Abstract
Interactions between P-selectin and P-selectin glycoprotein ligand-1 (PSGL-1) mediate the earliest “rolling” of leukocytes on the lumenal surface of endothelial cells at sites of inflammation. Previously, PSGL-1 has been shown to be the primary mediator of interactions between neutrophils and P-selectin, but studies on the ability of PSGL-1 to mediate interactions between P-selectin and other subsets of leukocytes have yielded variable and conflicting results. A novel IgG monoclonal antibody (MoAb) to human PSGL-1 was generated, and the specificity of this MoAb was confirmed by both flow cytometric analysis and Western blotting of cells transfected with human PSGL-1. This newly developed MoAb, KPL1, inhibited interactions between P-selectin expressing COS cells and either HL60 cells, neutrophils, or lymphocytes. Furthermore, KPL1 completely inhibited interactions between P-selectin and either purified CD4 T cells or neutrophils in a flow assay under physiological conditions, but had no effect on interactions of T cells or neutrophils with E-selectin. In addition, KPL1 blocked interactions between lymphoid cells transfected with L-selectin and COS cells expressing PSGL-1. The KPL1 epitope was mapped to a site within a consensus tyrosine sulfation motif of PSGL-1, previously shown to be essential for interaction with P-selectin and now shown to be essential for interaction with L-selectin, and to be distinct from the epitope identified by the PL1 function blocking anti-PSGL-1 MoAb. Two-color flow cytometry of normal leukocytes showed that while natural killer (NK) cells (CD16+), monocytes, CD4 and CD8 T cells, and α/β and γ/δ T cells were uniformly positive for PSGL-1, B cells expressed low levels of the KPL1 epitope. This low level of KPL1 staining was also observed immunohistologically in germinal centers, which had no detectable KPL1 staining, whereas T-cell areas (interfollicular region) were positive for KPL1. Interestingly, plasma cells in situ and interleukin-6–dependent myeloma cell lines were KPL1+. Thus, PSGL-1 is expressed on essentially all blood neutrophils, NK cells, B cells, T cells, and monocytes. Variation in tyrosine sulfation during B-cell differentiation may affect the ability of B cells to interact with P- and L-selectin.
THE SELECTIN FAMILY of adhesion molecules mediates the initial rolling of leukocytes on lumenal surfaces of vascular endothelium.1 L-selectin is constitutively expressed on all blood neutrophils, monocytes, the majority of T and B cells, eosinophils, and most bone marrow cells. E-selectin in expressed on endothelium in response to inflammatory mediators such as interleukin-1 (IL-1), tumor necrosis factor (TNF-α), or bacterial lipopolysaccharide (LPS). P-selectin is stored in α granules of platelets and in Weibel-Palade bodies of endothelial cells and is rapidly transported to the cell surface after stimulation with thrombin, histamine, or phorbol esters. All three selectins are structurally similar, with their C-terminal lectin domain playing the major role in recognition of and adhesion to their sialylated, fucosylated, and/or sulfated carbohydrate ligands.
A glycoprotein ligand for P-selectin (PSGL-1) has been identified and cloned.2,3 PSGL-1 is a mucinlike disulfide-linked homodimer consisting of two identical 120-kD glycoprotein chains. The sequence of human PSGL-1 contains a cleavage site for paired basic amino acid converting enzymes (PACE).3 Three potential tyrosine sulfation sites are located in a consensus sequence just downstream of the PACE cleavage site, followed by 15 decamer repeats high in proline, serine, and threonine. The extracellular portion of the molecule contains three potential N-linked glycosylation sites. The remaining C-terminal sequence consists of a single transmembrane spanning domain followed by a 69-residue cytoplasmic tail. PSGL-1 has numerous sialylated fucosylated O-linked oligosaccharides branches, many of which terminate in the sialyl Lewis x (sLex) epitope.4-6 In addition to fucose and sialic acid, interactions between PSGL-1 and P-selectin require at least one tyrosine sulfate located in the amino terminal consensus sequence.7-9
PSGL-1 is expressed by essentially all blood leukocytes including neutrophils, monocytes, and lymphocytes, and has been shown to mediate the rolling of human neutrophils on P-selectin.10 In addition, PSGL-1 can serve as a ligand for L-selectin to mediate neutrophil-neutrophil interactions.11,12 However, data describing the function and expression of PSGL-1 on peripheral T and B cells are conflicting.13-17 Although some of these disparate observations may be related to expression of nonfunctional PSGL-1 by the majority of T cells, some of these discrepancies may be related to the properties of different anti–PSGL-1 reagents or differences in posttranslational modifications characteristic of different cell lineages.
To approach these questions, a new monoclonal antibody (MoAb) to human PSGL-1, KPL1, was generated. This MoAb inhibited interactions between PSGL-1 and P-selectin and between PSGL-1 and L-selectin, but had no effect on leukocyte binding to E-selectin. The KPL1 epitope was mapped to the tyrosine sulfation consensus motif of PSGL-1. Using two-color flow cytometry, all T cells (CD4+, CD8+, γ/δ and α/β), monocytes, and natural killer (NK) (CD16+) cells were shown to express high levels of PSGL-1, whereas B cells expressed lower levels of the KPL1 epitope compared with other leukocyte subsets.
MATERIALS AND METHODS
Production of MoAbs to PSGL-1.
A cDNA for human PSGL-110 (kindly provided by Henri Lichenstein, Amgen Inc, Thousand Oaks, CA) was subcloned into a modified SRα vector18 containing a neoRcassette, and used to transfect the murine 300.19 pre B-cell line by electroporation. Transfectants were selected in medium containing 2 mg/mL G418 (geneticin; GIBCO-BRL, Grand Island, NY). Transfected cells expressing PSGL-1 were initially identified by flow cytometry with PL2, an MoAb which recognizes human PSGL-1 (kindly provided by Kevin Moore, Department of Medicine, University of Oklahoma Health Sciences Center), and subcloned by limiting dilution. Balb/c mice were tolerized with untransfected 300.19 cells followed 4 hours later by an intraperitoneal (IP) injection of 5 μg cyclophosphamide (Neosar; Pharmacia Inc, Columbus, OH). Starting 7 days later, mice were repeatedly immunized IP with 300.19/PSGL-1 cells. Three days after receiving an intravenous boost, a standard cell fusion procedure was performed, followed by selection in hypoxanthine, aminopterin, thymidine (HAT)-containing medium. Culture supernatants were screened via flow cytometry for staining of 300.19/PSGL-1 transfectants but not untransfected 300.19 cells, using goat anti-mouse IgG-FITC (Biosource, Camarillo, CA) as a second stage. Hybridomas were subcloned twice at 0.5 cells/well. After the second round of subcloning, one clone (KPL1) was rescreened by flow cytometry on 300.19 cells, 300.19/PSGL-1 transfectants, HL60 cells, and freshly isolated human neutrophils and lymphocytes. Ascites was generated from the KPL1 clone, and antibodies were purified using the Affi-Gel Protein A MAPS II kit (Bio-Rad Laboratories, Hercules, CA). Conjugation of KPL1 to FITC (fluorescein isothiocyanate; Sigma Chemical Co, St Louis, MO) and biotin (N-hydroxysuccinimido-biotin; Sigma Chemical Co) were carried out by standard procedures.
Flow cytometry.
For one-color analysis, 0.5 × 106 cells were incubated in 100 μL of phosphate-buffered saline (PBS)/1% fetal calf serum (FCS)/NaN3 containing pretitered amounts of the indicated PSGL-1 MoAb, washed, and incubated in goat anti-mouse IgG-FITC, diluted 1:100. For two-color flow cytometry, 0.5 × 106 cells were incubated in 100 μL of PBS/1% FCS/NaN3 containing anti-PSGL-1 MoAb conjugated to FITC or biotin plus a leukocyte subset specific antibody conjugated to either biotin or FITC. After washing, cells were stained with Streptavidin-PE (Phycoerythrin; Fischer Scientific Co, Pittsburgh, PA). Antileukocyte subset antibodies included MoAb directed against CD4, CD8, CD3, CD16, CD19, and CD14. MoAbs against T-cell receptor (TCR)-γ/δ and TCR-α/β (WT31) were obtained from Becton Dickinson (San Jose, CA). Samples were analyzed on a Becton Dickinson FACScan and data were collected on a total of 10,000 to 20,000 light scatter gated events. Data were analyzed with CELLQuest software (Becton Dickinson) and both fluorescence histograms and two-color contour plots were displayed on a 4-decade logarithmic scale.
Western blotting.
HL60, 300.19, 300.19/PSGL-1 cells, or COS cells transiently transfected with either human PSGL-1 cDNA or a control plasmid were washed twice in RPMI. HL60, 300.19, and 300.19/PSGL-1 cells were resuspended at 2 × 107 cells/mL in lysis buffer (1% Triton-X-100 [Sigma; St Louis, MO]; 150 mmol/L NaCl; 10 mmol/L Tris-HCl, pH 7.6; 1 mmol/L CaCl2; 1 mmol/L MgCl2; 1 mmol/L aprotinin; 1 mmol/L phenylmethylsulfonyl fluoride (PMSF); 1 mmol/L leupeptin; and 1 mmol/L pepstatin A) and incubated on ice for 30 minutes. COS cells were resuspended at 2 × 106 cells/mL in lysis buffer containing 2% Triton-X-100 on ice for 30 minutes. Samples were clarified by centrifugation at 14,000g for 30 minutes at 4°C, and supernatants were transferred to fresh tubes. For certain experiments, aliquots of lysates were treated overnight at 37°C with 1 U of Aerobacter aerogenes arylsulfatase (Sigma). Samples were boiled for 5 minutes in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer with or without 6.5% 2-mercaptoethanol, electrophoresed on a 6% polyacrylamide gel, and transferred to nitrocellulose. The nitrocellulose was blocked with 2% gelatin (Bio-Rad) in Tris-buffered saline-Tween (TBS-T; 20 mmol/L Tris HCl, pH 7.6; 137 mmol/L NaCl; and 0.1% Tween-20) for 2 hours at room temperature. Blots were probed with KPL1 or PL2 in TBS-T plus 2% gelatin for 30 minutes, washed five times in TBS-T, and then incubated with goat anti-mouse IgG conjugated to horseradish peroxidase (Biosource) for 30 minutes. After five washes in TBS-T, blots were visualized by chemiluminescence generated after the addition of enhanced chemiluminescence (Amersham, Arlington Heights, IL) and exposed to Hyperfilm (Amersham).
Generation and expression of full-length FFFE/PSGL-1 cDNA.
The plasmid pED.FFFE.148.Fc9 was kindly provided by Gray Shaw (Genetics Institute, Cambridge, MA). This chimeric protein contains the extracellular 148 amino acids of PSGL-1 fused to the heavy chain CH2-CH3 region of IgG, and replaces the three tyrosines at positions 5, 7, and 10 of PSGL-1 with phenylalanine and the aspartic acid at position 11 with glutamic acid.9 A 423-bp Xma I/Stu I insert containing the FFFE mutation was ligated into the full-length PSGL-1 cDNA in pBluescript, and the full-length FFFE/PSGL-1 was inserted into a modified SRα vector containing a neoR cassette. Approximately 15 μg of the FFFE or wild-type PSGL-1 were used to transiently transfect 1.5 × 106 300.19 cells by electroporation.
Neutrophil, peripheral blood mononuclear cell (PBMC), and CD4+ T-cell purification.
Human neutrophils were isolated from heparinized blood by dextran sedimentation, Ficoll density gradient centrifugation (Histopaque-1077; Sigma), and hypotonic lysis of the neutrophil-rich pellet. PBMCs were isolated from heparinized whole blood by dextran sedimentation followed by Ficoll density gradient centrifugation. CD4 T cells were purified from single donor human platelet pheresis residues by sequential density gradient centrifugation and elutriation followed by culture overnight and positive selection on Dynabeads (model M-450 CD4) and Dynal DETACHaBEAD (Dynal, Great Neck, NY) as described.16
Low shear force COS cell adhesion assay.
This assay was performed as previously described.19-22Briefly, COS cells were transfected with either P-selectin or E-selectin cDNA by the DEAE-dextran method in 100-mm tissue culture grade petri dishes. For binding of 300.19/L-selectin (300.19/L) or 300.19/P-selectin (300.19/P) cells,19 23 COS cells were cotransfected with plasmids encoding PSGL-1 or FFFE/PSGL-1, FucT-VII, and C2GnT. The following day, COS cells were replated on 35-mm dishes (assay plates), and allowed to readhere overnight. The next day, HL60 cells, freshly purified human neutrophils, or PBMC were washed twice in RPMI 1640 and resuspended at 2 × 106 cells in a total volume of 100 μL containing saturating amounts of either KPL1 ascites or control antibody and placed on ice for 15 minutes. Alternatively, for 300.19/L or 300.19/P cells binding to transfected COS cells, the COS cells were incubated with saturating amounts of KPL1 or control ascites. After cells had been washed and resuspended in 0.6 mL of RPMI, each petri dish was washed three times with unsupplemented RPMI 1640, followed by the addition of appropriate cells, and incubated on a constantly rocking platform for 15 minutes at 4°C. The plates were washed five times with RPMI 1640 followed by fixation with cold 0.37% formaldehyde/RPMI 1640. Mean number of cells bound per COS cell was determined by counting the number of cells bound/COS cell on ∼125 COS cells in multiple 40× fields using a standard inverted light microscope.
Parallel plate flow chamber adhesion assay.
The flow chamber apparatus used in these studies has been described previously.15,24,25 Chinese hamster ovary (CHO)/P-selectin or CHO/E-selectin monolayers were grown to confluence on glass coverslips and inserted into the flow chamber. A defined flow level of 1.8 dynes/cm2 was obtained by drawing media containing the desired cell population through the chamber using a syringe pump. Purified CD4+ T cells or neutrophils, 1 × 106, were assayed in each experiment. The flow chamber was mounted on an inverted microscope (Nikon Diaphot; Melville, NY) and each 5-minute perfusion period was recorded on videotape by a video camera and video cassette recorder. Leukocyte adhesion was determined as previously described.15,24,25 In some experiments, monolayers of CHO/P cells were preincubated with HDPG2/3, a blocking anti–P-selectin antibody3 at 10 μg/mL for 30 minutes at 37°C. Monolayers of CHO/E were incubated with HEL3/2, a blocking anti–E-selectin antibody.26 W6/32 was used as an isotype control for both monolayers. In other experiments, cells (1 × 106) were incubated with either KPL1 ascites or control ascites at a 1:200 dilution for 15 minutes at 4°C before use in assays.
Immunohistology.
Immunohistology was performed using an indirect biotin-streptavidin method.27 Incubation with primary antibody was followed by incubation with a biotinylated goat anti-mouse second stage antibody (Jackson Immunoresearch Laboratories, Inc, West Grove, PA) followed by peroxidase-conjugated streptavidin (Jackson Immunoresearch Laboratories). Diaminobenzedene was used as a substrate for the horseradish peroxidase. Frozen sections were fixed in acetone for 10 minutes at 4°C before staining. Formalin-fixed deparaffinized sections were stained without pretreatment or microwaved for 15 minutes in 0.01 mol/L citrate buffer at pH 6.0 before staining.
RESULTS
Development and characterization of a new MoAb to human PSGL-1.
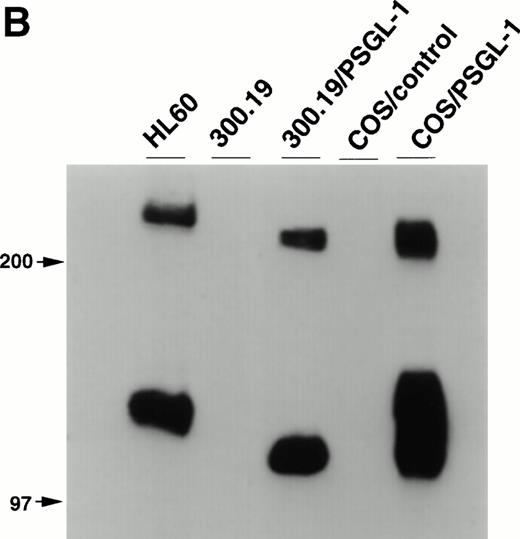
Balb/c mice were immunized with 300.19/PSGL-1 cells as described in Materials and Methods. After routine immunization and fusion protocols, hybridoma supernatants were screened by flow cytometry for negative staining on untransfected 300.19 cells and positive staining on 300.19/PSGL-1 cells (Fig 1A). One MoAb, KPL1, was identified. The KPL1 MoAb stained the 300.19/PSGL-1 cells at high levels but did not stain untransfected 300.19 cells (Fig 1A). To further confirm the specificity of KPL1, Western blot analysis of cells known to express PSGL-1 or transfected with PSGL-1 cDNA was performed. PSGL-1 is a homodimer of ∼240 to 250 kD under nonreducing conditions and ∼120 kD under reducing conditions.2 10 PSGL-1 is difficult to completely reduce, allowing visualization of both reduced and nonreduced forms of PSGL-1. KPL1 reacted with bands of ∼120 and ∼240 kD in lysates from HL60 cells (a myeloid cell line known to express PSGL-1), 300.19/PSGL-1 transfectants, or COS/PSGL-1 transfectants, but not in lysates from untransfected 300.19 cells or COS cells transfected with a control plasmid (Fig 1B). The slight differences in molecular weight between PSGL-1 expressed by HL60 and the two transfectants were most likely caused by differences in glycosylation in these different cell types. Finally, KPL1 stained HL60 cells, freshly purified neutrophils, and peripheral blood lymphocytes by flow cytometry (see below).
Specificity of KPL1 for PSGL-1 by flow cytometry and Western blotting. (A) Untransfected 300.19 cells or 300.19 cells transfected with human PSGL-1 cDNA were stained with a negative control (top) or KPL1 (bottom) as described in Materials and Methods. KPL1 does not stain untransfected 300.19 cells, whereas it stains 300.19/PSGL-1 transfectants. (B) Western blotting of whole cell lysates made from HL60 cells (lane 1), 300.19 cells (lane 2), 300.19 cells transfected with human PSGL-1 cDNA (lane 3), COS cells transfected with a control plasmid (lane 4), or COS cells transfected with human PSGL-1 cDNA (lane 5) with the KPL1 antibody. Bands of the appropriate molecular weight (∼120 and ∼240 KD) were seen only in lysates from cells expressing PSGL-1 endogenously (HL60 cells in lane 1) or transfected with human PSGL-1 cDNA (lanes 3 and 5).
Specificity of KPL1 for PSGL-1 by flow cytometry and Western blotting. (A) Untransfected 300.19 cells or 300.19 cells transfected with human PSGL-1 cDNA were stained with a negative control (top) or KPL1 (bottom) as described in Materials and Methods. KPL1 does not stain untransfected 300.19 cells, whereas it stains 300.19/PSGL-1 transfectants. (B) Western blotting of whole cell lysates made from HL60 cells (lane 1), 300.19 cells (lane 2), 300.19 cells transfected with human PSGL-1 cDNA (lane 3), COS cells transfected with a control plasmid (lane 4), or COS cells transfected with human PSGL-1 cDNA (lane 5) with the KPL1 antibody. Bands of the appropriate molecular weight (∼120 and ∼240 KD) were seen only in lysates from cells expressing PSGL-1 endogenously (HL60 cells in lane 1) or transfected with human PSGL-1 cDNA (lanes 3 and 5).
KPL1 recognizes a unique epitope within the tyrosine sulfation motif of PSGL-1.
Because of apparent differences in staining results with different anti-PSGL-1 antibodies (see Introduction and below), the epitope defined by KPL1 was explored in some detail. Neuraminidase treatment of HL60 cells did not affect expression of the KPL1 epitope, but removed all surface sLex carbohydrates as measured by lack of HECA452 staining (data not shown). In addition, KPL1 stained cells regardless of the presence (neutrophils, HL60 cells) or absence (300.19/PSGL-1 cells) of either FucT-VII or C2GnT (Fig 1 and data not shown; by RT-PCR,21,22 28 300.19 cells do not detectably express mRNA for either FucT-VII or C2GnT). Thus, the KPL1 epitope appears to be independent of carbohydrate modifications, including terminal sialic acid residues, branched O-linked glycans, or fucosylation.
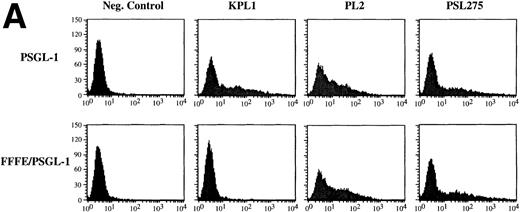
We have shown elsewhere that KPL1 fails to recognize a PSGL-1 mutant in which amino acid residues 5-11 (YEYLDYD), comprising a consensus tyrosine sulfation motif,29 have been deleted.30 To more precisely evaluate the potential contribution of residues comprising the tyrosine sulfate motif to the KPL-1 epitope, we analyzed binding of KPL1 to a PSGL-1 mutant in which all three tyrosines were simultaneously replaced by phenylalanines (FFFE9). 300.19 cells were transiently transfected with cDNA encoding wild-type PSGL-1 or the FFFE mutant, and analyzed by flow cytometry. Replacement of these three tyrosines at positions 5, 7, and 10 with phenylalanine resulted in complete loss of the KPL1 epitope (Fig 2A). However, epitopes recognized by two other anti–PSGL-1 antibodies were still present (Fig 2A), confirming surface expression of the mutated form of PSGL-1. Thus, KPL1 interacts with the tyrosine sulfation motif of PSGL-1.
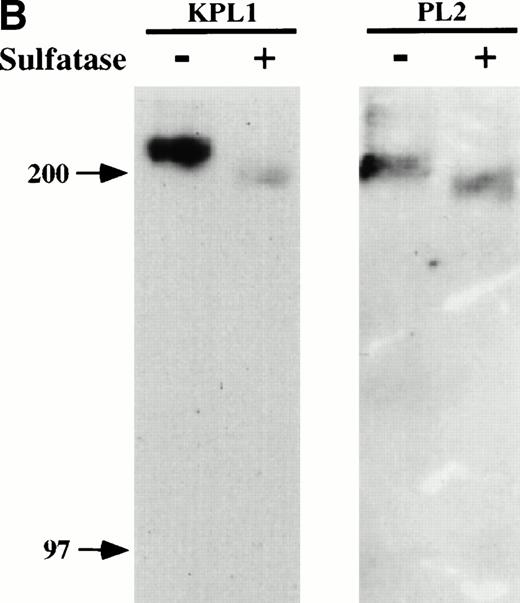
KPL1 recognizes the tyrosine sulfation motif of PSGL-1. (A) 300.19 cells were transiently transfected with either full length PSGL-1 (top) or a mutated form of PSGL-1 referred to as FFFE/PSGL-1 (bottom) in which the tyrosines at positions 5, 7, and 10 were replaced with phenylalanine. Twenty-four hours posttransfection, the 300.19 cells were stained with either a negative control antibody, KPL1, PL2, or PSL275. PL2 and PSL275 recognize both the full length and mutated form of PSGL-1, whereas KPL1 was able to interact with full length PSGL-1 but did not recognize FFFE/PSGL-1. (B) Arylsulfatase treatment of whole cell lysates abrogates binding of KPL1. Whole cell lysates were prepared as described in Materials and Methods and treated with 1U of arylsulfatase, and Western blotting was performed as described for Fig 1, using KPL1 (left) or PL2 (right).
KPL1 recognizes the tyrosine sulfation motif of PSGL-1. (A) 300.19 cells were transiently transfected with either full length PSGL-1 (top) or a mutated form of PSGL-1 referred to as FFFE/PSGL-1 (bottom) in which the tyrosines at positions 5, 7, and 10 were replaced with phenylalanine. Twenty-four hours posttransfection, the 300.19 cells were stained with either a negative control antibody, KPL1, PL2, or PSL275. PL2 and PSL275 recognize both the full length and mutated form of PSGL-1, whereas KPL1 was able to interact with full length PSGL-1 but did not recognize FFFE/PSGL-1. (B) Arylsulfatase treatment of whole cell lysates abrogates binding of KPL1. Whole cell lysates were prepared as described in Materials and Methods and treated with 1U of arylsulfatase, and Western blotting was performed as described for Fig 1, using KPL1 (left) or PL2 (right).
To determine whether binding of KPL1 required sulfation of PSGL-1, cell lysates were treated with a bacterial arylsulfatase, which cleaves sulfates from tyrosine residues within proteins but does not digest sulfated carbohydrates, or with buffer alone, and Western blotting was performed with either KPL1 or PL2. As shown in Fig 2B, treatment of cell lysates with arylsulfatase nearly abrogated recognition with KPL1, but did not affect recognition by PL2. Therefore, binding of KPL1 to PSGL-1 is dependent on sulfates within the PSGL-1 tyrosine sulfate motif.
Other investigators have suggested that at least some B cells express non–PACE-processed PSGL-1, and therefore that PSGL-1 on B cells is expressed in a form that is not recognized by antibodies generated against or specific for PACE-cleaved PSGL-1.14 After a 24-hour incubation with PACE secreting or control CHO cells,14 several B-cell lines were analyzed by flow cytometry with MoAb PSL-275, which recognizes only PACE-cleaved PSGL-1.14 One cell line showed a significant increase in PSL-275 staining, but these cells stained identically with KPL1 whether they were incubated with control or CHO-PACE cells (data not shown). Therefore, KPL1 binding is independent of PACE cleavage of PSGL-1.
KPL1 MoAb inhibits binding of normal leukocytes and HL60 cells to P-selectin but not E-selectin.
Adhesion assays were performed to determine if KPL1 could inhibit interactions between normal leukocytes or HL60 cells and P- or E-selectin. In the absence of antibody, HL60 cells, PBMCs, and neutrophils each bound well to COS cells transfected with either P- or E-selectin, with neutrophils binding slightly better to both P- and E-selectin compared with PBMC or HL60 cells (Fig3). Consistent with a requirement for tyrosine sulfation of PSGL-1 for interaction with P-selectin, preincubation of leukocytes with KPL1 virtually completely blocked (>95%) interactions of HL60 cells and neutrophils with P-selectin and almost completely (>90%) inhibited interactions of PBMCs with P-selectin (Fig 3A). In contrast, KPL1 had no effect on binding to E-selectin (Fig 3B).
KPL1 inhibits the binding of HL60 cells, neutrophils, and PBMCs to P-selectin but not E-selectin expressing COS cells in a low shear adhesion assay. (A) Adhesion of HL60 cells (▪), neutrophils (▧), and PBMC (□) to COS cells transiently transfected with P-selectin was performed as described in Materials and Methods. All three cell types adhered well to P-selectin. KPL1 almost completely blocked these interactions, whereas a control antibody had no effect on adhesion. (B) HL60 cells (▪), neutrophils (▧) and PBMC (□) bound well to E-selectin, but neither KPL1 nor a control antibody inhibited adhesion to E-selectin expressing COS cells. Values are mean ±SD; one of at least five experiments.
KPL1 inhibits the binding of HL60 cells, neutrophils, and PBMCs to P-selectin but not E-selectin expressing COS cells in a low shear adhesion assay. (A) Adhesion of HL60 cells (▪), neutrophils (▧), and PBMC (□) to COS cells transiently transfected with P-selectin was performed as described in Materials and Methods. All three cell types adhered well to P-selectin. KPL1 almost completely blocked these interactions, whereas a control antibody had no effect on adhesion. (B) HL60 cells (▪), neutrophils (▧) and PBMC (□) bound well to E-selectin, but neither KPL1 nor a control antibody inhibited adhesion to E-selectin expressing COS cells. Values are mean ±SD; one of at least five experiments.
KPL1 MoAb inhibits rolling of freshly isolated human neutrophils and CD4 T cells on P-selectin but not E-selectin.
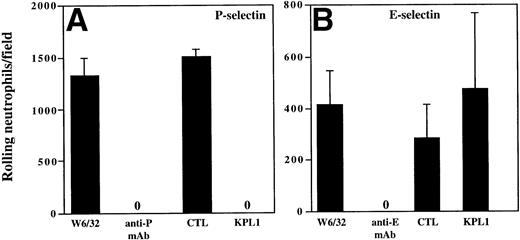
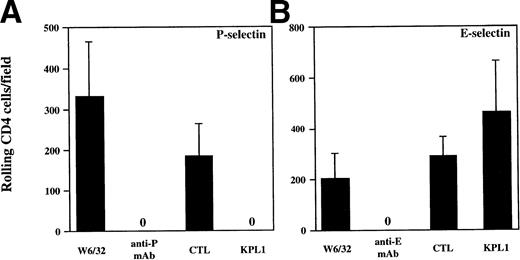
The results from the low shear stress COS cell adhesion assay were extended to analysis of rolling under defined shear stress (Figs 4 and5). Purified human neutrophils or purified CD4 T cells were incubated with media containing either KPL1 or control antibody, or the monolayers were incubated with either HDPG2/3, an anti–P-selectin antibody (CHO/P monolayer); HEL3/2, an anti–E-selectin antibody (CHO/E monolayer); or an isotype control antibody (W6/32). Preincubation of the monolayers with MoAb to the appropriate selectin completed blocked rolling of neutrophils. Preincubation of neutrophils with KPL1 also completely inhibited rolling of neutrophils on P-selectin (Fig 4A), but had no effect on interactions with E-selectin (Fig 4B). Similarly, preincubation of transfected monolayers with the appropriate anti-selectin antibody completely inhibited rolling of CD4 T cells, and, as observed with neutrophils, KPL1 completely inhibited rolling of CD4 T cells on CHO/P but had no effect on CHO/E (Fig 5A and B).
KPL1 inhibits rolling of neutrophils on CHO cells transfected with P-selectin but not E-selectin under defined shear stress. (A) The rolling of neutrophils on CHO cells expressing P-selectin in the presence of W6/32 (isotype control), HDPG2/3 (a blocking anti–P-selectin antibody), KPL1, or a control antibody for KPL1 was performed as described in Materials and Methods. Both KPL1 and HDPG2/3 completely blocked rolling of neutrophils on P-selectin. No effect on rolling was observed when the control antibodies were used. (B) The ability of freshly isolated neutrophils to roll on E-selectin transfected COS cells in the presence of W6/32, HEL3/2 (a blocking anti–E-selectin antibody), KPL1, or control antibody was performed as described in (A). Only the anti–E-selectin antibody inhibited rolling of neutrophils on E-selectin. No inhibition of rolling was observed when neutrophils were pre-incubated with KPL1 or control antibody, or when monolayers were exposed to W6/32. Values are mean ±SD; one of at least three experiments.
KPL1 inhibits rolling of neutrophils on CHO cells transfected with P-selectin but not E-selectin under defined shear stress. (A) The rolling of neutrophils on CHO cells expressing P-selectin in the presence of W6/32 (isotype control), HDPG2/3 (a blocking anti–P-selectin antibody), KPL1, or a control antibody for KPL1 was performed as described in Materials and Methods. Both KPL1 and HDPG2/3 completely blocked rolling of neutrophils on P-selectin. No effect on rolling was observed when the control antibodies were used. (B) The ability of freshly isolated neutrophils to roll on E-selectin transfected COS cells in the presence of W6/32, HEL3/2 (a blocking anti–E-selectin antibody), KPL1, or control antibody was performed as described in (A). Only the anti–E-selectin antibody inhibited rolling of neutrophils on E-selectin. No inhibition of rolling was observed when neutrophils were pre-incubated with KPL1 or control antibody, or when monolayers were exposed to W6/32. Values are mean ±SD; one of at least three experiments.
KPL1 inhibits rolling of all CD4+ T cells on CHO cells transfected with P-selectin but not E-selectin under defined shear stress. Freshly isolated CD4 T cells were assayed for their ability to roll on CHO cells transfected with either P- or E-selectin. (A) Rolling was assessed as described in Materials and Methods after exposure to either W6/32 (isotype control antibody), HDPG2/3 (a blocking anti–P-selectin antibody), KPL1, or a control antibody. Rolling of freshly isolated CD4 T cells was completely inhibited by pre-incubation of CD4 cells with KPL1 or monolayers with HDPG2/3. (B) Rolling of CD4 T cells on CHO cells transfected with E-selectin after exposure to either W6/32, HEL3/2 (a blocking anti–E-selectin antibody), KPL1, or a control antibody for KPL1 was performed as described for (A). Rolling of CD4 T cells was completely inhibited by preincubation of the E-selectin expressing monolayer with HEL3/2, whereas incubation of CD4 cells with either KPL1 or control antibodies had no effect. Values are mean ±SD; one of at least three experiments.
KPL1 inhibits rolling of all CD4+ T cells on CHO cells transfected with P-selectin but not E-selectin under defined shear stress. Freshly isolated CD4 T cells were assayed for their ability to roll on CHO cells transfected with either P- or E-selectin. (A) Rolling was assessed as described in Materials and Methods after exposure to either W6/32 (isotype control antibody), HDPG2/3 (a blocking anti–P-selectin antibody), KPL1, or a control antibody. Rolling of freshly isolated CD4 T cells was completely inhibited by pre-incubation of CD4 cells with KPL1 or monolayers with HDPG2/3. (B) Rolling of CD4 T cells on CHO cells transfected with E-selectin after exposure to either W6/32, HEL3/2 (a blocking anti–E-selectin antibody), KPL1, or a control antibody for KPL1 was performed as described for (A). Rolling of CD4 T cells was completely inhibited by preincubation of the E-selectin expressing monolayer with HEL3/2, whereas incubation of CD4 cells with either KPL1 or control antibodies had no effect. Values are mean ±SD; one of at least three experiments.
KPL1 inhibits interactions between L-selectin and PSGL-1.
PSGL-1 has recently been identified as a leukocyte ligand for L-selectin, and appears to mediate neutrophil-neutrophil interactions which may be important in amplifying an inflammatory response.11,12 Because normal neutrophils express ligands for L-selectin in addition to PSGL-1,11 31 we analyzed the effect of KPL1 on L-selectin/PSGL-1 interactions using L-selectin transfectants binding to PSGL-1 transfectants, which isolates the molecular interaction of interest. COS cells were cotransfected with individual plasmids containing cDNA encoding PSGL-1, FucT-VII, and C2GnT. For the assay, these COS cells were preincubated with either KPL1 or control antibody, washed, and incubated with 300.19 cells stably transfected with either L-selectin (300.19/L) or P-selectin (300.19/P). Preliminary studies (data not shown) demonstrated that binding of either 300.19/P or 300.19/L was absolutely dependent on expression of both PSGL-1 and FucT-VII, and that binding of 300.19/L was enhanced by cotransfection with C2GnT cDNA. Binding of 300.19/P was not significantly enhanced by transfection with C2GnT cDNA, presumably because of the endogenous expression of C2GnT by COS cells.
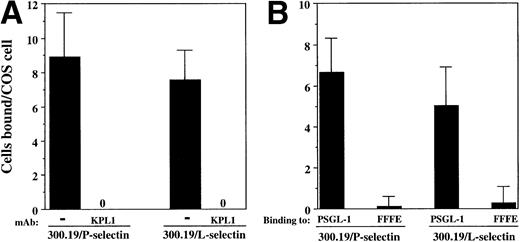
Both 300.19/P and 300.19/L cells bound to COS cells coexpressing PSGL-1, FucT-VII, and C2GnT (Fig 6A). Binding of both 300.19/P and 300.19/L was completely blocked in the presence of KPL1 (Fig 6A). In addition, no binding of either 300.19/L or 300.19/P to the FFFE/PSGL-1 mutant was detected (Fig 6B). These data strongly indicate that interaction between PSGL-1 and either P-selectin or L-selectin involves an identical or overlapping region of PSGL-1, which includes the KPL1 epitope within the tyrosine sulfation motif.
Interactions between PSGL-1 and L-selectin depend on the tyrosine sulfate motif. (A) COS cells were cotransfected with cDNA encoding PSGL-1, FucT-VII, and C2GnT. COS cells were preincubated with either KPL1 or control antibody for 15 minutes. After extensive washing, 300.19/P or 300.19/L cells were added to the COS cells and the assay was performed as described in Materials and Methods. Adhesion of both cell types was completely inhibited by preincubation of transfected COS cells with KPL1, but not control MoAb (not shown). (B) COS cells were cotransfected with cDNA encoding FucT-VII, C2GnT, and either PSGL-1 or FFFE/PSGL-1. Binding of 300.19/L or 300.19/P cells to the FFFE mutant was undetectable. Values are mean ±SD; one of three experiments.
Interactions between PSGL-1 and L-selectin depend on the tyrosine sulfate motif. (A) COS cells were cotransfected with cDNA encoding PSGL-1, FucT-VII, and C2GnT. COS cells were preincubated with either KPL1 or control antibody for 15 minutes. After extensive washing, 300.19/P or 300.19/L cells were added to the COS cells and the assay was performed as described in Materials and Methods. Adhesion of both cell types was completely inhibited by preincubation of transfected COS cells with KPL1, but not control MoAb (not shown). (B) COS cells were cotransfected with cDNA encoding FucT-VII, C2GnT, and either PSGL-1 or FFFE/PSGL-1. Binding of 300.19/L or 300.19/P cells to the FFFE mutant was undetectable. Values are mean ±SD; one of three experiments.
The KPL1 epitope is expressed at high levels on circulating T cells, monocytes, and natural killer (NK) cells, but at low levels on B cells.
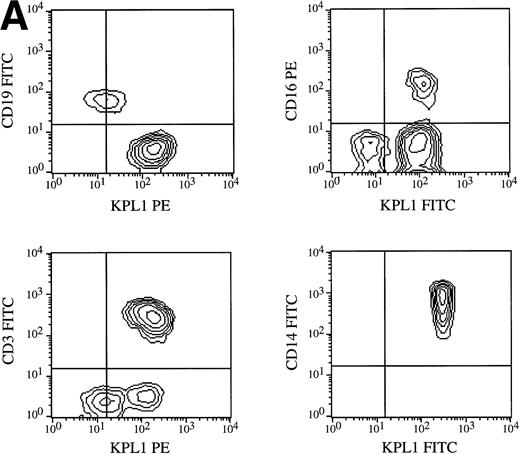
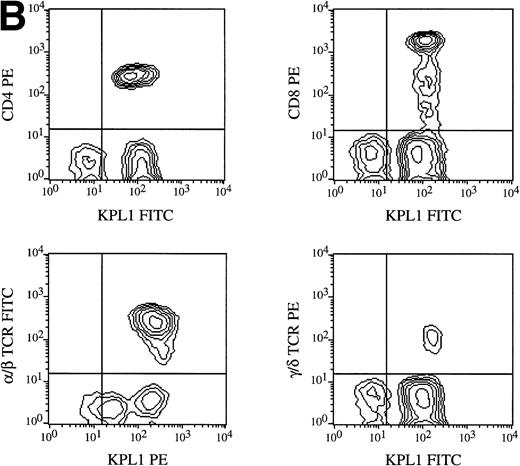
All circulating leukocyte subsets have been reported to express PSGL-1, although different researchers have reported variable levels of expression, or lack of expression, on specific subsets. Some of these differences may be caused by properties of different PSGL-1 specific antibodies. Staining of PBMCs with KPL1 in one-color flow cytometry showed a small but distinct subpopulation expressing lower levels of PSGL-1 (data not shown; see below), whereas most anti–PSGL-1 MoAb exhibit a single peak on lymphocytes10 17 (data not shown). To identify this KPL1lo subpopulation, two-color flow cytometry was performed with a leukocyte lineage marker in one color and KPL1 in a second color. Appropriate electronic scatter gates were set for either monocytes or lymphocytes. All NK cells (CD16+), monocytes (CD14+ ), and T cells (CD3+) (Fig 7A), including all CD4+, CD8+, α/β, and γ/δ T cells (Fig 7B), were uniformly positive for PSGL-1 (Fig 7A). The KPL1lo cells were identified as B cells by virtue of their staining with CD19; all detectable KPL1lo cells were CD19+, and no CD19− cells were KPL1lo (Fig 7A). These findings were observed with B cells analyzed from four different donors. This relatively low level of KPL1 staining was also observed on a panel of Epstein-Barr virus–transformed B-cell lines (data not shown).
Expression of PSGL-1 on leukocyte subsets. (A) Human PBMC were isolated by Ficoll density gradient centrifugation and stained for two-color flow cytometry as described in Materials and Methods. Electronic scatter gating was used to select specific subpopulations for detailed analysis. All NK cells (CD16+), monocytes (CD14+), and T cells (CD3+) were positive for PSGL-1 as measured by KPL1, whereas B cells (CD19+) expressed low levels of the KPL1 epitope. (B) All CD4, CD8, α/β (WT31 positive), or γ/δ (anti-TCR-γ/δ-1 positive) cells express uniform levels PSGL-1 as measured by KPL1. Horizontal and vertical lines delineating positive and negative staining were set with appropriate negative control MoAb.
Expression of PSGL-1 on leukocyte subsets. (A) Human PBMC were isolated by Ficoll density gradient centrifugation and stained for two-color flow cytometry as described in Materials and Methods. Electronic scatter gating was used to select specific subpopulations for detailed analysis. All NK cells (CD16+), monocytes (CD14+), and T cells (CD3+) were positive for PSGL-1 as measured by KPL1, whereas B cells (CD19+) expressed low levels of the KPL1 epitope. (B) All CD4, CD8, α/β (WT31 positive), or γ/δ (anti-TCR-γ/δ-1 positive) cells express uniform levels PSGL-1 as measured by KPL1. Horizontal and vertical lines delineating positive and negative staining were set with appropriate negative control MoAb.
Expression of PSGL-1 in lymphoid tissues.
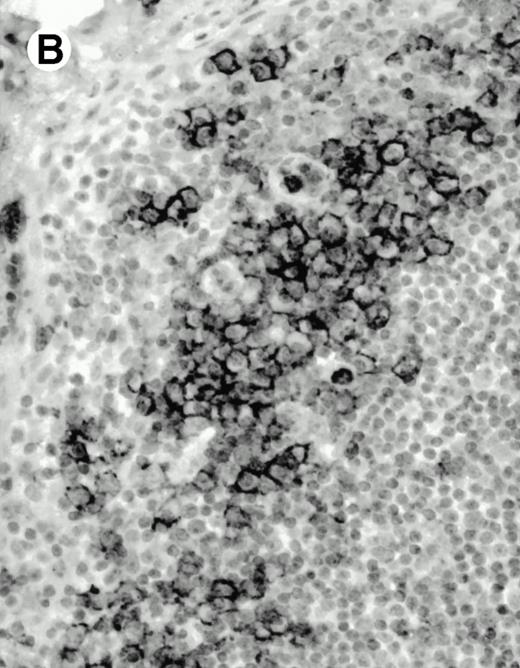
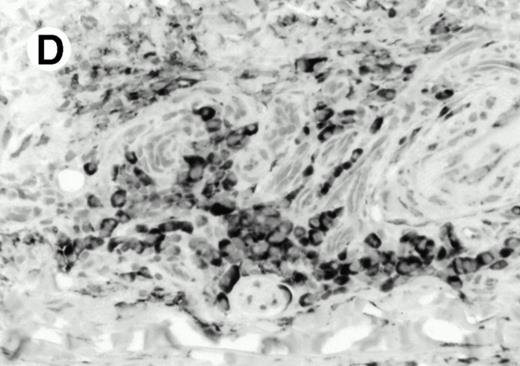
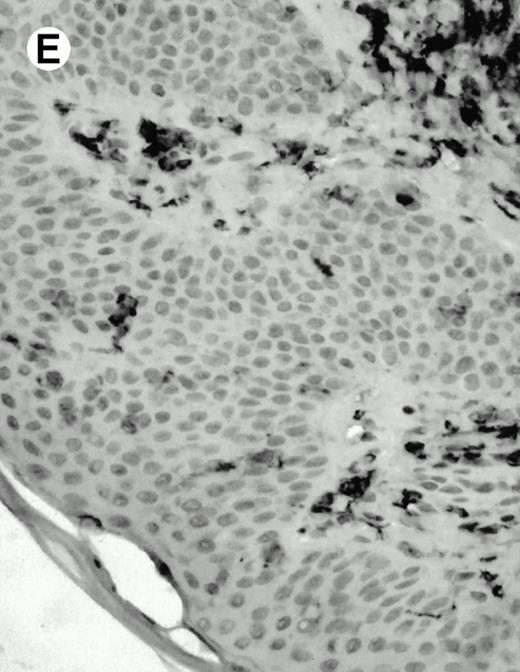
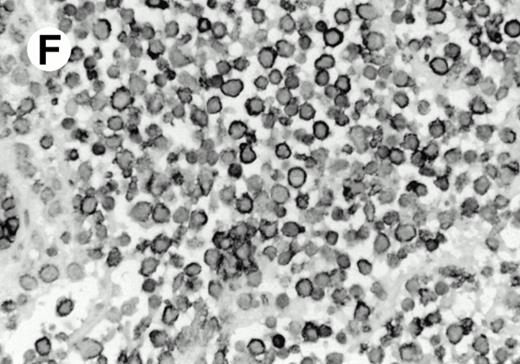
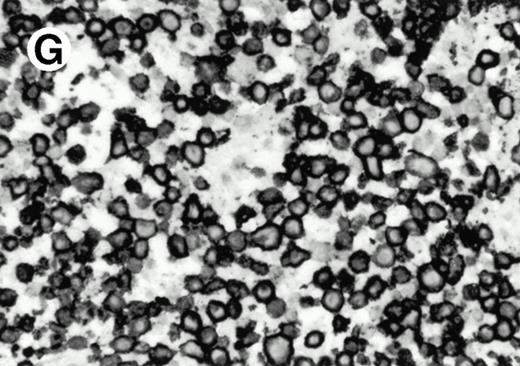
Both paraffin-embedded tissues and frozen sections (with or without heat antigen retrieval) were prepared for immunohistology as described in Materials and Methods. KPL1 stained numerous T cells in the T-zone (TZ) of a human tonsil (Fig 8A), but did not stain B cells in the mantle zone (M) or germinal center (GC) of the secondary follicles. Macrophages in the germinal center stained but follicular dendritic cells did not (Fig 8A). Just outside the germinal center, intense KPL1 staining of overlying plasma cells was observed. A high-power magnification (Fig 8B) shows this intense staining of plasma cells which surround the germinal centers. Subepithelial plasma cells also stained with KPL1 (data not shown), as did plasma cells around vessels in the skin (Fig 8C); these plasma cells costained with the plasma cell marker VS38 (Fig 8D). Thus, B cells located in germinal centers either lack the KPL1 epitope, or express it at levels which are not detected by these methods, whereas plasma cells in numerous sites express high levels. Similarly, four IL-6–dependent human myeloma cell lines expressed high levels of the KPL1 epitope (Diane Jelinek, personal communication, March 1997). Both cortical and medullary lymphocytes in the thymus stained with KPL1 (data not shown). Langerhans cells, bone marrow–derived antigen presenting cells which reside in the suprabasilar region of the epidermis, were also positive for KPL1 (Fig 8E). Langerhans cells in the tonsillar epithelium also stained with KPL1 (data not shown). Langerhans cells in soft tissue also stain with both KPL1 (Fig 8F) and CD1a (Fig 8G).
Expression of PSGL-1 in human tissues. (A) Paraffin tonsil stained with KPL1. The vast majority of the lymphocytes in the T-cell zone (TZ) show dark staining, while the B-lymphocytes in the mantle (M) and germinal center (GC) do not stain. (B) A high magnification of paraffin embedded tonsil stained with KPL1. Plasma cells adjacent to the overlying epithelium show dark membrane staining. The mantle B lymphocytes in the right lower corner do not stain. (C) Plasma cells near vessels in skin stain well with KPL1. (D) Costaining of the cells in (C) with VS38 confirms their identification as plasma cells.
(E) Paraffin section of skin stained with KPL1. The Langerhans cells in epidermis and superficial dermis stain intensely in this example of Langerhans cell histiocytosis. The stained dendritic processes are apparent in the epidermis. (F) Langerhans cells in soft tissues also stain with KPL1. (G) Identification of Langerhans cells in (F) is confirmed by costaining with CD1a (immunoperoxidase with hematoxylin counterstain).
Expression of PSGL-1 in human tissues. (A) Paraffin tonsil stained with KPL1. The vast majority of the lymphocytes in the T-cell zone (TZ) show dark staining, while the B-lymphocytes in the mantle (M) and germinal center (GC) do not stain. (B) A high magnification of paraffin embedded tonsil stained with KPL1. Plasma cells adjacent to the overlying epithelium show dark membrane staining. The mantle B lymphocytes in the right lower corner do not stain. (C) Plasma cells near vessels in skin stain well with KPL1. (D) Costaining of the cells in (C) with VS38 confirms their identification as plasma cells.
(E) Paraffin section of skin stained with KPL1. The Langerhans cells in epidermis and superficial dermis stain intensely in this example of Langerhans cell histiocytosis. The stained dendritic processes are apparent in the epidermis. (F) Langerhans cells in soft tissues also stain with KPL1. (G) Identification of Langerhans cells in (F) is confirmed by costaining with CD1a (immunoperoxidase with hematoxylin counterstain).
Finally, an extensive tissue survey failed to detect KPL1 staining on stomach, colon, liver, pancreas, uterus (endometrium and myometrium), ovary, prostate, testis, brain, lung, heart, thyroid, parathyroid, and skeletal muscle (data not shown). However, leukocytes, including dendritic cells and macrophages, reacted in many of the tissues, eg, the Kupffer cells in the liver. PSGL-1 expression, as assessed by KPL1 staining, therefore appears to be limited to the hematopoietic system, at least in humans.
DISCUSSION
We have generated a novel MoAb against PSGL-1, designated KPL1. The specificity of the KPL1 MoAb for PSGL-1 was confirmed by both flow cytometry and Western blotting of multiple PSGL-1–expressing transfectants. The epitope defined by KPL1 is independent of carbohydrate modifications, including the presence or absence of sialylation, fucosylation, or branched O-linked structures, because KPL1 stains cells equivalently regardless of whether the cells express no or high levels of FucT-VII or C2GnT, and KPL1 staining was not affected by treatment of cells with neuraminidase. In addition, this epitope is independent of PACE processing. However, the KPL1 epitope requires sulfation of at least one tyrosine contained within a consensus tyrosine sulfation motif (YEYLDYD)29 at the extreme amino terminus of PSGL-1. Deletion of these seven amino acids eliminated binding of KPL1,30 as did simultaneous mutation of all three tyrosines to phenylalanine (Fig 2A) or sulfatase treatment of PSGL-1 (Fig 2B). Thus, the KPL1 epitope maps to the tyrosine sulfation motif of PSGL-1.
Consistent with the KPL1 epitope mapping to the tyrosine sulfate motif, and the requirement for tyrosine sulfation of PSGL-1 in recognition of P-selectin,7-9 the KPL1 MoAb essentially completely inhibited adhesion between P-selectin and HL60 cells, neutrophils, or PBMCs in a low shear adhesion assay, and between P-selectin and CD4 T cells or neutrophils in a parallel plate flow assay. In contrast, KPL1 had no effect on interactions between these different cell types and E-selectin. Although PSGL-1 is not required for adhesion to E-selectin,21 PSGL-1 appears capable of interacting with both P- and E-selectin,9,32,33 with different (or multiple) regions of PSGL-1 mediating adhesion to E-selectin.30 It is currently not known if an identical site or closely overlapping site within the first 19 amino acids of PSGL-1 mediates interaction with both P- and E-selectin, or whether this or another site on PSGL-1 is functional during normal leukocyte interactions with E-selectin.
PSGL-1 also interacts with L-selectin, an interaction likely to be of importance in neutrophil-neutrophil interactions during inflammation.11,12,34 Because normal neutrophils express ligands for L-selectin in addition to PSGL-1,11,31 we analyzed whether KPL1 could inhibit this molecular interaction in a “pure” system, consisting of 300.19/L-selectin transfectants binding to COS cells cotransfected with cDNA encoding PSGL-1, FucT-VII, and C2GnT. All three of these gene products are required for binding to P-selectin.3,10,21,26,33,35 Untransfected 300.19 cells do not bind to COS cells cotransfected with cDNA encoding PSGL-1, FucT-VII, and C2GnT, and binding of 300.19/L or 300.19/P cells is completely dependent on transfection of COS cells with cDNA encoding both PSGL-1 and FucT-VII. We found that KPL1 virtually completely blocked interactions between L-selectin and PSGL-1 (Fig 6A). Binding of L-selectin, like binding of KPL1 (Fig 2) or P-selectin,7-9was also blocked by mutation of all three tyrosines in the PSGL-1 tyrosine sulfation motif to phenylalanine (Fig 6B). P-selectin and L-selectin (and KPL1) therefore recognize identical or closely overlapping sites on PSGL-1, and PSGL-1 does not appear to have multiple L-selectin binding sites.
Both PSGL-1 and at least some L-selectin ligands require sulfation,7-9,36 and for PSGL-1 this sulfation occurs exclusively on tyrosine.8,9 In contrast, sulfation of L-selectin HEV ligands36 occurs on specific carbohydrate side chains of CD34, GlyCAM-1, and possibly sgp200.37-39However, recognition of PSGL-1 by L-selectin is dependent on sulfation of tyrosines within the PSGL-1 tyrosine sulfation motif (Fig 6). Taken together, these data show that L-selectin recognizes an array of distinct sulfated ligands whose sulfate moiety can be attached to either carbohydrate or protein.
Our findings clearly establish that PSGL-1 is the principal or sole ligand for P-selectin on T cells. Disparate results were previously found using a polyclonal antiserum against recombinant PSGL-1 (termed Rb 3026). Alon et al13 found that this reagent inhibited T-cell interaction with P-selectin, whereas Vachino et al14failed to detect inhibition. This same reagent previously was found to inhibit monocyte adhesion to P-selectin.15 Rb 3026 also failed to detect a subset of γ,δ T cells.40 In contrast, we and others17 have clearly shown that all subsets of blood T cells express PSGL-1; PSGL-1− T cells were never detected in blood from any donor. However, that same study17 failed to detect PSGL-1 on T cells in tissues, whereas we observed significant staining with KPL1 in T cells areas of tonsil (Fig 8). PSGL-1 levels do not change significantly with T-cell activation in vitro,14 16 making it unlikely that activation within tissues can explain differences in staining between blood (by FACS) and tissues (by immunohistology), especially by the same MoAb. The basis for this variation remains unknown.
B cells in peripheral blood stained at lower levels with KPL1 than other leukocytes, but were clearly positive (Fig 7). We also found PSGL-1 on 7/7 EBV B-cell lines (data not shown). These results contrast with those of Vachino et al,14 who detected PSGL-1 expression on only ∼30% of blood B cells. Laszik et al17also found lower levels of PSGL-1 on B cells, but whether B cells expressed uniformly lower levels or whether a subpopulation of B cells do not express PSGL-1 was not specified in that study.17 In contrast, Moore et al10 previously found uniform expression of PSGL-1 on essentially all lymphocytes. The basis for these discrepancies is unclear. B cells within mantle zones or germinal centers of tonsil failed to stain detectably with KPL1. The basis for the relatively low level of KPL1 staining on B cells is not known, but may relate to the state of differentiation of the B cell, as both plasma cells in situ (Fig 8) and cultured, IL-6–dependent myeloma cells (Diane Jelinek, personal communication, March 1997) stain brightly with KPL1. These results, in combination with the localization of the KPL1 epitope to the tyrosine sulfation motif, suggest that B cells may regulate sulfation of PSGL-1 in a manner distinct from the apparently constitutive sulfation characteristic of other types of leukocytes. These putative differences in sulfation of PSGL-1 may affect the migratory capacity of B cells, and may explain why B cells are rarely found at most sites of inflammation.
A recent report documented strong PSGL-1 mRNA expression in most tissues of the mouse,41 implying that PSGL-1 is expressed on some nonhematopoietic cells. However, extensive immunohistochemical analysis by ourselves and others17 failed to detect PSGL-1 on any nonhematopoietic cells in any tissues examined. In addition, we failed to detect PSGL-1 staining on cultured normal endothelium, smooth muscle cells, fibroblasts, or keratinocytes (data not shown). The precise cellular source of the strong mRNA signals from multiple murine tissues is presently unknown, and the apparent species difference between humans and mice is unexpected.
The previously described function-blocking anti–PSGL-1 MoAb PL110 and the KPL1 MoAb described in the present report share a number of functional characteristics, including essentially complete inhibition of leukocyte binding to P-selectin10(Figs 3-5) and L-selectin binding to PSGL-111 (Fig6). However, these two MoAbs map to distinct epitopes near the amino terminus of PSGL-1. In particular, KPL1 recognizes the tyrosine sulfation motif comprising residues 5-11 (Fig 2), whereas PL1 sees the nontyrosine-sulfated sequence centered around residues 13-17 (LPETE).42 Although these data do not exclude the possibility that each MoAb sterically obscures the neighboring site as well, the data are most consistent with the idea that these two MoAbs are recognizing distinct components of physiological ligands for both L- and P-selectin. Thus, KPL1 prevents recognition of the tyrosine sulfates, whereas PL1 probably blocks recognition of sialylated and fucosylated carbohydrates attached to threonine 16, mutation of which inhibits recognition of P-selectin.8 9 Therefore, these observations provide support for the idea of a “discontiguous carbohydrate epitope”5,43 as a physiological recognition structure for selectins.
Generation of MoAb against human cell surface glycoproteins by immunizing with 300.19 transfectants stably expressing the molecule of interest has previously produced large numbers of MoAbs, many of which recognize evolutionarily conserved epitopes present of the homologues in other mammalian species.44,45 In contrast, we produced only a single MoAb to PSGL-1 with this approach, and this MoAb does not recognize leukocytes from any mammalian species thus far tested, including cat, dog, horse, cow, or sheep (Mark Jutila, personal communication, April 1997). Similarly, the PL1 MoAb does not see leukocytes from any other mammalian species tested (Kevin Moore, personal communication, March 1997). In part, this may relate to the relatively low conservation of amino acid sequence at the amino terminus of PSGL-1: only 7 of 16 residues are identical or similar between human and mouse PSGL-1, and the tyrosine sulfation motifs of the two species are at dissimilar relative positions.3 41
In summary, using a novel MoAb directed against the functionally essential tyrosine sulfation motif of human PSGL-1, we show that PSGL-1 is expressed on all circulating leukocytes, including neutrophils, monocytes, all subsets of T cells, NK cells, and B cells, and is the principal or sole ligand for P-selectin on at least T cells and neutrophils. The expression and function of PSGL-1 on B cells requires further investigation, as these cells may uniquely regulate tyrosine sulfation of PSGL-1.
ACKNOWLEDGMENT
The authors gratefully acknowledge Kevin Moore (University of Oklahoma Health Sciences Center, Oklahoma City, OK) for supplying PL2 MoAb; Henri Lichenstein (Amgen, Inc, Boulder, CO) for PSGL-1 cDNA; and Gray Shaw, Ray Camphausen, and Gloria Vachino (Genetics Institute, Inc, Cambridge, MA) for PSL275 MoAb, CHO/PACE cells, and the FFFE PSGL-1 mutant and helpful discussions; and M. Snapp for inspiration.
Supported by the American Cancer Society Grant No. CB-204 (G.S.K.) and HL36028 (F.W.L.) and National Institutes of Health Grant No. CA34233 (R.W.). G.S.K. is an Established Investigator of the American Heart Association.
Address reprint requests to Geoffrey S. Kansas, PhD, Department of Microbiology-Immunology, Northwestern Medical School, 303 E Chicago Ave, Chicago, IL 60611.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal