Abstract
Previously, we found the first congenital deficiency of histidine-rich glycoprotein (HRG) in a Japanese woman with thrombosis. To elucidate the genetic basis of this deficiency, we first performed Southern blot analysis and found no gross deletion or insertion in the proband's HRG gene. We then examined the nucleotide sequences of all seven exons of the proband's HRG gene. A single nucleotide substitution, G to A at nucleotide position 429, which mutates Gly85 to Glu in the first cystatin-like domain, was found in exon 3 in 13 of 22 amplified clones. This mutation generates a unique Taq I site. Exon 3 was amplified from the proband, her family members, and 50 unrelated normal Japanese individuals, and Taq I fragmentation was examined. Fragmentation of exon 3 was observed in one allele of the genes from the proband and the family members who also have decreased plasma levels of HRG. Fifty unrelated normal Japanese individuals had a normal HRG gene, indicating that the G to A mutation is not a common polymorphism. To elucidate the identified mutation as a cause for the secretion defect of HRG in the proband's plasma, we constructed and transiently expressed the recombinant Tokushima-type HRG mutant (Gly85 to Glu) in baby hamster kidney (BHK) cells, and examined an intracellular event of the mutant protein. The results showed that only about 20% of the Tokushima-type HRG was secreted into the culture medium, and intracellular degradation of the mutant was observed. Thus, the present study strongly suggests that the HRG deficiency is caused by intracellular degradation of the Gly85 to Glu mutant of HRG in the proband.
HUMAN HISTIDINE-RICH glycoprotein (HRG) is a single-chain glycoprotein (Mr 67,000) composed of 507 amino acid residues that circulates in blood at a relatively high concentration (∼100 mg/L).1-3 The HRG gene spans approximately 11 kb on chromosome 3q28-q29 and consists of seven exons and six introns.4,5 Platelets store HRG in the α-granules and secrete HRG upon thrombin activation.3 HRG has an unusually high content of proline and histidine residues in the carboxy-terminal region.2 By structural similarity, it belongs to the cystatin superfamily, eg, cystatin SN, SA, and C and high–molecular weight kininogen.4 The histidine-rich region of HRG is homologous to the corresponding area of high molecular weight kininogen, a modulator in the contact activation system of blood coagulation.2
HRG has been shown to interact with plasma proteins involved in blood coagulation and fibrinolysis such as fibrinogen,6 factor XIIIa,7 plasminogen,8 and heparin.9It also interacts with several other blood components: heme,10 metal ions,10 activated platelets,11 thrombospondin,12 T lymphocytes,13 and several complement factors.14 HRG diminishes heparin activity in plasma; removal of HRG from the plasma increased the heparin effect on thrombin inhibition by antithrombin9 and activated protein C inhibition by protein C inhibitor.15 However, HRG is reported not to interfere with interactions between antithrombin and heparan-sulfate on vascular endothelial cells.16 Although HRG interacts with plasminogen, it probably has no function in the fibrinolytic system, because a high or low concentration of HRG did not affect plasminogen activation in the plasma milieu.17 Thus, the physiologic significance of these molecular interactions of HRG remains unclear.18
In 1993, we discovered the first case of familial deficiency of HRG in a 43-year-old Japanese woman with cerebral sinus thrombosis.19 Laboratory tests for platelet count and procoagulant activity were normal. The concentrations of various coagulation factors and fibrinolytic agents were also in the normal range in the patient's plasma, except for HRG. The patient had only 21% of the normal level of HRG, and the affected family members also had low levels of HRG. Recently, Souto et al20 have discovered a second family with this deficiency. The patient developed pulmonary embolism on two occasions at age 36. Her father had thrombosis in the central artery of the retina at age 59. These two family cases suggest that HRG plays an antithrombotic role to prevent thrombotic disorders.
To elucidate the molecular and cellular mechanism for the first case of congenital HRG deficiency, which we refer to as “HRG Tokushima,” we identified a single Gly to Glu mutation in the proband's HRG gene and examined the secretion of the recombinant Tokushima-type HRG mutant using transiently transfected baby hamster kidney (BHK) cells and compared it against the wild-type HRG.
MATERIALS AND METHODS
Subject.
Clinical and laboratory data relevant to the congenital HRG deficiency in this report have been previously presented.19 In brief, the proband was a 43-year-old Japanese woman with right transverse sinus thrombosis while taking contraceptive medication. Her plasma HRG level was only 21% of the normal level of 109.5% ± 51.5% (mean ± 2 SD). The HRG concentration in her plasma determined on four occasions over 6 months was consistently low. She showed no clinical signs of liver function abnormality or sepsis. Low levels of plasma HRG were also found in her aunt, uncle, and two daughters. These results suggest that congenital HRG deficiency is inherited in this family.
Enzymes and reagents.
All restriction enzymes and the Wizard Plus Minipreps DNA Purification System were purchased from Promega (Madison, WI). T7 Taq DNA polymerase was obtained from Boehringer Mannheim (Indianapolis, IN). The Ultrapure dNTP set and Sequenase Version 2 Deaza-dCTP kit were purchased from US Biomedical (Cleveland, OH). X-gal and agarose were obtained from Life Technologies Inc (Gaithersburg, MD). The GeneClean II kit was obtained from Bio 101 (La Jolla, CA). The TA cloning kit including the pCR 2.1 vector and INVαF′ Escherichia coli cells was purchased from Invitrogen (San Diego, CA). The Multiprime DNA-labeling kit, deoxycytidine 5′-[α-32P]triphosphate ([α-32P]dCTP), deoxyadenosine 5′-[α-35S]triphosphate ([α-35S]dATP), and Hybond-N transfer membrane were obtained from Amersham Life Science (Arlington Heights, IL). ZMB3 expression vector was kindly provided by Dr Don Foster (ZymoGenetics, Seattle, WA).
DNA isolation and Southern blot analysis.
Blood was drawn from the patient or donors after obtaining informed consent according to the Declaration of Helsinki. Genomic DNA was isolated from leukocytes by a standard technique.21 A plasmid containing the full-length cDNA for human HRG, λ HHRG3,2 was digested with EcoRI. Approximately 2.0-kb EcoRI fragments were isolated using the GeneClean II kit and radiolabeled with [α-32P]dCTP. Ten micrograms of genomic DNA was digested with either EcoRI, HincII,Pst I, or Xba I, and the fragments were isolated by electrophoresis on a 0.7% agarose gel. The DNA fragments were then transferred to a Hybond-N membrane and hybridized with the32P-labeled 2.0-kb EcoRI fragments according to a standard procedure. The membrane was exposed to Kodak X-Omat AR film (Eastman Kodak, Rochester, NY) at −80°C with an intensifying screen.
Polymerase chain reaction and DNA sequence analysis.
Oligonucleotide primers were synthesized based on the intron sequences approximately 50 bp apart from the intron/exon boundaries of the HRG gene (Wakabayashi S, Koide T, submitted) using the 380B DNA Synthesizer (Applied Biosystems, Foster City, CA). Exon 7 was divided into five segments (7-1 to 7-5) separately amplified using the primers synthesized similarly as before. The sequences of all primers used for polymerase chain reaction (PCR) are listed in Table1. For TaqI fragmentation of exon 3, pair 12 primers were used for amplification. The PCR mixtures contained 1X PCR buffer (10 mmol/L Tris hydrochloride, pH 8.4, 50 mmol/L KCl, 1.5 mmol/L MgCl2, and 0.01% gelatin), 1 μg genomic DNA, and 2 U Taq polymerase. Amplification was performed as follows: denaturation at 94°C for 1 minute, annealing at 55°C for 1 minute, and extension reaction at 72°C for 1.5 minutes for 35 cycles. The amplified DNA fragments were directly subcloned into pCR 2.1 vector supplied with the TA Cloning System kit. The DNA sequence was determined by the dideoxy chain-termination method using T7 DNA polymerase and [α-35S]dATP.
PCR Primers for Amplification of Exons in the HRG Gene
| Primer . | Region . | Direction . | Sequence . | Length of Product (bp) . |
|---|---|---|---|---|
| Pair 1 | Exon 1 | Upstream | 5′-CGGAATTCTCTGCAGTGGCAGATC-3′ | 256 |
| Downstream | 5′-CGGGATCCTCTGCCATTGGCAATTCC-3′ | |||
| Pair 2 | Exon 2 | Upstream | 5′-CGGAATTCACATGTTGCTTTTGGC-3′ | 184 |
| Downstream | 5′-CGGGATCCAGAGTGAAGGTGCCCTC-3′ | |||
| Pair 3 | Exon 3 | Upstream | 5′-CGGAATTCCTCAAGAGCCTCTTTC-3′ | 149 |
| Downstream | 5′-CGGGATCCAATTTAACAGATTACC-3′ | |||
| Pair 4 | Exon 4 | Upstream | 5′-CGGAATTCACTGCTATCTTCTTTC-3′ | 224 |
| Downstream | 5′-CGGGATCCGAACCTTAGTGGAGAC-3′ | |||
| Pair 5 | Exon 5 | Upstream | 5′-CGGAATTCTTGAAACTATTTGATCCC-3′ | 155 |
| Downstream | 5′-CGGGATCCAACGATCACAGACATTTC-3′ | |||
| Pair 6 | Exon 6 | Upstream | 5′-CGGAATTCTAACAGCTCCTCATTCC-3′ | 169 |
| Downstream | 5′-CGGGATCCATGACAAAGTCTGCTTAGAC-3′ | |||
| Pair 7 | Exon 7-1 | Upstream | 5′-GATGATAGGCACTTTTCTGTGACC-3′ | 239 |
| Downstream | 5′-AGGGCCTTGTGGAAGTGGGGGTCC-3′ | |||
| Pair 8 | Exon 7-2 | Upstream | 5′-CCTCCTCCAGATGAAAGAGATCAC-3′ | 243 |
| Downstream | 5′-CTGTCTATGGGTATCATGTTCATG-3′ | |||
| Pair 9 | Exon 7-3 | Upstream | 5′-CACGGACACCATCCCCATGCACAC-3′ | 213 |
| Downstream | 5′-GCCGTGGCCATGGCAACAGTGACC-3′ | |||
| Pair 10 | Exon 7-4 | Upstream | 5′-CCTTGTGACCCACCACCCCATAAC-3′ | 201 |
| Downstream | 5′-CAATGGGAAGCTGGGAAAATTGGC-3′ | |||
| Pair 11 | Exon 7-5 | Upstream | 5′-AAAAGGTGAGGTGCTGCCACTTCC-3′ | 211 |
| Downstream | 5′-CATTTTCCTCTTCAAAGGAATCAC-3′ | |||
| Pair 12 | Exon 3 | Upstream | 5′-AGTAGGAGGATGTTGCATCAC-3′ | 283 |
| Downstream | 5′-CATAGATGAGACAAATACCAC-3′ |
| Primer . | Region . | Direction . | Sequence . | Length of Product (bp) . |
|---|---|---|---|---|
| Pair 1 | Exon 1 | Upstream | 5′-CGGAATTCTCTGCAGTGGCAGATC-3′ | 256 |
| Downstream | 5′-CGGGATCCTCTGCCATTGGCAATTCC-3′ | |||
| Pair 2 | Exon 2 | Upstream | 5′-CGGAATTCACATGTTGCTTTTGGC-3′ | 184 |
| Downstream | 5′-CGGGATCCAGAGTGAAGGTGCCCTC-3′ | |||
| Pair 3 | Exon 3 | Upstream | 5′-CGGAATTCCTCAAGAGCCTCTTTC-3′ | 149 |
| Downstream | 5′-CGGGATCCAATTTAACAGATTACC-3′ | |||
| Pair 4 | Exon 4 | Upstream | 5′-CGGAATTCACTGCTATCTTCTTTC-3′ | 224 |
| Downstream | 5′-CGGGATCCGAACCTTAGTGGAGAC-3′ | |||
| Pair 5 | Exon 5 | Upstream | 5′-CGGAATTCTTGAAACTATTTGATCCC-3′ | 155 |
| Downstream | 5′-CGGGATCCAACGATCACAGACATTTC-3′ | |||
| Pair 6 | Exon 6 | Upstream | 5′-CGGAATTCTAACAGCTCCTCATTCC-3′ | 169 |
| Downstream | 5′-CGGGATCCATGACAAAGTCTGCTTAGAC-3′ | |||
| Pair 7 | Exon 7-1 | Upstream | 5′-GATGATAGGCACTTTTCTGTGACC-3′ | 239 |
| Downstream | 5′-AGGGCCTTGTGGAAGTGGGGGTCC-3′ | |||
| Pair 8 | Exon 7-2 | Upstream | 5′-CCTCCTCCAGATGAAAGAGATCAC-3′ | 243 |
| Downstream | 5′-CTGTCTATGGGTATCATGTTCATG-3′ | |||
| Pair 9 | Exon 7-3 | Upstream | 5′-CACGGACACCATCCCCATGCACAC-3′ | 213 |
| Downstream | 5′-GCCGTGGCCATGGCAACAGTGACC-3′ | |||
| Pair 10 | Exon 7-4 | Upstream | 5′-CCTTGTGACCCACCACCCCATAAC-3′ | 201 |
| Downstream | 5′-CAATGGGAAGCTGGGAAAATTGGC-3′ | |||
| Pair 11 | Exon 7-5 | Upstream | 5′-AAAAGGTGAGGTGCTGCCACTTCC-3′ | 211 |
| Downstream | 5′-CATTTTCCTCTTCAAAGGAATCAC-3′ | |||
| Pair 12 | Exon 3 | Upstream | 5′-AGTAGGAGGATGTTGCATCAC-3′ | 283 |
| Downstream | 5′-CATAGATGAGACAAATACCAC-3′ |
Construction of expression vectors.
A region of nucleotides 118 to 2067 of human HRG cDNA (HHRG3)2 was used for the expression experiment. Nucleotide 118 is just four bases upstream from the initiation codon ATG, and this cDNA was subcloned in pUC19 plasmid. A BamHI site was introduced into this DNA at the 3′ end by PCR using a primer, 5′-TTGGATCCCTCTTCTCAGGC-3′, complementary to nucleotides 1843 to 1862 in which two bases were replaced. The amplified fragment was digested with EcoRI and BamHI and inserted into ZMB3 expression vector.22 To avoid incorporation of an unexpected mutation, the EcoRI/Bal I fragment (nucleotides 118 to 1411) was replaced by that of the original clone. The 3′ region from theBal I site to the BamHI site was sequenced to verify the absence of mutation. The cDNA carrying the mutation found in the proband was amplified with the above-mentioned primer and the mutated primer, 5′-GTGATCGAACAATGTAAGGT-3′. This PCR product was used as one of the primers for successive second-round PCR. The other was a universal −40 primer. The whole cDNA fragment carrying the mutation thus amplified was digested with EcoRI and HincII, and subcloned into pUC19 plasmid to verify incorporation of the mutation without any other unexpected replacements. The whole cDNA was then constructed on pUC19 plasmid, released by EcoRI andBamHI, and ligated into ZMB3 vector.
Cell culture and pulse-chase experiments.
To examine secretion of the Tokushima-type HRG mutant, pulse-chase experiments were performed using transiently transfected BHK cells as described previously.23 Briefly, about 5 × 104 cells in 35-mm dishes were transfected with an expression plasmid (7.5 μg) by the calcium phosphate method. After transfection, cells were cultured under a 3% CO2atmosphere for 16 hours, transferred to 5% CO2, and further cultured for 24 hours. Then, the cells were incubated in the medium without Met and Cys for 30 minutes, labeled with 35 μCi EXPRE35S35S (NEN-DuPont, Tokyo, Japan) per dish for 1 hour, washed with phosphate-buffered saline, and chased with Dulbecco's modified Eagle's medium/10% fetal calf serum containing 2 mmol/L Met. At selected time intervals, the medium was harvested and cells were lysed. The labeled HRG was immunoprecipitated using affinity-purified rabbit anti-human HRG IgG and Staphylosorb (Mercian, Tokyo, Japan). After washing, immunoadsorbed proteins were dissociated by heating at 85°C for 5 minutes, and then electrophoresed on 8% polyacrylamide gel in the presence of sodium dodecyl sulfate (SDS) and β-mercaptoethanol.24 After fixing the gels, radioactivity on dried gels was measured quantitatively by the Fujix BAS2000 Bio-Imaging Analyzer system (Fuji Photo Film, Tokyo, Japan). Autoradiographs were made by exposure to Kodak XAR films at −80°C.
RESULTS
Southern blot analysis.
To investigate if any gross structural alternation is present in the HRG gene of the proband, the genomic DNA digested with EcoRI,HincII, Pst I, or Xba I was subjected to Southern blot analysis as described earlier. The digestion pattern of the proband's DNA was indistinguishable from that of normal individuals (data not shown), indicating that neither gross deletion nor gross insertion is present in the proband's HRG gene.
DNA sequencing.
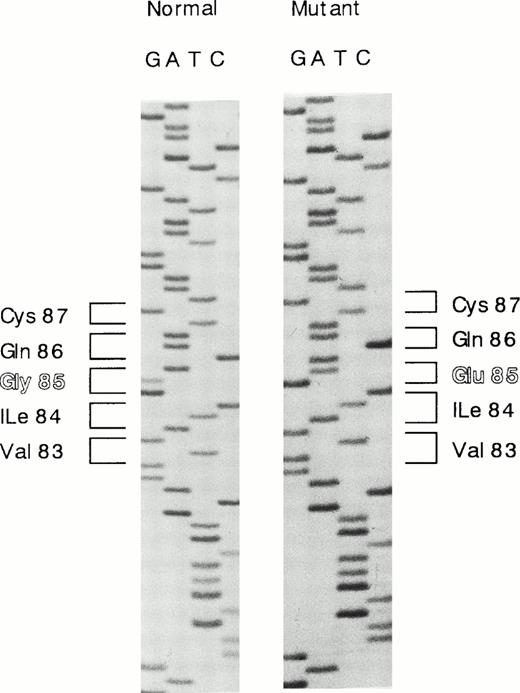
The results of Southern blot analysis indicated that a small structural alternation or a point mutation(s) occurs in the proband's HRG gene. We then examined the nucleotide sequences of all seven exons of the proband's DNA. Each exon was amplified by PCR, subcloned into pCR2.1 vector, and transfected into INVΙNVαF′ cells. At least 10 independent clones were isolated for each exon and sequenced. One single-base substitution was found in exon 3, and no substitution was found in the other exons. The amino acid residues of five polymorphic sites shown by Hennis et al25,26 were identified as Ile162, Pro186, His322, Arg430, and Asn475, all of which agree with those originally reported by Koide et al,2 and no polymorphisms were found in the proband's HRG gene. A single G to A substitution at nucleotide position 429, which mutates Gly85 to Glu in the first cystatin-like domain, was found in 13 of 22 different clones analyzed, and nine clones contained the normal sequence (Fig1). These results indicate that the proband carries a heterozygous deficiency.
Nucleotide sequence showing the G to A substitution in exon 3 of the HRG gene. G to A substitution at nucleotide position 429 mutates Gly85 to Glu in the first cystatin-like domain.
Nucleotide sequence showing the G to A substitution in exon 3 of the HRG gene. G to A substitution at nucleotide position 429 mutates Gly85 to Glu in the first cystatin-like domain.
Taq I digestion of exon 3.
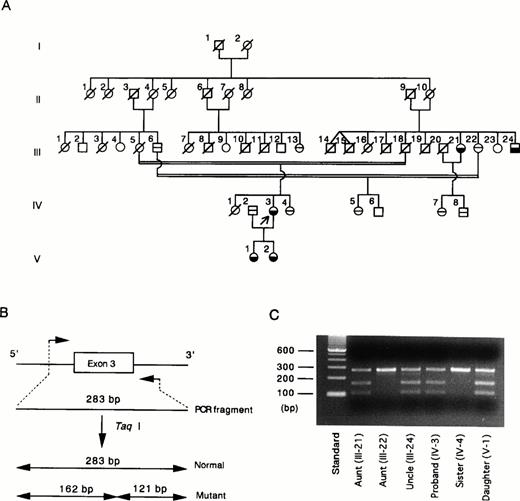
The G429 to A mutation creates a unique Taq I restriction site (TCGA) in exon 3. Taq I digestion facilitates detection of the G to A mutation in exon 3. We studied Taq I cleavage of exon 3 of the proband (Fig 2A, IV-3), her family members, and 50 normal Japanese individuals. The normal exon 3 is composed of 283 bp, whereas the G to A–mutated exon 3 yields two fragments, 162 and 121 bp, after digestion with Taq I (Fig 2B). As predicted, the proband's gene exhibited a composite digestion pattern indicating that the mutation occurs in one allele of the HRG gene (Fig 2C). To confirm the genetic basis for the phenotype observed in this family, we studied Taq I digestion of exon 3 amplified from other available family members. The aunt (III-21), uncle (III-24), and daughter (V-1) have been found to have approximately half the normal plasma level of HRG. These individuals had the same TaqI digestion pattern of exon 3 as observed for the proband (Fig 2C). A younger sister (IV-4) and another aunt (III-22) had a normal plasma level of HRG, and Taq I did not cleave exon 3 amplified from these individuals. A third uncle (III-6), a parent's cousin (III-13), and two of the proband's cousins (IV-5 and IV-7) with normal levels of HRG also had normal exon 3 (data not shown). These results clearly show that the G429 to A substitution found in exon 3 of the proband's gene is inherited in this family. We then studied the Taq I digestion with 50 unrelated healthy Japanese individuals and did not find any mutations in exon 3 in the population. This study eliminates the possibility that this mutation is a common polymorphism.
(A) Pedigree of the family with congenital HRG deficiency.19 Arrow, the proband; £ {, affected subjects with reduced HRG levels; ⊖, subjects with normal HRG levels; □ ∅︀, deceased family members; □ ○, unexplored subjects. (B) PCR-Taq I digestion analysis of normal and mutated exon 3. Normal exon 3 is composed of 283 bp and is not cleaved by Taq I. Mutated exon 3 with 1 Taq I restriction site (TCGA) is cleaved by the enzyme to form 2 fragments, 162 bp and 121 bp. (C) For each family member, exon 3 amplified by PCR was digested with Taq I and subsequently analyzed by electrophoresis on a 2% agarose gel. Lane 1, molecular weight standard; lanes 2 to 7, Taq I digests of exon 3 derived from the proband and 5 of her family members. {/CAPT;;;left;stack}
(A) Pedigree of the family with congenital HRG deficiency.19 Arrow, the proband; £ {, affected subjects with reduced HRG levels; ⊖, subjects with normal HRG levels; □ ∅︀, deceased family members; □ ○, unexplored subjects. (B) PCR-Taq I digestion analysis of normal and mutated exon 3. Normal exon 3 is composed of 283 bp and is not cleaved by Taq I. Mutated exon 3 with 1 Taq I restriction site (TCGA) is cleaved by the enzyme to form 2 fragments, 162 bp and 121 bp. (C) For each family member, exon 3 amplified by PCR was digested with Taq I and subsequently analyzed by electrophoresis on a 2% agarose gel. Lane 1, molecular weight standard; lanes 2 to 7, Taq I digests of exon 3 derived from the proband and 5 of her family members. {/CAPT;;;left;stack}
Secretion of wild-type and Tokushima-type HRG in transiently transfected BHK cells.
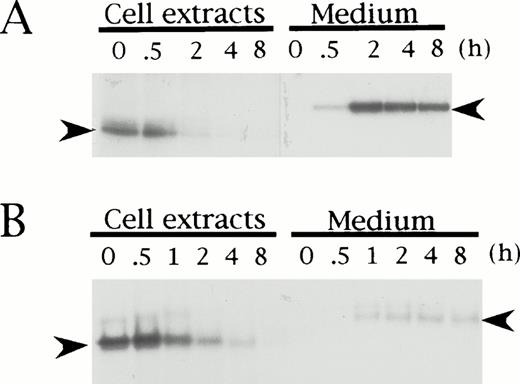
To elucidate if the mutation in HRG Tokushima is responsible for the secretion defect of the mutant protein in the proband, we performed pulse-chase experiments of Tokushima-type HRG using transiently transfected BHK cells, and compared the results with those obtained for wild-type HRG. Both wild-type and Tokushima-type HRG were detected as a band of 62 kD (Fig 3, left arrowhead) and 72 kD (right arrowhead) in the cell extracts and culture medium, respectively, and they were synthesized at nearly equal levels in BHK cells as detected in cell extracts at time 0 of the chase. During the chase period, wild-type HRG was secreted into the culture medium, and essentially no HRG band was detected in the 2-hour chased cell extracts (Fig 3A). However, the total amount of radioactivity was maintained during the chase period. On the other hand, only a small amount (20%) of Tokushima-type HRG was secreted into the culture medium even after an 8-hour chase, and a decrease in total radioactivity was observed, suggesting intracellular degradation of the mutant HRG (Fig 3B). The time to 50% disappearance of total radioactivity from the cell extracts was estimated to be 1 hour.
Pulse-chase analysis of recombinant wild-type (A) and Tokushima-type (B) HRG expressed in BHK cells. Transiently transfected BHK cells were pulse-labeled for 1 hour and chased for 0, 0.5, 1, 2, 4, and 8 hours. Labeled HRG in cell extracts and culture medium was immunoprecipitated and analyzed on SDS-PAGE. Left arrowheads indicate HRG bands within cells, and right arrowheads indicate HRG bands secreted into culture medium.
Pulse-chase analysis of recombinant wild-type (A) and Tokushima-type (B) HRG expressed in BHK cells. Transiently transfected BHK cells were pulse-labeled for 1 hour and chased for 0, 0.5, 1, 2, 4, and 8 hours. Labeled HRG in cell extracts and culture medium was immunoprecipitated and analyzed on SDS-PAGE. Left arrowheads indicate HRG bands within cells, and right arrowheads indicate HRG bands secreted into culture medium.
DISCUSSION
In this study, we showed that a single missense mutation (nucleotide G429 to A in exon 3) leading to a mutation of Gly85 to Glu in the first cystatin-like domain on the HRG molecule is a genetic cause for the first case of congenital HRG deficiency. This is the first report on the molecular and cellular basis of congenital HRG deficiency.
Hennis et al25,26 discovered five amino acid polymorphisms in HRG in Dutch families. These mutations occur in both alleles: Ile162 to Thr, Pro186 to Ser, His322 to Arg, Arg430 to Cys, and Asn475 to Ile. The Pro to Ser mutation changes both the mature HRG mass and the plasma HRG concentration. The mutated Ser generates one additional carbohydrate chain in HRG at a newly created carbohydrate-attaching motif, resulting in a higher–molecular mass HRG (77 kD) than the normal protein (75 kD on SDS–polyacrylamide gel electrophoresis [PAGE]). The mean plasma HRG level was highest (156%) in individuals homozygous for Ser and lowest (93%) in homozygotes for Pro, whereas it was intermediate (121%) in heterozygotes. Hennis et al also reported that a dinucleotide polymorphism is located in the intron between the last two exons of the HRG gene5 and is associated with a higher level of plasma HRG in a Dutch family.27
To elucidate if the Gly85 to Glu mutation identified in HRG Tokushima is a cause of congenital I HRG deficiency, we expressed the mutant HRG in transiently transfected BHK cells and examined the secretion rate and intracellular events. The results showed that only about 20% of total Tokushima-type HRG was secreted into the culture medium and most of the molecules (80%) were degraded within the cells, while essentially all of the wild-type HRG was secreted within a 2-hour chase. Thus, the low plasma levels of HRG observed in the proband and the affected family members are most likely the result of intracellular degradation of the mutant HRG.
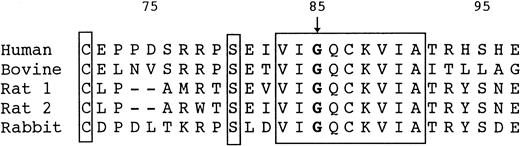
Gly85 and the surrounding sequence in HRG are strictly conserved among species, including human,2 bovine,28rabbit,29 and rat 1 and 2 (Wakabayashi S, et al, manuscript in preparation; Fig 4). This may suggest that Gly85 is indispensable for correct folding of HRG. In general, glycine is known to be one of the conservative amino acids and is important to support the correct conformation of proteins.30 The glycine located in the turning points of the backbone of polypeptide chains is irreplaceable by any other amino acids. Therefore, it is conceivable that the Gly85 to Glu mutation causes a misfolding of HRG, leading to the degradation. Intracellular degradation of genetic mutants of plasma proteins causing a type I deficiency has been demonstrated for α1-antitrypsin Z-variant31,32 and null-variants33 and the antithrombin ΔGlu (deletion of Glu313) mutant.34 The sites for intracellular degradation of such misfolded proteins have been shown to be the endoplasmic reticulum to cis-Golgi compartment35-37 or the cytosol through the ubiquitin-proteasome pathway.38-40 To confirm this, cellular experiments are under way.
Amino acid sequences surrounding Gly85 of human, bovine, rat, and rabbit HRG. Residues that are identical in all 5 proteins are blocked. Numbering starts at the N-terminus of human HRG.2
Amino acid sequences surrounding Gly85 of human, bovine, rat, and rabbit HRG. Residues that are identical in all 5 proteins are blocked. Numbering starts at the N-terminus of human HRG.2
ACKNOWLEDGMENT
The authors are grateful to Dr E.W. Davie for support throughout this study, Dr D.W. Chung for valuable advice, K. Takeshima for technical help, J. Harris for synthesis of oligonucleotides, and Dr Don Foster for supplying ZMB3 expression vector.
Supported by a Grant-in-Aid for Scientific Research (06671097) and a Grant-in-Aid for Scientific Research on Priority Areas (Intracellular Proteolysis) from the Ministry of Education, Science, Sports, and Culture of Japan, and Research Grant No. HL16919 from the US National Institutes of Health.
Address reprint requests to Toshio Shigekiyo, MD, First Department of Internal Medicine, School of Medicine, The University of Tokushima, 3-18-15 Kuramoto, Tokushima 770, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal