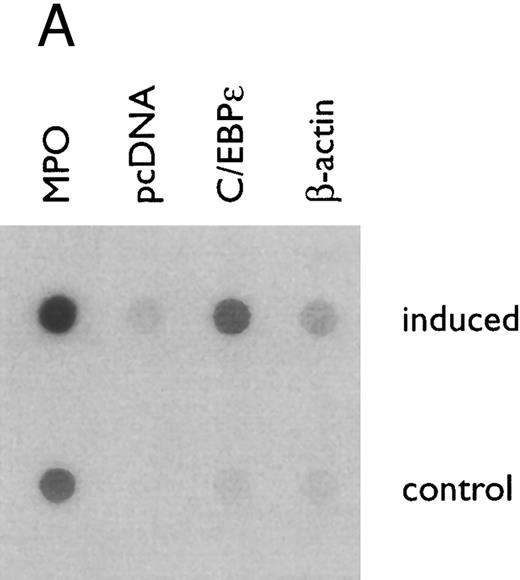
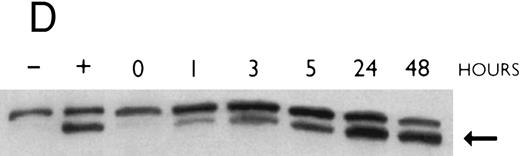
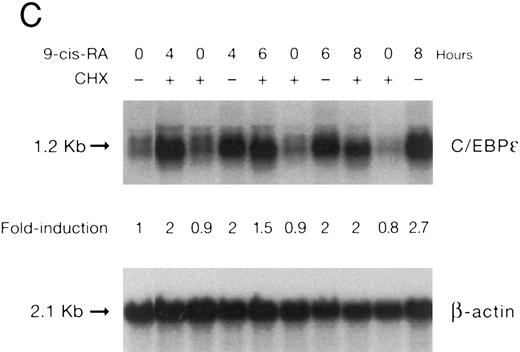
(A) Rate of transcription of C/EBPε in untreated NB4 cells (control) and NB4 cells induced for 12 hours with 9-cis RA (5 × 10−7 mol/L). Nuclei were isolated for run-off experiments as described in the Materials and Methods. Equivalent counts of 32P-labeled RNA transcripts were hybridized to plasmids immobilized on nitrocellulose membranes as indicated (MPO, full-length MPO cDNA in PUC vector; pcDNA, empty pcDNA vector; C/EBPε, full-length C/EBPε antisense cDNA in pcDNA vector; β-actin, 3′ UT β-actin in PBR322 vector). (B) Half-life of mature C/EBPε RNA in NB4 cells. Northern blot of total RNA (30 μg/lane) of NB4 cells exposed for 0 to 4 hours to actinomycin (10 μg/mL) with or without prior treatment with 9-cis RA as indicated. (C) Effect of a protein synthesis inhibitor on C/EBPε RNA expression. Northern blot with total RNA (30 μg/lane) of NB4 cells treated either with 10 μg/mL CHX alone, with CHX (10 μg/mL) and 9-cis RA (5 × 10−7 mol/L), or with 9-cis RA (5 × 10−7 mol/L) alone. Cells were harvested at 0, 4, 6, and 8 hours. The top panel shows the hybridization with full-length C/EBPε cDNA probe. The bottom panel shows the hybridization with β-actin probe as assessment of RNA quantities on each lane. Fold-inductions were calculated by the ratio of densitometry readings of C/EBPε to β-actin. (D) Effect of ATRA on C/EBPε protein expression in NB4 cells. NB4 cells were cultured with ATRA (5 × 10−7 mol/L) for different durations. Lysates were made from the cells and Western blotted. COS cells transfected with either a human C/EBPε cDNA expression vector (lane +) or empty vector (lane −) were used as positive and negative controls, respectively. The arrow denotes C/EBPε protein as seen by Western blot.
(A) Rate of transcription of C/EBPε in untreated NB4 cells (control) and NB4 cells induced for 12 hours with 9-cis RA (5 × 10−7 mol/L). Nuclei were isolated for run-off experiments as described in the Materials and Methods. Equivalent counts of 32P-labeled RNA transcripts were hybridized to plasmids immobilized on nitrocellulose membranes as indicated (MPO, full-length MPO cDNA in PUC vector; pcDNA, empty pcDNA vector; C/EBPε, full-length C/EBPε antisense cDNA in pcDNA vector; β-actin, 3′ UT β-actin in PBR322 vector). (B) Half-life of mature C/EBPε RNA in NB4 cells. Northern blot of total RNA (30 μg/lane) of NB4 cells exposed for 0 to 4 hours to actinomycin (10 μg/mL) with or without prior treatment with 9-cis RA as indicated. (C) Effect of a protein synthesis inhibitor on C/EBPε RNA expression. Northern blot with total RNA (30 μg/lane) of NB4 cells treated either with 10 μg/mL CHX alone, with CHX (10 μg/mL) and 9-cis RA (5 × 10−7 mol/L), or with 9-cis RA (5 × 10−7 mol/L) alone. Cells were harvested at 0, 4, 6, and 8 hours. The top panel shows the hybridization with full-length C/EBPε cDNA probe. The bottom panel shows the hybridization with β-actin probe as assessment of RNA quantities on each lane. Fold-inductions were calculated by the ratio of densitometry readings of C/EBPε to β-actin. (D) Effect of ATRA on C/EBPε protein expression in NB4 cells. NB4 cells were cultured with ATRA (5 × 10−7 mol/L) for different durations. Lysates were made from the cells and Western blotted. COS cells transfected with either a human C/EBPε cDNA expression vector (lane +) or empty vector (lane −) were used as positive and negative controls, respectively. The arrow denotes C/EBPε protein as seen by Western blot.
Preexposure of NB4 cells to 5 × 10−7 mol/L ATRA for 30 minutes potentiated differentiation of NB4 by nonretinoids.26,28 For example, ATRA-priming (5 × 10−7 mol/L) and then the addition of either HMBA (2 mmol/L) or DMSO (1.25%, vol/vol) resulted in greater than 50% and greater than 90% NBT+ and CD11b+ cells, respectively (data not shown). RA-priming alone resulted in 14% NBT+ and 31% CD11b+ cells, respectively. This ATRA-priming alone increased the abundance of C/EBPε fourfold after washing and culturing in regular medium for 3 days as compared with the control (Fig 2). Nonretinoids used alone did not alter the low levels of C/EBPε transcripts. However, cells primed with ATRA and then treated with either HMBA or DMSO, both potent granulocytic-inducing agents,26,29 resulted in a greater than 8- to 10-fold increase in C/EBPε transcript level. In contrast, both TPA and vitamin D3 induced NB4 cells to differentiate down the monocytic pathway.26,29 30 NB4 cells pulsed with ATRA for 30 minutes and then treated with either TPA (5 × 10−8 mol/L) or vit D3 (10−7 mol/L) for 3 days became adherent (>90% and >80%, respectively), as consistent with monocytic differentiation. ATRA-primed cells treated with TPA (5 × 10−8 mol/L) or 1,25(OH)2D3 (10−7 mol/L) for 3 days showed an approximately 10-fold reduction of C/EBPε mRNA levels.
Regulation of C/EBPε mRNA expression in NB4 cells treated with 9-cis RA. To determine if 9-cis RA affected C/EBPε expression at the level of initiation of transcription, nuclei both from NB4 induced with 9-cis RA (5 × 10−7 mol/L) for 12 hours and from untreated NB4 (as control) were isolated and subjected to run-off transcription assays. The rate of transcription of C/EBPε genes in vivo, as shown by the amount of labeled RNA hybridizing to the full-length C/EBPε cDNA on the dot blot (Fig 3A), was increased more than threefold in NB4 cells treated with 9-cis RA than the untreated NB4 control cells, as shown by radioactive counts (normalized with β-actin).
The half-life of C/EBPε mRNA was measured by calculating the decay of RNA accumulation after transcription was blocked with actinomycin D (Fig 3B). Cumulative radioactive counting showed that the half-life of NB4 cells was approximately 2 hours in both the presence and absence of 9-cis RA treatment.
To examine whether C/EBPε mRNA induction by 9-cis RA was dependent on ongoing protein synthesis, NB4 cells were preincubated with cycloheximide (10 μg/mL) for 30 minutes and then treated with 9-cis RA (5 × 10−7 mol/L for 4, 6, and 8 hours). The data show that upregulation of the C/EBPε mRNA by 9-cis RA occurred even in the absence of protein synthesis (Fig 3C).
To examine whether the induction of C/EBPε mRNA by 9-cis RA in NB4 cells is matched by the induction of the C/EBPε protein, NB4 cells were cultured with ATRA (5 × 10−7 mol/L) for different durations. Cells were harvested and lysates were examined by Western blotting. Parallel to the increase of C/EBPε mRNA, the C/EBPε protein levels began to increase by 1 hour (Fig 3D). The maximum induction occurred by 24 hours (>5-fold).
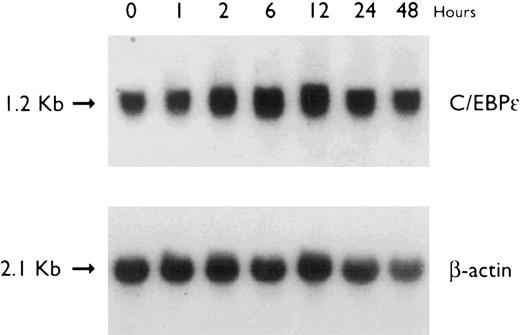
Accumulation of C/EBPε mRNA during induction of differentiation of U937 cells to granulocytes. The U937 cells differentiated along the granulocytic pathway after exposure to ATRA.31 32 Radioactive counting showed that the level of C/EBPε mRNA in U937 cells increased approximately twofold after 48 hours of ATRA-induction (Fig 4).
Regulation of C/EBPε expression in U937 cells by ATRA. Northern blot of total RNA (30 μg/lane) of U937 cells exposed for various lengths of time to ATRA (5 × 10−7 mol/L). The top panel shows the hybridization with full-length C/EBPε cDNA probe, and the bottom panel shows the hybridization with β-actin cDNA probe as assessment of RNA quantities in each lane.
Regulation of C/EBPε expression in U937 cells by ATRA. Northern blot of total RNA (30 μg/lane) of U937 cells exposed for various lengths of time to ATRA (5 × 10−7 mol/L). The top panel shows the hybridization with full-length C/EBPε cDNA probe, and the bottom panel shows the hybridization with β-actin cDNA probe as assessment of RNA quantities in each lane.
Accumulation of C/EBPε mRNA during induction of differentiation of HL-60 cells to granulocytes. The HL-60 cells differentiated along the granulocytic pathway after exposure to DMSO and ATRA.22,33 34 For example, either ATRA (10−7 mol/L for 5 days) or DMSO (1.25% vol/vol for 5 days) induced NBT reduction (>60%) of HL-60 cells as compared with control HL-60 cells (5% NBT+; data not shown). The C/EBPε mRNA level increased approximately twofold by 6 hours of ATRA-induction (Fig 5A). In contrast, C/EBPε mRNA levels decreased after 2 hours of DMSO-induction and then increased to baseline levels after 24 to 72 hours (Fig 5B).
(A) Regulation of C/EBPε expression in HL60 cells by ATRA. Northern blot of total RNA (20 μg/lane) of HL60 cells exposed for various lengths of time to ATRA (5 × 10−7 mol/L). The top panel shows the hybridization with full-length C/EBPε cDNA probe, and the bottom panel displays the hybridization with β-actin cDNA probe as assessment of RNA quantities in each lane. (B) Regulation of C/EBPε expression in HL60 cells by DMSO. Northern blot of total RNA (20 μg/lane) of HL60 cells exposed for various lengths of time to DMSO (1.25%). The top panel shows the hybridization with full-length C/EBPε cDNA probe, and the bottom panel shows the hybridization with β-actin cDNA probe as assessment of RNA quantities in each lane.
(A) Regulation of C/EBPε expression in HL60 cells by ATRA. Northern blot of total RNA (20 μg/lane) of HL60 cells exposed for various lengths of time to ATRA (5 × 10−7 mol/L). The top panel shows the hybridization with full-length C/EBPε cDNA probe, and the bottom panel displays the hybridization with β-actin cDNA probe as assessment of RNA quantities in each lane. (B) Regulation of C/EBPε expression in HL60 cells by DMSO. Northern blot of total RNA (20 μg/lane) of HL60 cells exposed for various lengths of time to DMSO (1.25%). The top panel shows the hybridization with full-length C/EBPε cDNA probe, and the bottom panel shows the hybridization with β-actin cDNA probe as assessment of RNA quantities in each lane.
DISCUSSION
Promyelocytic NB4, HL-60, and myelomonoblastic U937 cells were used to study the regulation of expression of C/EBPε gene during their differentiation. We found that RA treatment increased the abundance of C/EBPε mRNA approximately twofold in HL-60 and U937 cells and approximately fivefold in NB4 cells. Accumulation of C/EBPε mRNA paralleled the increased granulocytic differentiation of the cells. This occurred most prominently in the NB4 cells. These findings paralleled our data33 and the data of others34 using fresh human hematopoietic cells. The early, normal hematopoietic stem cell (CD34+/CD33−) had very weak expression of C/EBPε; the CD34+/CD33+ more mature myeloid stem cell expressed slightly more C/EBPε. As the cells differentiated to promyelocytes, myelocytes, metamyelocytes, and granulocytes, levels of C/EBPε mRNA obtained a plateau elevated level. In contrast, monocyte and macrophages had very low levels of expression; and in erythroid cells, C/EBPε was undetectable. Taken together, these results suggest an important function for C/EBPε at the myeloblast/promyelocyte stage of granulocyte differentiation. Analysis of C/EBPε deletional mice will further help determine the role of C/EBPε in hematopoiesis.
The contribution of transcription and mRNA stability to the increase in steady-state levels of C/EBPε observed in retinoid-stimulated NB4 cells was examined. Stability of the C/EBPε transcripts as assayed by measuring the decay of message in the presence of actinomycin D showed no change during 9-cis RA–mediated differentiation of NB4 cells to granulocytes. Rate of transcription of the C/EBPε gene was also measured from nuclei isolated from both untreated NB4 and NB4 cells induced with 9-cis RA. The retinoid (5 × 10−7 mol/L) increased the rate of transcription of C/EBPε by approximately threefold at 12 hours, consistent with the level of induction of C/EBPε mRNA observed by Northern analysis. Also, ATRA-induction of NB4 cells resulted in a peak fivefold induction of C/EBPε protein at 24 hours, paralleling the increase in the transcript levels. The fact that they stimulated accumulation of C/EBPε mRNA in the absence of new protein synthesis is further evidence to suggest that retinoids have a direct effect on transcription of C/EBPε. The enhancement in accumulation of C/EBPε mRNA by retinoids may not merely be a reflection of granulocytic differentiation, because HL-60 cells cultured with DMSO undergo granulocytic differentiation,35 36 but levels of C/EBPε mRNA at least initially decreased rather than increased. These studies suggest strongly for transcriptional control of C/EBPε via a retinoid acid response element.
Could the prominent induction of expression of C/EBPε mNRA observed in NB4 be unique to this cell type? The cell line NB4 was established from the leukemia cells of a patient with acute promyelocytic leukemia27 and the cells contain the pathognomic chromosomal rearrangement t(15; 17) resulting in the PML-RAR fusion gene.37-39 This PML-RAR fusion product has been suggested to block or interfere with transcription of genes critical to myeloid differentiation, perhaps by sequestering the RARα in nonfunctional locations as shown by a nuclear speckled pattern of the fusion protein.40 High concentrations of retinoic acid allow the RAR fusion protein to assume a more typical RAR nuclear location. It is possible that the retinoid-PML/RAR complex more readily binds and transactivates the C/EBPε gene because retinoids had only a moderate enhancement of C/EBPε in HL-60 and U937 cells, even though the ligand induced granulocytic differentiation of these cells similar to NB4. Another explanation for the prominent effect of retinoids in NB4 could relate to expression of other transcriptional factors that combinatorially interact with the RA/RAR to activate C/EBPε. Furthermore, cells that normally cannot express C/EBPε, such as early erythroblastic K562 cells, cannot be induced by RA to express C/EBPε,18 suggesting that modulation of the gene in the cells requires more than retinoid ligand and its receptor. Further studies using NB4 and HL-60 cell lines that are resistant to retinoids as well as functional analysis of a potential retinoic acid response element in the C/EBPε gene are required.
ACKNOWLEDGMENT
The authors thank Carl W. Miller for his helpful technical advice.
Supported by National Institutes of Health grants and also in part by the Concern Foundation and the Parker Hughes Trust. H.P.K. is a member of the UCLA Jonsson Comprehensive Cancer Center and holds an endowed Mark Goodson Chair of Oncology Research at Cedars-Sinai Medical Center/UCLA School of Medicine. D.Y.C. is a recipient of a Medical Scientist Training Grant (No. GM08042) from the National Institutes of Health.
Address reprint requests to H. Phillip Koeffler, MD, Director, Division of Hematology/Oncology, Professor of Medicine, UCLA, 8700 Beverly Blvd, B-213, Los Angeles, CA 90048.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal