Abstract
Retroviral vectors constitute the most efficient system to deliver and integrate foreign genes into mammalian cells. We have developed a producer cell line that yields high titers of amphotropic retroviral vectors carrying the enhanced green fluorescent protein (EGFP) gene, a codon humanized, red-shifted variant of the green fluorescent protein (GFP) gene, which can be used as a selectable marker. We have used a hybrid vector that has been shown to efficiently drive gene expression in hematopoietic cells. Virtually all murine and human cell lines and primary human hematopoietic cells tested were transduced with varying efficiency after incubation with vector-containing supernatants. Human CD34+ cells obtained from cord blood or aphereses products were transduced using a protocol that involves daily addition of vector-containing supernatants for 6 consecutive days. At day 6, up to 16% of the cells expressed EGFP, as assessed by flow cytometry. Sorted EGFP-expressing cells were able to produce fluorescent hematopoietic colonies. EGFP's main advantages are its fast flow cytometry determination and the possibility of cell sorting and simultaneous evaluation of the transduction efficiency along with other phenotypic markers.
GENE TRANSFER into hematopoietic stem cells is often hampered by the low transduction efficiency and the limitations of the vectors used. Reporter genes have been used extensively for studies on gene expression, gene transfer research, and clinical gene-marking protocols. In recent years, the green fluorescent protein (GFP) from the jellyfish Aequorea victoria has emerged as a very useful tool for these and other purposes.1 GFP is a 238-amino acid polypeptide with a molecular weight of 27 kD. The cDNA encoding GFP was cloned and sequenced in 1992,2 opening the way for expression of the protein in heterologous systems. GFP has been expressed in a variety of species, including bacteria, plants, Drosophila melanogaster, Caenorhabditis elegans, zebra fish, and mammals.3,4 GFP fluorescence is very stable and does not require any substrate or cofactor.5 GFP absorbs blue light (absorption wavelength peak, 395 nm) and emits green light (emission wavelength peak, 508 nm) that is detectable using a fluorescence microscope or a fluorescence-activated cell sorter (FACS).
Enhanced GFP (EGFP) is a recently developed, red-shifted variant of GFP that contains two mutations (S65T and P64L). In addition, EGFP has been re-engineered with more than 190 silent base changes corresponding to human codon-usage preferences.6-8 This GFP variant exhibits higher expression and fluorescence intensity than wild-type (wt) GFP.9 The combined effect of these changes results in an increased sensitivity of up to 350 times over wt GFP when excited at 488 nm.10 The major excitation peak wavelength of the red-shifted variants encompasses the excitation wavelength of commonly used filter sets, so the resulting signal is much brighter than that of the wt GFP. The fluorescent characteristics of EGFP make this system especially attractive for gene expression and gene transfer research. It allows analysis by fluorescence microscopy or flow cytometric techniques in real time11 and cell sorting without additional steps, unlike other marker systems such as drug-resistance genes, which require tedious and time-consuming selection methods and clonogenic assays for the determination of transduction efficiency.
Retroviruses constitute the only vectors that integrate their genes efficiently into the genome of the target cells and can provide long-term expression of the transgenes. For this reason, several groups have attempted to generate stable cell lines producing high titers of retroviral vectors containing the GFP or EGFP genes.12-14 We have developed a retroviral vector carrying a fluorescent protein (EGFP) gene and investigated its use as a reporter and as a selectable marker in cell lines and in primary hematopoietic cells. Here we describe the generation of a stable, amphotropic cell line, PA317/EGFP1, which produces high titers of a retroviral vector containing the EGFP gene. Using supernatants from this cell line, we have transduced a variety of cell lines and primary human hematopoietic cells. We have used a hybrid retroviral vector created and characterized by Baum et al,15 in which cis elements determining transcriptional activity have been designed to overcome transcriptional inefficiency and silencing associated with retroviral gene transfer into myeloid progenitors and hematopoietic stem cells.
MATERIALS AND METHODS
Cell lines and hematopoietic cell culture.K562 (human chronic myelogenous leukemia cell line), HeLa (human cervical carcinoma cell line), J774 A.1 (murine macrophage cell line), and TF-1 (human bone marrow pluripotent CD34+ myeloid progenitor cell line) were obtained from the American Type Culture Collection (Rockville, MD). NIH/3T3 murine fibroblasts, the PA317/LN producer cell line, and the PA317 amphotropic and PE501 echotropic packaging cell lines were kindly provided by A.D. Miller (Fred Hutchinson Cancer Research Center, Seattle, WA). K562, J774 A.1, He-La, and TF-1 cells were grown in RPMI 1640, whereas murine fibroblasts were grown in Dulbecco's Modified Eagle's Medium (DMEM) + 4.5 g/L glucose. All media was supplemented with 10% heat-inactivated fetal calf serum (FCS), 50 IU/mL penicillin, 50 μg/mL streptomycin, and 2 mmol/L L-glutamine. TF-1 cell cultures were also supplemented with granulocyte-macrophage colony-stimulating factor (GM-CSF) at 4 ng/mL.
Human CD34+ cells were cultured at a cell density of 5 × 104 cells/mL in Iscove's modified Eagle's medium (IMEM) supplemented with 12.5% FCS, 12.5% heat-inactivated horse serum (HS), 1 mmol/L sodium pyruvate, 1 μmol/L hydrocortisone, 0.1 mmol/L 2-mercaptoethanol, 50 IU/mL penicillin, 50 μg/mL streptomycin, 2 mmol/L L-glutamine, recombinant human stem cell factor (rhSCF) and rhFlt-3 ligand (both kindly provided by Amgen, Thousand Oaks, CA), and rh interleukin-3 (rhIL-3) and rhIL-6 (both kindly provided by Sandoz, Basel, Switzerland). All cultures were grown in a humidified incubator with 5% CO2 at 37°C.
CD34+ hematopoietic cell samples.Hematopoietic cells were obtained from human cord blood and aphereses samples. CD34+ progenitor cells were positively selected by an indirect immunomagnetic method.16 After selection, CD34+ cells were either resuspended in the previously described medium to be transduced or stored in liquid nitrogen.
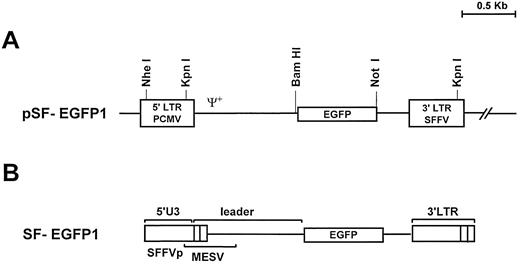
Vector construction.For the retroviral vector construction we isolated the EGFP gene as a 741-bp fragment from the plasmid pEGFP-1 (Clontech Laboratories Inc, Palo Alto, CA) by BamHl and Xba l endonuclease digestion. From plasmid pSF-MDR, kindly provided by C. Baum (Heinrich-Pette-Institut für Experimentelle Virologie und Immunologie an der Universität Hamburg) et al,15 the mdr-1 cDNA was removed by digestion with BamHl and Xba l restriction enzymes. The EGFP insert was cloned into this vector to generate pSF-EGFP (Fig 1A).
(A) Structure of the pSF-EGFP1 retroviral vector. EGFP, enhanced green fluorescent protein; LTR, long terminal repeat; Ψ+ indicates the presence of the retroviral packaging sequence; PCMV, PCC4-cell-passaged mutant virus; SFFVp, polycythemic strain of the spleen focus-forming virus. (B) Proviral form of the pSF-EGFP1 vector following retroviral integration. PBS, primer binding site; MESV, murine embrionic stem cell virus.
(A) Structure of the pSF-EGFP1 retroviral vector. EGFP, enhanced green fluorescent protein; LTR, long terminal repeat; Ψ+ indicates the presence of the retroviral packaging sequence; PCMV, PCC4-cell-passaged mutant virus; SFFVp, polycythemic strain of the spleen focus-forming virus. (B) Proviral form of the pSF-EGFP1 vector following retroviral integration. PBS, primer binding site; MESV, murine embrionic stem cell virus.
Producer cell lines.PA317 packaging cells were transduced with 0.2 μm-filtered supernatants from PE501 cell cultures transfected by calcium phosphate precipitation with 10 μg of the proviral pSF-EGFP plasmid. EGFP expression was analyzed by fluorescence-activated cell sorter (FACS) analysis and, using an autoclone device, single cells were seeded in 96-well cell-culture plates containing 70 μL of a mixture of 50% fresh culture medium and 50% of the cell culture conditioned medium (presorting). The transduction efficiency of about 500 clone supernatants was tested on NIH/3T3 cells by FACS analysis.
To test the integrity of the provirus and the clonality of the producer cell line PA317/EGFP1, genomic DNA was isolated from 107 cells by standard procedures.17 Five micrograms of DNA was digested separately with Nhe I and Kpn I restriction enzymes, electrophoretically resolved in a 0.7% agarose gel, and transferred onto a positively charged nylon membrane. After fixing at 80°C, the membrane was probed with the 0.8-kb BamHl-Xba l fragment from the pEGFP plasmid which was radiolabelled by random priming.
FACS analysis and sorting.All cytometric analyses were done on an EPICS Elite-ESP cytometer (Coulter Electronics, Hialeah, FL) equipped with an argon ion laser tuned at 488 nm. Cells were sorted according to light scatter and fluorescence properties by an autoclone device coupled to the cytometer able to clone a single cell per well on flat-bottom 96-well plates.
Retroviral transduction of cell lines and CD34+ cells.Different volumes of 0.2 μm filtered supernatant were added, with protamine sulfate to a final concentration of 4 μg/mL, to a known number of target cells seeded 24 hours before. Infection of cell lines was performed for 48 hours at 37°C (5% CO2 in humidified air).
Viral titers were estimated indirectly by transducing a known number of NIH/3T3 cells with different volumes of retroviral containing supernatant. We plotted increasing volumes of supernatant (range, 5 to 1,000 μL) against percentage of fluorescent target cells. Transduced cells were analyzed after 48 hours by FACS, and the titer was calculated from the volumes corresponding to the linear slope of the curve according to the following formula:
CD34+ cells were transduced by replacing 90% of their culture medium after centrifugation (1,600g for 5 minutes) with filtered PA317/EGFP1 vector containing supernatants (same medium) daily for 6 consecutive days. CD34+ cells fed with culture supernatants from the PA317/LN cell line were used as a control. Protamine sulfate (4 μg/mL) and the above-mentioned cytokines (50 ng/mL) were added each day. Transduction efficiency was analyzed directly by FACS on 200,000 target cells resuspended in phosphate-buffered saline (PBS) with 1% bovine serum albumin and 0.1% sodium azide.
To test whether the supernatants from the PA317/EGFP1-producing cell line contained helper viruses, we used the marker rescue assay on transduced NIH/3T3 cells passaged for 2 weeks as previously described.18
Colony assays for hematopoietic progenitors.Colony-forming unit cells (CFU-Cs) were grown and analyzed using a commercial kit that contains methylcellulose and a mixture of recombinant cytokines including rhSCF, rhIL-3, rhIL-6, rh erythropoietin (rhEpo), rhG-CSF, and rhGM-CSF (MethoCult GF H4434; StemCell Technologies Inc, Vancouver, BC, Canada). Five hundred to 7,000 sorted cells were seeded in each well in a volume of 250 μL using 4-well multidish plates (cat. no. 176740; Nunc, Roskilde, Denmark) and grown in triplicate. Colonies were counted and analyzed after 10 days of incubation.
Fluorescence microscopy.Transduced human and murine hematopoietic cell lines were grown on appropriate media and 5 × 105 cells were laid on poly-L-lysine–coated slides by cytocentrifugation. Adherent cells were grown to confluence on chamber slides. For detection, slides were mounted in 20 μL of antifade solution (Vectashield; Vector Laboratories, Burlingame, CA) containing 150 ng/mL of DAPI. Cell preparations were visualized under a fluorescence microscope (Vanox AH-3; Olympus Optical Co, Tokyo, Japan) equipped with the appropriate filter set. Hematopoietic colonies were analyzed under an inverted fluorescence microscope (1 × 70; Olympus).
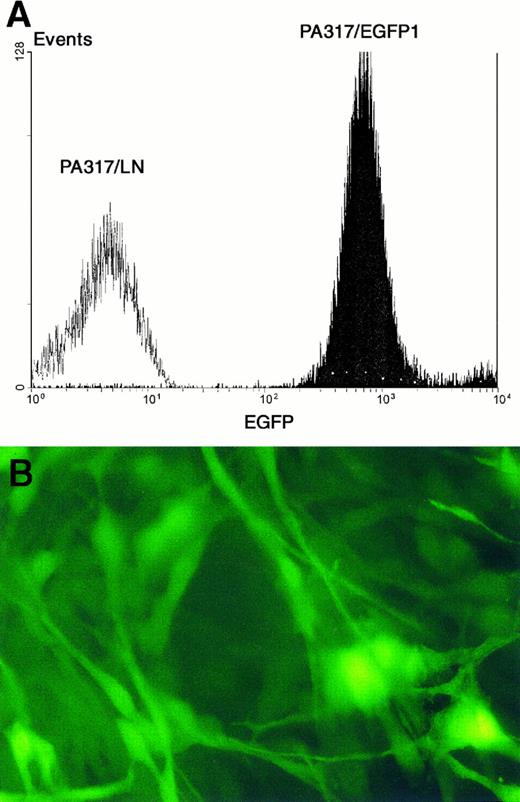
(A) FACS analysis of PA317/EGFP1 vector producing cells. PA317/LN cells (carrying the neomycin resistance gene) were used as a control. Fluorescence intensity is more than 100 times above the control cells. (B) Picture shows PA317/EGFP1 cells under a fluorescence microscope (400× optical magnification).
(A) FACS analysis of PA317/EGFP1 vector producing cells. PA317/LN cells (carrying the neomycin resistance gene) were used as a control. Fluorescence intensity is more than 100 times above the control cells. (B) Picture shows PA317/EGFP1 cells under a fluorescence microscope (400× optical magnification).
Polymerase chain reaction analysis.CFU-Cs grown in semisolid medium were individually picked and lysed as described by Fruehauf et al.19 The PCR was performed on 10-μL lysates in a final volume of 50 μL containing 2 U of Taq polymerase (Boehringer Mannheim GmbH, Mannheim, Germany), 1X reaction buffer supplied with the enzyme, 200 μmol/L of each dNTP, 1.25 mmol/L MgCl2 , and 10 pmol of each primer. Sequences of the 20-mer amplimers used in the amplification of EGFP were: 5′GCCACAAGTTCAGCGTGTCC3′ and 5′AGCTCGATGCGGTTCACCAG3′, which amplify a 304-bp DNA fragment. A β-actin sequence of 838 bp was amplified simultaneously as a control for the DNA yield from 10 μL of each lysate in a separate tube with primers: 5′ATCTGGCACCACACCTTCTACATTGAGCTGCG3′ and 5′CGTCATACTCCTGCTTGCTGATCCACATCTGC3′ (Clontech, Palo Alto, CA). Amplification conditions were as follows: 95°C for 5 minutes, then 40 cycles of 95°C for 45 seconds, 60°C for 45 seconds, 72°C for 2 minutes, followed by extension at 72°C for 10 minutes. Ten microliters of each PCR product was separated by electrophoresis on a 1.5% agarose gel and visualized in UV light after ethidium-bromide staining.
The viral titer of the PA317/EGFP1 cell line was also estimated by semi-quantitative PCR. Vector copy number per cell standards were prepared by diluting PA317/EGFP1 cells, which contain a single copy of the provirus, with PA317/LN cells (100%, 50%, 25%, 5%, and 0% of PA317/EGFP1 cells). DNA was extracted from the cell mixtures and used for the PCR reaction. One microgram of genomic DNA from both NIH/3T3 cells transduced with different volumes of viral supernatant and from the standards was amplified for 24 cycles, under the above-described conditions, except that 1 U of Taq polymerase was used. Duplex PCR for EGFP and the β chain of the murine platelet-derived growth factor receptor (PDGF-R) sequences was performed. Amplification of the PDGF β receptor DNA with the specific primers 5′CATTGGCTCCATCCTGCATA3′ and 5′GGATAAGCCTCGAACACCAC3′20 was used to normalize EGFP PCR products. Twenty microliters of the amplified products was separated on a 1.5% agarose gel, visualized under UV light after ethidium bromide staining, and quantified by densitometric analysis of the negatives.
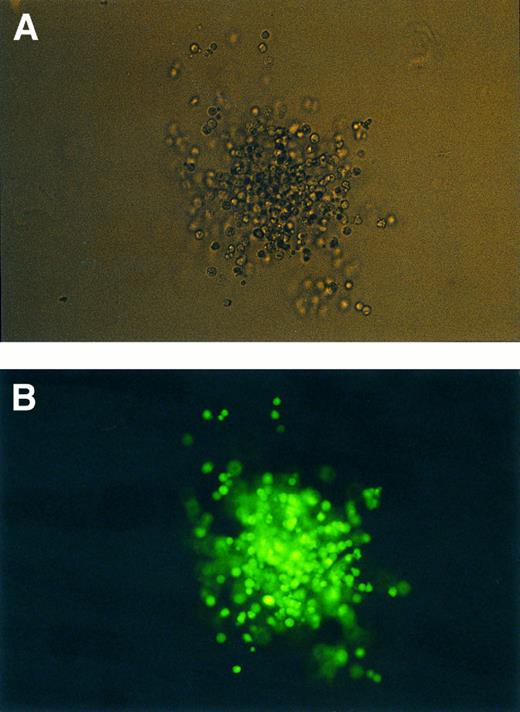
Transduced CFU-GM under normal (A) and UV light (B) (400× optical magnification).
Transduced CFU-GM under normal (A) and UV light (B) (400× optical magnification).
RESULTS
Selection of producer cell lines.Single EGFP transduced PA317 cells expressing high and intermediate levels of green fluorescence were sorted on 96-well culture plates. Out of 500 supernatants tested, the one exhibiting the highest viral titer was selected according to its ability to transduce NIH/3T3 cells. This clone, named PA317/EGFP1, as well as all the other clones obtained with high vector titers, were derived from the cell population expressing intermediate fluorescence levels (data not shown). Interestingly, most supernatants from clones expressing higher levels of fluorescence failed to infect NIH/3T3 cells. The PA317/EGFP1 cell line grows normally, and EGFP expression has been stable for more than 3 months (Fig 2). Its viral titer, as calculated by the flow cytometric method described above, is about 2 × 106 infectious particles/mL.
Analysis of the provirus integrity in the PA317/EGFP1 cell line by genomic DNA digestion with Kpn I followed by Southern blot showed a single band of the expected size (2.5 kb), which indicates that the provirus was not rearranged (data not shown). In addition, clonality was demonstrated by the presence of a single band detected after Nhe I genomic DNA digestion and Southern blot (data not shown).
Transduction efficiency on cell lines.To investigate the range of infection of the PA317/EGFP1 vector and its transduction capability, we infected a variety of murine and human hematopoietic and nonhematopoietic cell lines (Table 1). In all cases the transduced cells could be distinguished from the nontransduced cells by their fluorescence level.
Transduction Efficiency and Peak Fluorescence Ratio of Different Cell Lines Transduced With PA317/EGFP1 Retroviral Vectors
| Cell Line . | Transduction Efficiency (%)* . | Peak Fluorescence Ratio† . |
|---|---|---|
| NIH/3T3 | 73 | 560 |
| HeLa | 9 | 48 |
| J744 A.1 | 65 | 64 |
| K562 | 16 | 489 |
| TF-1 | 7 | 374 |
| Cell Line . | Transduction Efficiency (%)* . | Peak Fluorescence Ratio† . |
|---|---|---|
| NIH/3T3 | 73 | 560 |
| HeLa | 9 | 48 |
| J744 A.1 | 65 | 64 |
| K562 | 16 | 489 |
| TF-1 | 7 | 374 |
All transductions were made by seeding 4 × 104 cells on 1.6-cm culture dishes for 24 hours and then incubating them with 100 μL of vector-containing supernatant and 4 μg/mL protamine sulphate for 48 hours more.
The ratio obtained by dividing the peak fluorescence channel of the transduced cells by the control cell peak channel.
To validate the transduction efficiency as determined by FACS analysis, we analyzed the samples of transduced NIH/3T3 cells used for titer determination both by flow cytometry and semiquantitative PCR. For this purpose, a standard curve with different proportions of PA317/EGFP1 and PA317/LN cells was made (see Materials and Methods). Transduction efficiency by PCR on our transduced cell populations was estimated by comparing the optical density of the PCR bands with those obtained from the standards. Transduction efficiency as determined by both methods showed good agreement (data not shown).
Transduced cell lines have been maintained in culture to assess the stability of EGFP expression. They grow as fast as nontransduced cells, with the proportion and intensity of fluorescence of the transduced cells stable for at least 8 weeks.
Supernatants from transduced NIH/3T3 cells passaged for 2 weeks were not able to infect NIH/3T3 cells, indicating the absence of helper virus.
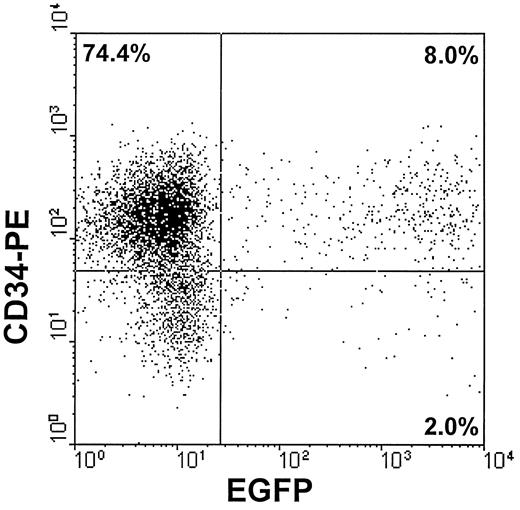
Transduction of CD34+ cells.Human CD34+ cells transduced with vector-containing supernatants were evaluated by FACS analysis. On day 4, after 4 cycles of infection, up to 9.2% of the cells showed green fluorescence (mean of five experiments, 7.4%; range, 3.1% to 9.2%). The fluorescent peak of these cells was 100 to 1,000 times higher than the peak of the nontransduced cells, allowing simultaneous evaluation of the transduction efficiency and CD34 expression by FACS analysis (Fig 3).
A representative FACS analysis of human cord blood hematopoietic cells after transduction with PA317/EGFP1 supernatants for 4 consecutive days. Cells were stained at day 4 with phycoerythrin-labeled anti-CD34 monoclonal antibody (Clone 8G12; Becton Dickinson, San Jose, CA). The dot plot represents CD34 versus EGFP expression in the target cells. Percentages of analyzed events are shown for each cell subpopulation.
A representative FACS analysis of human cord blood hematopoietic cells after transduction with PA317/EGFP1 supernatants for 4 consecutive days. Cells were stained at day 4 with phycoerythrin-labeled anti-CD34 monoclonal antibody (Clone 8G12; Becton Dickinson, San Jose, CA). The dot plot represents CD34 versus EGFP expression in the target cells. Percentages of analyzed events are shown for each cell subpopulation.
Transduction efficiency on CFU-Cs was also analyzed by direct microscopic observation and in individual colonies by PCR from a particular experiment which yielded 8.1% of green fluorescent cells by FACS analysis at day 6. CFU-Cs from three different populations of cells sorted at day 6 were analyzed (Fig 4). The overall population, a second one gated on positivity for CD34 and green fluorescent events, and a third one gated on positivity for CD34 but not for green fluorescence. Table 2 shows the results in these three cell populations.
Clonogenic Assays of Human CD34+ Cells and PCR Results
| Type of Cell Sorted . | Green Fluorescent . | PCR (+) CFU-Cs . |
|---|---|---|
| . | CFU-Cs*/Total CFU-Cs . | . |
| Overall cell population | 4/57 (7%) | 2/40 (5%) |
| CD34+/green fluorescence (+) | 83/118 (70.3%) | 3/4 (75%) |
| CD34+/green fluorescence (−) | 1/78 (1.28%) | 0/20 (0%) |
| Type of Cell Sorted . | Green Fluorescent . | PCR (+) CFU-Cs . |
|---|---|---|
| . | CFU-Cs*/Total CFU-Cs . | . |
| Overall cell population | 4/57 (7%) | 2/40 (5%) |
| CD34+/green fluorescence (+) | 83/118 (70.3%) | 3/4 (75%) |
| CD34+/green fluorescence (−) | 1/78 (1.28%) | 0/20 (0%) |
CFU-Cs observed under a fluorescent microscopy.
DISCUSSION
One of the most promising strategies to overcome the low transduction efficiency of retroviral vectors in the hematopoietic system in vivo is the selection of transduced cells. For this purpose, selectable genes must be transduced along with the therapeutic genes. Competition by nontransduced cells for the homing sites in the bone marrow can be eliminated by infusing only the selected transduced cells, ensuring better engraftment of such cells. Strategies used to select the transduced cells can be classified broadly into either metabolic or cell-surface mediated estrategies.21 Fluorescent markers are becoming very powerful tools in gene transfer studies.3 22 GFP or EGFP based systems do not require additional gene products, cofactors or substrates, whereas most current genetic marker systems do.
Because the objective of our work was to achieve efficient gene transfer and expression in hematopoietic cells, we chose the SFFVp/MESV hybrid vector, which has been selected to drive increased transgene expression into myeloid progenitors and stem cells.15 We have cloned the EGFP cDNA in the retroviral pSF plasmid backbone (Fig 1A). Due to the mechanism of reverse transcription, the integrated provirus contains the leader sequence of the murine embryonic stem cell virus (MESV) and a modified LTR of the SFFVp in its 5′ end (Fig 1B). This LTR has been re-engineered to avoid recombination of the provirus and minimize the risk of producing helper viruses. The presence of both retroviral cis elements allows efficient and long-term EGFP expression in the myeloid compartment.15
We have obtained a vector-producing cell line (PA317/EGFP1) yielding a high-titer, amphotropic retroviral vector containing the EGFP gene. Using this vector we have been able to efficiently transduce murine and human hematopoietic and nonhematopoietic cell lines as well as primary human CD34+ cells. The viral titer of the producer cell line (about 2 × 106 infectious particles/mL) has been stable for several months. Analysis by flow cytometry showed that transduced cells expressing EGFP exhibit a fluorescent peak 100 to 1,000 times higher than the nonexpressing cells peak, allowing a clear threshold that facilitates FACS multianalysis and cell sorting. In general, hematopoietic cell lines showed a higher fluorescence level than the nonhematopoietic cell lines, with the exception of NIH/3T3 cells, indicating an increased level of EGFP expression, as expected by the relative higher tissue-specific activity of the promoter used.15
An important issue is whether all transduced cells express high enough levels of EGFP to be detectable by FACS or fluorescent microscopy. To address this question we have analyzed samples of a transduced cell line both by FACS and PCR. Comparison of the results suggests that transduction efficiency as determined by both methods is similar. In addition, the level of transduction of hematopoietic colonies as determined by direct fluorescent microscopic observation was similar to that detected by PCR. Altogether, these results indicate that most transduced cells express the transgene and can be detected by flow cytometry.
Previous attempts to generate retroviral vectors containing GFPs have had only partial success. First experiments with wt GFP failed,23 probably because of its poor quantum efficiency, its short emission wavelength, and its low expression level in mammalian cells. Once the newer human codon adjusted and mutant GFP variants became available, several investigators reported positive results with retroviral vectors.12-14 As far as we know, three stable producing cell lines have been reported in the literature. Two of them used the LNCX vector, containing the neomycin resistance (neo) gene driven by the retroviral LTR, and two different variants of a humanized, red-shifted GFP gene driven by the cytomegalovirus promoter.12,14 The third one used a fusion gene containing the hygromycin resistance gene and the GFPS65T gene,13 which contains a single amino acid mutation. Supernatants of these three cell lines conferred fluorescence to the target cells, but a neomycin or hygromycin selection process was needed to allow an easy detection of the transduced cells, indicating that viral titers were probably low.
Some investigators have suggested that expression of high levels of a red-shifted GFP can be toxic for the cells.24 However, EGFP could overcome this problem because it provides a higher protein expression level and has a lower threshold for detection than previous versions of GFP (30 nmol/L v 1 μmol/L of wt GFP in HeLa cells),10 which can account for the reduction in the number of EGFP molecules the cell needs to fluoresce in comparison with previous versions of GFP. On the other hand, the existence of viable transgenic mice expressing EGFP25 indicates that expression of this protein does not produce significant toxicity. In this regard, green fluorescent CD34+ cells were able to generate a variety of fluorescent colonies (CFU-E, CFU-Mix, and CFU-GM), and had similar clonogenic ability than nonfluorescent CD34+ cells (data not shown). Failure to detect 100% of the colonies generated by the double-positive sorted cells using fluorescent microscopy and PCR may be related to the sorting purity (about 80%, data not shown) and to changes in gene expression associated with differenciation of the CD34+ cells.
A hypothetical disadvantage of this system, the putative immunogenicity of GFP or EGFP for in vivo applications, has not yet been evaluated. Multiparameter flow cytometry sorting on EGFP-transduced cells could become a very valuable tool for future gene therapy protocols.
ACKNOWLEDGMENT
The authors thank Gregorio Martı́n-Henao and Gemma Capmany for providing the selected CD34+ cells; Adela Miralles for assistance with cell cryopreservation; Maria Julià for assistance with the molecular techniques; Pura Muñoz, Michael Lynch, Anna Bigas, and, very specially, to Prof Donnall Thomas, Dottie Thomas, and Dusty Miller from the Fred Hutchinson Cancer Research Center (Seattle, WA) for their critical reviews.
Supported by Grants No. CICYT SAF96-0130, FIS 94/0019-05, and DGICYT PB93-1011 and by Conselleria de Sanitat de la Generalitat de Catalunya, Spain. T.P. is a recipient of a PhD grant from CIRIT no. 1997FI 00828.
Address reprint requests to Jordi Barquinero, MD, Department of Cryobiology and Cell Therapy, Institut de Recerca Oncològica, Gran Via Km 2.7 L'Hospitalet 08907, Barcelona, Spain.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal