Abstract
Death-inducing ligands (DILs) such as tumor necrosis factor α (TNFα) or the cytotoxic drug doxorubicin have been shown to activate a nuclear factor κB (NFκB)-dependent program that may rescue cells from apoptosis induction. We demonstrate here that TRAIL (TNF-related apoptosis-inducing ligand), a recently identified DIL, also activates NFκB in lymphoid cell lines in a kinetic similar to TNFα. NFκB activity is independent from FADD, caspases, and apoptosis induction. To study the influence of NFκB activity on apoptosis mediated by TRAIL, CD95, TNFα, or doxorubicin, NFκB activation was inhibited using the proteasome inhibitor N-acetyl-L-leucinyl-L-leucinyl-L-norleucinal or transient overexpression of mutant IκBα. Sensitivity for induction of apoptosis was markedly increased by these treatments in apoptosis sensitive cell lines. Moreover, both in cell lines and in primary leukemia cells that are resistant towards induction of apoptosis by DILs and doxorubicin, antagonization of NFκB activity partially restored apoptosis sensitivity. These data suggest that inhibition of NFκB activation may provide a molecular approach to increase apoptosis sensitivity in anticancer treatment.
RESISTANCE OF TUMOR cells towards induction of apoptosis is a main reason for failure of anticancer treatment.1 Recent studies suggest that Nuclear factor κB (NFκB) may mediate a survival pathway in many cells.2 In the present study, we suggest that inhibition of NFκB activation may decrease apoptosis resistance for death-inducing ligands (DILs) and cytostatic drugs in lymphoid cells. NFκB is a DNA binding protein that augments transcription of various genes involved in cell proliferation, such as growth factors.3 NFκB plays an important role for cell development,4 survival, and oncogenesis,5 eg, in the immune system,2 and mediates its function through homodimers or heterodimers formed by NFκB/Rel family members (RelA/p65, RelB, c-rel, NFκB1/p50, and NFκB2/p52).6 In quiescent cells, cytosolic NFκB is bound to inhibitory IκB-proteins.3,7 Upon activation, IκB becomes phosphorylated, ubiquitinated, and subsequentially degraded by proteasomes,8-12 enabling nuclear translocation and DNA binding activity of activated NFκB.13
A variety of different stimuli activate NFκB, such as oxidative stress, cytokines, and apoptosis-inducing stimuli including drugs used in anticancer treatment.14,15 Most importantly, direct triggering of death receptors, eg, by tumor necrosis factor α (TNFα), may activate a NFκB dependent pathway that antagonizes apoptosis. Induction of apoptosis by TNFα involves signaling through adapter molecules such as FADD that bind to the intracellular death domain of the respective receptor via TRADD to activate downstream caspases and apoptosis effectors. In contrast, activation of NFκB seems to be the consequence of a pathway that is independent from FADD-mediated death signaling and involves different adapter molecules such as TRAF/NIK.16 TNFα activates NFκB in a broad spectrum of cells and activation of NFκB was shown to modify induction of apoptosis.17,18 Thus, in RelA −/− cells19 or cells overexpressing IκB,20induction of apoptosis by TNFα is markedly enhanced. Apart from TNFα, two additional DILs are characterized so far, all members of the TNF superfamily: CD95-ligand (CD95-L) and TRAIL (TNF-related apoptosis-inducing ligand). CD95-L activates NFκB in a cell-type–specific manner and independently from its cytotoxic function.16,21 Recently, a new DIL was found, TRAIL,22-24 that mediates apoptosis by its receptors DR4/TRAIL-R1/TRICK125 and DR5/TRAIL-R2/TRICK2,26-31 eg, in leukemic cells.32 We report here that TRAIL activates NFκB in lymphoid cell lines. Inhibition of NFκB activation strongly increases apoptosis sensitivity in some cell lines. In addition, we found that inhibition of NFκB activation enables induction of apoptosis by several DILs as well as cytostatic drugs in cells that are cross-resistant for apoptosis induction by DILs and drugs.
MATERIALS AND METHODS
Cell lines and culture.
All cell lines were cultured in RPMI medium with 10% fetal calf serum (Conco, Wiesbaden, Germany), 100 U/mL penicilin, 100 μg/mL streptomycin, and 10 mmol/L HEPES (all GIBCO, Life Technologies, Paisley, Scotland). For all assays cells were seeded at a density of 106 cells/mL. The following cell lines were used: CEM and Jurkat, acute T-cell leukemia; BJAB, Burkitt lymphoma; BOE and PD31, premature B-cell leukemia; CEM-R and Jurkat-R, derivative cell lines resistant to doxorubicin-induced apoptosis and cross-resistant against DIL-induced apoptosis. CEM-R cells were generated by culturing parental CEM cells in increasing doses of doxorubicin for several months and Jurakt-R cells were cultivated in anti–APO-1, respectively.
Stimulation of cells.
Doxorubicin (Sigma, Deisenhofen, Germany) was dissolved in water, supplemented with ethanol to a 96% ethanol stock solution (1 mg/mL), and stored in aliquots at −80°C. TNFα was purchased from Boehringer (Mannheim, Germany). N-acetyl-L-leucinyl-L-leucinyl-L-norleucinal (LLnL; Sigma) was dissolved in dimethylsulfoxide (DMSO) to a 25 mmol/L stock solution. N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl-ketone (zVAD) was purchased from Enzyme System Products (Dublin, CA) and dissolved in DMSO to a 50 mmol/L stock solution. Phorbol myristic acetate (PMA; Sigma) was dissolved in DMSO to a 100 μg/mL stock solution. Ionomycin (Sigma) was dissolved in 96% ethanol to a 5 mg/mL stock solution. TNFα, LLnL, zVAD, PMA, and Ionomycin were stored in aliquots at −20°C. TRAIL was produced as described.32Briefly, TRAIL cDNA was cloned into the vector pPIC3 and transfected into Pichia Pastoris strain GS115 (Invitrogen, NY Leek, The Netherlands). Protein production was induced by culturing yeast in 2.5% methanol. Cells were lysed and TRAIL purified by nickel-histidine interaction (Qiagen, Hamburg, Germany). LLnL and zVAD were preincubated in stimulation assays for 1 hour.
Electrophoresis mobility shift assay.
Cells (2 × 106) were lysed in 600 mmol/L KCl, 20 mmol/L HEPES, 200 μmol/L EDTA (all Merck, Darmstadt, Germany), 1 mmol/L Phenylmethylsulfonyl fluoride (PMSF), 2 μg/mL Aprotinin, and 2 μg/mL Leupeptin (all Sigma) for 45 minutes on ice in 5 packed cell volumes and centrifuged at 12,000g for 5 minutes. Five micrograms of supernatant was incubated with poly dIdC (1 μg; Pharmacia, Uppsala, Sweden) in binding buffer (10 mmol/L Tris-HCl, pH 7.45, 50 mmol/L NaCl, 0.5 mmol/L EDTA, 1 mmol/L dithiothreitol [DTT], 50 μmol/L PMSF, 2 vol% glycerol, 2% Ficoll 400) for 20 minutes at room temperature. Lysats were exposed for 15 minutes at room temperature to double-stranded oligonucleotides for the consensus binding sites of NFκB (5′AGT TGA GGG GAC TTT CCC AGG C 3′) that had been labaled with [32P]-ATP (Amersham, Braunschweig, Germany) by polynucleotide kinase (Boehringer). Protein-oligo mixtures were separated on a nondenaturing 6% polyacrylamide gel in 0.5× TBE. Gels were dried and exposed to an x-ray film. For specific competition, proteins were preincubated with unlabeled NFκB binding oligonucleotide in 100-fold excess; for supershift, preincubation was performed using 2 μL anti-p50, 5 μL anti-p65, or 1.5 μL anti-c-rel antibody (all Santa Cruz, Heidelberg, Germany) for 1 hour on ice.
Western blotting.
Cells (2 × 106) were washed twice in phosphate-buffered saline (PBS) and lysed for 5 minutes on ice in 10 packed cell volumes using 30 mmol/L Tris-HCl, pH 7.4, 150 mmol/L NaCl, 1 mmol/L EDTA, 0,5% Triton X-100, 0.5% Na-Desoxycholate, 1 mmol/L DTT, 1 mmol/L PMSF, 2 μg/mL Aprotinin, and 2 μg/mL Leupeptin. Cell debris were removed, and 15 μg of proteins was run on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto a nylon membrane. Filter was hybridized with anti-IκBα antibody and subsequentially with antirabbit antibody coupled to horseradish peroxidase (both Santa Cruz). For detection, an Enhanced Chemilumiscent System (Amersham) was used. Equal protein loading was proven by subsequential Ponceau red staining of the filter.
Measurement of apoptosis.
Apoptosis was measured by forward side scatter analysis in FACScan flow cytometer (Becton Dickinson) equipped with the Cell Quest 2.0 Software. Specific apoptosis was calculated as described.32
Transfections.
Cells (3 × 107) were washed twice in PBS and resuspended in 200 μL PBS. Twenty micrograms of specific plasmid DNA (mock or IκBα) and 2 μg plasmid DNA containing pEGFP DNA for transfection control (Clontech, Heidelberg, Germany) was added. The IκBα mutant construct contains alanines in positions 32 and 36 instead of serines to disable phosphorylation and degradation of the protein.11 Electroporation was performed (240 V, 975 μF) and cells were resuspended in 10 mL fresh medium. After 48 hours, living cells were separated by Ficoll gradient and subsequentially stimulated for another 12 hours. Forward side scatter FACScan analysis was performed gating on green fluorescent protein (GFP)-positive cells.
Measurement of apoptosis in primary leukemic cells.
At the time of diagnosis of acute leukemia, mononuclear cells were isolated from 6 patients' bone marrow cells using Ficoll gradient centrifugation. Staining of membrane antigens showed the percentage of blast cells to be greater than 90% in all patients. Cells were seeded at a density of 106 cells/mL and incubated at 37°C, 5% CO2 in humidity. Spontaneous apoptosis was measured continuously using Trypan blue staining. As soon as 35% of cells or more stained positive (6 to 12 hours, respectively), the experiment was stopped and apoptosis was measured in a FACScan using forward side scatter analysis.
RESULTS
Activation of NFκB by TRAIL.
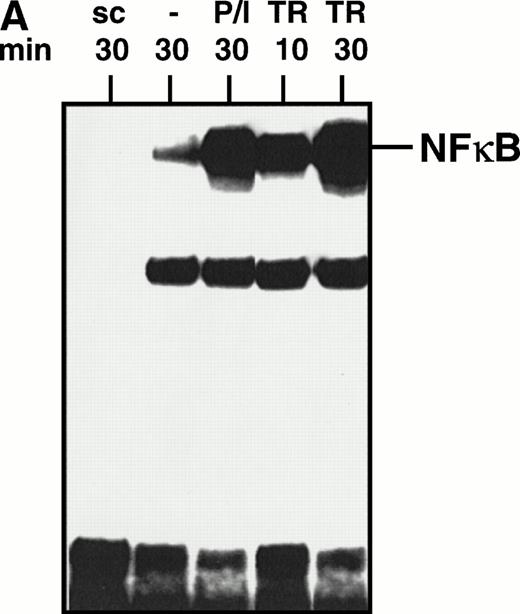
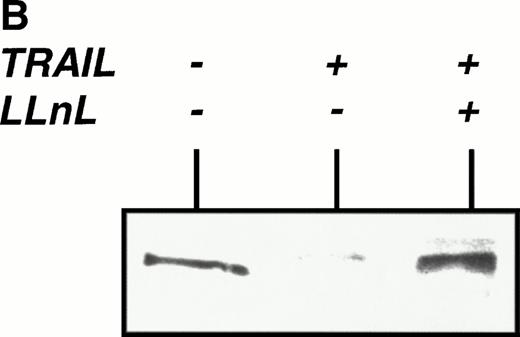
We used electromobility shift assay (EMSA) to investigate whether TRAIL is able to activate NFκB in lymphoid cells. As shown in Fig 1A, enhanced NFκB binding activity was found as early as 10 minutes after stimulation of Jurkat cells with TRAIL (0.3 μg/mL). Activation of NFκB was further enhanced after 30 minutes and was comparable with stimulation of Jurkat cells with PMA/Ionomycin (Fig 1A). Activation of NFκB by TRAIL was also observed in CEM cells and in the murine pre-B–cell line PD31 (data not shown). TNFα also activated NFκB in most cell lines tested, whereas CD95-triggering did not consistently induce NFκB activation33 (and data not shown). TRAIL-induced NFκB binding activity was mediated by a complex of dimers containing p50 and p65 proteins as demonstrated by supershift experiments (data not shown). NFκB activation was accompanied by degradation of its cytosolic inhibitor IκBα, which could be blocked by the proteasome inhibitor LLnL, as detected in Western blot (Fig 1B).
Activation of NFκB by TRAIL. (A) Jurkat cells were incubated with PMA (50 ng/mL)/Ionomycin (2 μg/mL) (P/I) or TRAIL (0.3 μg/mL) (TR) for the time periods indicated. Cells were harvested, cellular proteins were isolated, and EMSA was performed. sc, specific competition with unlabeled oligonucleotide. Similar results were obtained in three independent experiments. (B) Jurkat cells were stimulated with TRAIL (0.3 μg/mL) for 30 minutes in the presence or absence of LLnL (6.25 μmol/L, 1 hour of pretreatment). Fifteen micrograms of protein extract was run on a 12% polyacrylamide gel. Proteins were transferred onto a nylon filter that was subsequentially hybridized with an anti-IκBα antibody. Similar results were obtained in three independent experiments. Similar results were obtained with CEM cells.
Activation of NFκB by TRAIL. (A) Jurkat cells were incubated with PMA (50 ng/mL)/Ionomycin (2 μg/mL) (P/I) or TRAIL (0.3 μg/mL) (TR) for the time periods indicated. Cells were harvested, cellular proteins were isolated, and EMSA was performed. sc, specific competition with unlabeled oligonucleotide. Similar results were obtained in three independent experiments. (B) Jurkat cells were stimulated with TRAIL (0.3 μg/mL) for 30 minutes in the presence or absence of LLnL (6.25 μmol/L, 1 hour of pretreatment). Fifteen micrograms of protein extract was run on a 12% polyacrylamide gel. Proteins were transferred onto a nylon filter that was subsequentially hybridized with an anti-IκBα antibody. Similar results were obtained in three independent experiments. Similar results were obtained with CEM cells.
Independence of TRAIL-mediated NFκB activation from FADD and caspases.
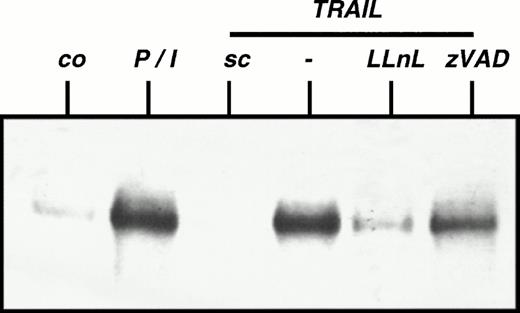
Because TRAIL mediates two effects, namely induction of apoptosis and activation of NFκB, we examined whether both signals use the same intracellular signaling pathway. After stimulation with TRAIL, BJAB cells stably overexpressing a dominant negative FADD mutant protein displayed similar NFκB activation compared with mock-transfected cells, suggesting that TRAIL-mediated NFkB activation is indepedent from FADD signaling (data not shown). To test whether TRAIL-induced activation of NFκB involves known downstream effector molecules for apoptosis mediated by TRAIL, we examined NFκB activation in the presence of inhibition of caspase activity. ZVAD, a broad spectrum inhibitor of caspases that is known to block TRAIL-induced apoptosis,32 had no significant influence on activation of NFκB by TRAIL (Fig 2), suggesting that both intracellular signaling pathways are at least partially independent.
Independence of TRAIL-mediated NFκB activation from caspases. CEM cells were preincubated with LLnL (2.5 μmol/L) or zVAD (50 μmol/L) for 1 hour. TRAIL (0.3 μg/mL) or PMA (50 ng/mL)/Ionomycin (2 μg/mL) (P/I) was added for another 30 minutes. Cells were harvested and EMSA was performed. sc, specific competition with unlabeled oligo. Similar results were obtained in two independent experiments. Similar results were obtained using Jurkat cells.
Independence of TRAIL-mediated NFκB activation from caspases. CEM cells were preincubated with LLnL (2.5 μmol/L) or zVAD (50 μmol/L) for 1 hour. TRAIL (0.3 μg/mL) or PMA (50 ng/mL)/Ionomycin (2 μg/mL) (P/I) was added for another 30 minutes. Cells were harvested and EMSA was performed. sc, specific competition with unlabeled oligo. Similar results were obtained in two independent experiments. Similar results were obtained using Jurkat cells.
Increase in apoptosis sensitivity of CEM cells by inhibition of NFκB activation.
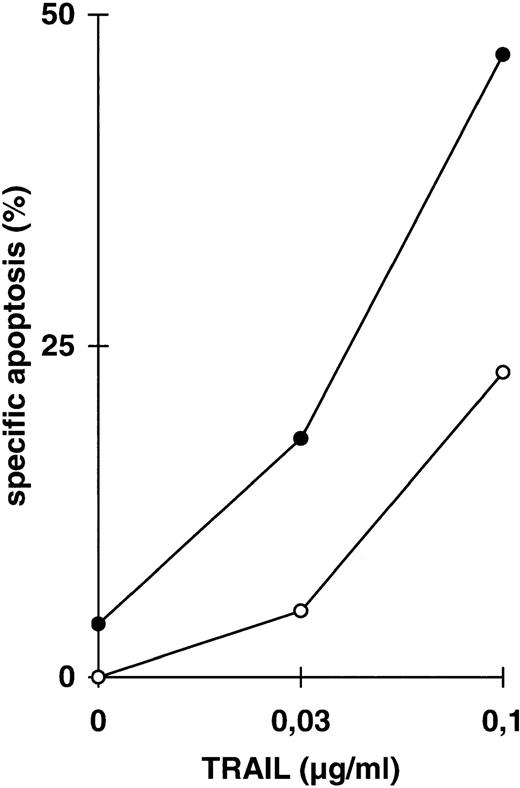
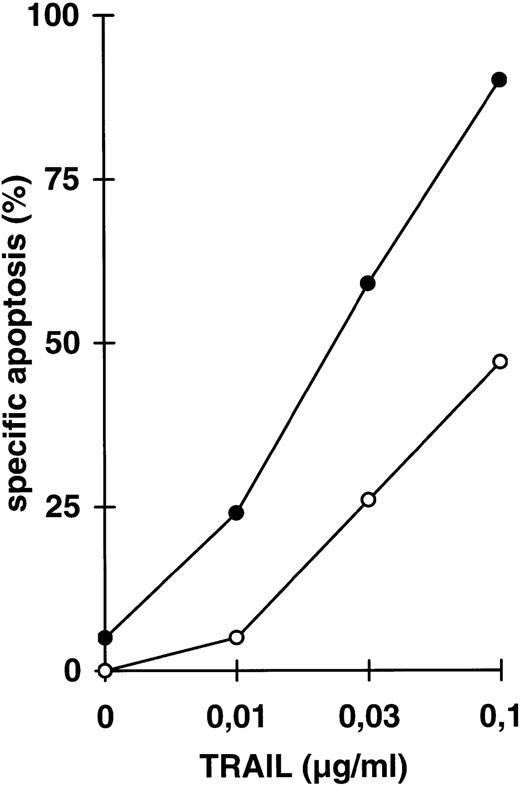
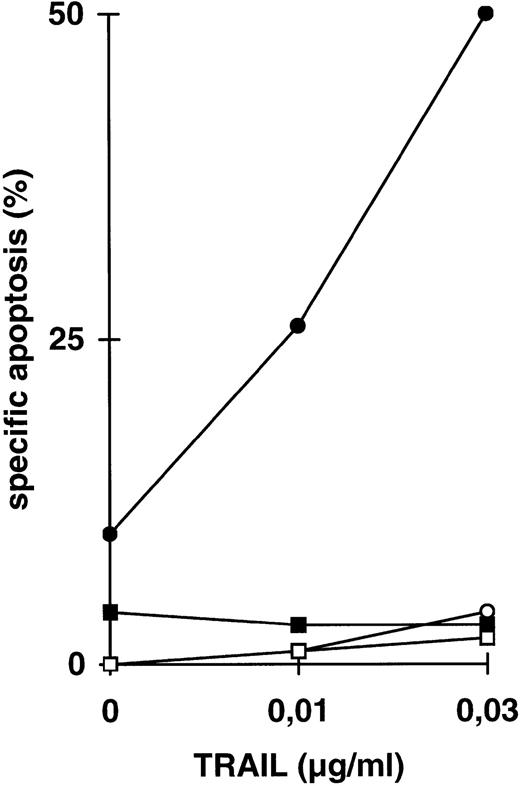
Because the two signals mediated by TRAIL use different intracellular signaling transmission, we further examined the influence of one signal on the other, here of coactivated NFκB activity on DIL and drug-induced apoptosis. We used parental CEM cells (Fig 3) and CEM-R cells, a derivative cell line resistant to doxorubicin-induced apoptosis and cross-resistant for DILs such as TRAIL (Fig 4). Both cell lines are resistant towards TNFα-induced apoptosis in the absence of cycloheximide. Treatment of CEM cells with LLnL in concentrations that block NFκB activation (compare Fig 2) strongly enhanced apoptosis induced by low doses of TRAIL (Fig 3A). This increase in apoptosis sensitivity was additionally found in the acute T-cell leukemia line Jurkat (Fig 3B), the Burkitt lymphoma line BJAB (Fig 3C), and in response to anti–APO-1, TNFα, and doxorubicin (data not shown). In contrast, BJAB cells stably overexpressing a dominant negative mutant FADD protein that are resistant towards TRAIL-induced apoptosis34-36 did not acquire sensitivity in the presence of LLnL, suggesting that the FADD-mediated signaling pathway is indepedent from NFκB (Fig 3C). To see more specifically whether the underlying mechanism of LLnL function is dependent on NFκB blockade, we used transient overexpression of a nondegradable IκBα mutant protein. Cotransfection with the gene for the GFP marker protein enabled separate forward side scatter FACScan analysis of transfected cells. Using this treatment, apoptosis induced by all three DILs was markedly enhanced in the presence of mutant IκBα in comparison to mock-transfected cells (Fig 3D and E).
Increase in apoptosis sensitivity of CEM cells by inhibition of NFκB activation. (A) CEM cells were incubated with TRAIL in the concentrations indicated in the presence (•) or absence (○) of LLnL (2.5 μmol/L, 1 hour of preincubation) for 24 hours. Apoptosis was measured using forward side scatter analysis in FACScan. Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in five independent experiments. (B) Jurkat cells were treated as in (A) using LLnL at 6.25 μmol/L. Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in two independent experiments. (C) BJAB cells, either mock-transfected (○, •) or overexpressing a dominant negative FADD mutant protein (□, ▪) were treated as in (A) using LLnL at 25 μmol/L. Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in two independent experiments. (D) CEM cells (3 × 107) were transfected with 20 μg of empty vector pPRB or pPRB containing the cDNA for mutant IκBα (serines on position 32 and 36 were replaced by alanines) and cotransfected with 2 μg of pEGFP. After 48 hours, living cells were separated by Ficoll gradient and stimulated with different concentrations of TRAIL for another 24 hours. Apoptosis was measured by forward side scatter analysis in FACScan cytometer gating on GFP-positive cells. Specific apoptosis was calculated using as control unstimulated, GFP-positive cells transfected with mock or IκBα, respectively. Data are the mean of triplicates with a standard deviation less than 10%. Similar results were obtained in two independent experiments. (E) CEM cells were transfected as in (D) and subsequentially stimulated with TRAIL (0.03 μg/mL), anti–APO-1 (0.3 μg/mL), or TNFα (0.3 μg/mL) for another 24 hours. Specific apoptosis was calculated as in (D). Data are the mean of triplicates with a standard deviation less than 10%. Similar results were obtained in three independent experiments. Similar results were obtained using Jurkat cells.
Increase in apoptosis sensitivity of CEM cells by inhibition of NFκB activation. (A) CEM cells were incubated with TRAIL in the concentrations indicated in the presence (•) or absence (○) of LLnL (2.5 μmol/L, 1 hour of preincubation) for 24 hours. Apoptosis was measured using forward side scatter analysis in FACScan. Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in five independent experiments. (B) Jurkat cells were treated as in (A) using LLnL at 6.25 μmol/L. Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in two independent experiments. (C) BJAB cells, either mock-transfected (○, •) or overexpressing a dominant negative FADD mutant protein (□, ▪) were treated as in (A) using LLnL at 25 μmol/L. Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in two independent experiments. (D) CEM cells (3 × 107) were transfected with 20 μg of empty vector pPRB or pPRB containing the cDNA for mutant IκBα (serines on position 32 and 36 were replaced by alanines) and cotransfected with 2 μg of pEGFP. After 48 hours, living cells were separated by Ficoll gradient and stimulated with different concentrations of TRAIL for another 24 hours. Apoptosis was measured by forward side scatter analysis in FACScan cytometer gating on GFP-positive cells. Specific apoptosis was calculated using as control unstimulated, GFP-positive cells transfected with mock or IκBα, respectively. Data are the mean of triplicates with a standard deviation less than 10%. Similar results were obtained in two independent experiments. (E) CEM cells were transfected as in (D) and subsequentially stimulated with TRAIL (0.03 μg/mL), anti–APO-1 (0.3 μg/mL), or TNFα (0.3 μg/mL) for another 24 hours. Specific apoptosis was calculated as in (D). Data are the mean of triplicates with a standard deviation less than 10%. Similar results were obtained in three independent experiments. Similar results were obtained using Jurkat cells.
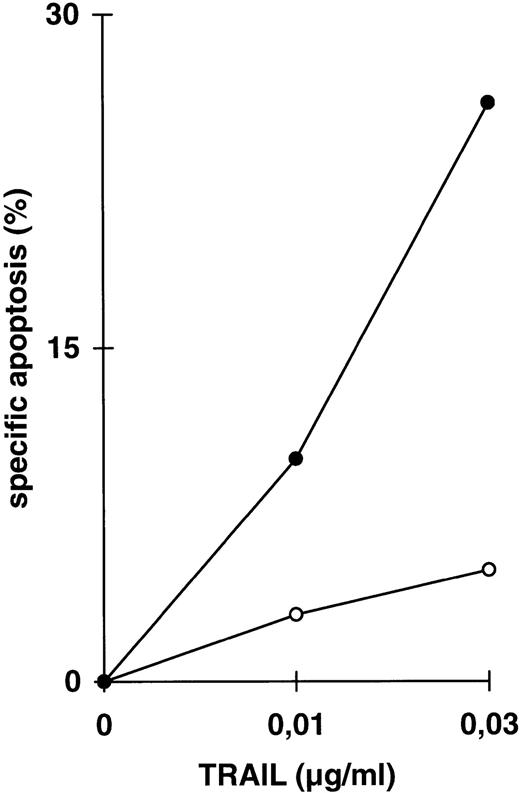
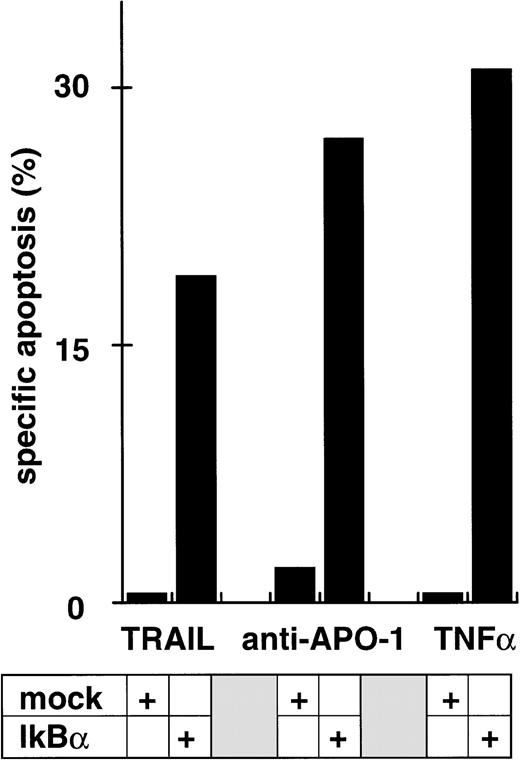
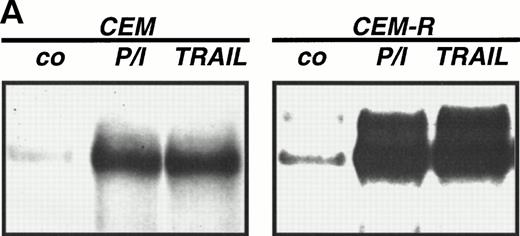
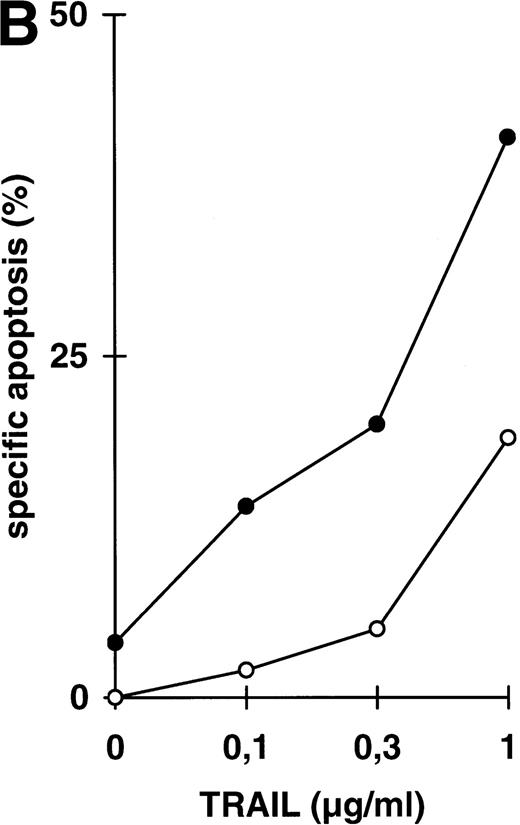
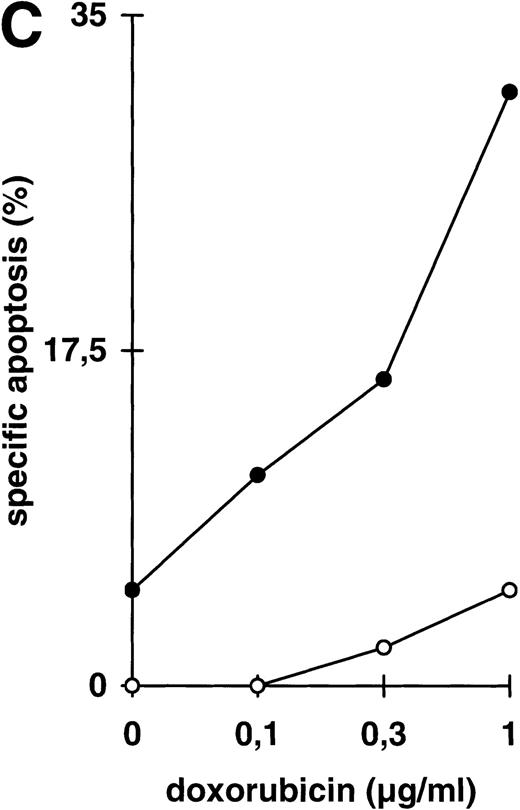
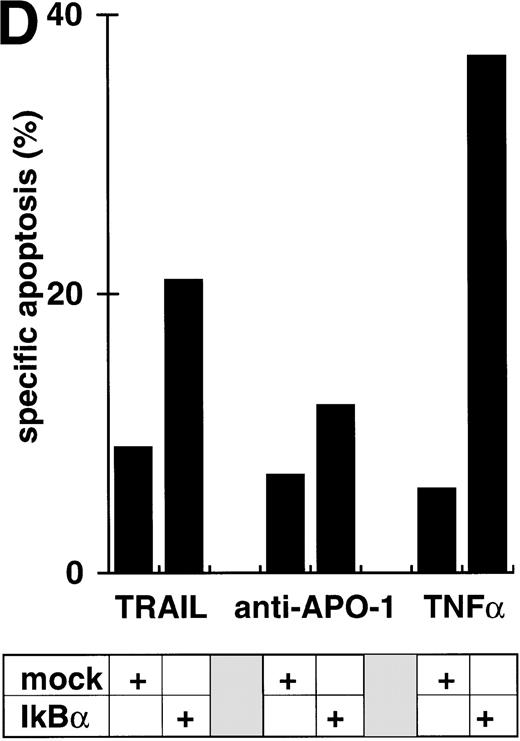
Abrogation of apoptosis resistance in CEM-R cells by inhibition of NFκB activation. (A) CEM and CEM-R cells were stimulated with TRAIL (0.3 μg/mL) or PMA (50 ng/mL)/Ionomycin (2 μg/mL) (P/I) for 30 minutes and EMSA was performed. Similar results were obtained in two independent experiments. (B) CEM-R cells were stimulated with TRAIL at the concentrations indicated in the presence (•) or absence (○) of LLnL (1.5 μmol/L, 1 hour of preincubation). After 12 hours, apoptosis was measured by forward side scatter analysis and specific apoptosis was calculated. Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in three independent experiments. (C) CEM-R cells were incubated with doxorubicin in concentrations indicated in the presence (•) or absence (○) of LLnL (1.5 μmol/L, 1 hour of preincubation) for 48 hours. Apoptosis was measured as in (A). Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in three independent experiments. (D) CEM-R cells (5 × 107) were transfected as in (Fig 3D). Stimulation was performed using TRAIL (1 μg/mL), anti–APO-1 (0.01 μg/mL), or TNFα (0.3 μg/mL) and apoptosis was measured after 12 hours of incubation. Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in two independent experiments.
Abrogation of apoptosis resistance in CEM-R cells by inhibition of NFκB activation. (A) CEM and CEM-R cells were stimulated with TRAIL (0.3 μg/mL) or PMA (50 ng/mL)/Ionomycin (2 μg/mL) (P/I) for 30 minutes and EMSA was performed. Similar results were obtained in two independent experiments. (B) CEM-R cells were stimulated with TRAIL at the concentrations indicated in the presence (•) or absence (○) of LLnL (1.5 μmol/L, 1 hour of preincubation). After 12 hours, apoptosis was measured by forward side scatter analysis and specific apoptosis was calculated. Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in three independent experiments. (C) CEM-R cells were incubated with doxorubicin in concentrations indicated in the presence (•) or absence (○) of LLnL (1.5 μmol/L, 1 hour of preincubation) for 48 hours. Apoptosis was measured as in (A). Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in three independent experiments. (D) CEM-R cells (5 × 107) were transfected as in (Fig 3D). Stimulation was performed using TRAIL (1 μg/mL), anti–APO-1 (0.01 μg/mL), or TNFα (0.3 μg/mL) and apoptosis was measured after 12 hours of incubation. Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in two independent experiments.
Abrogation of apoptosis resistance in CEM-R cells by inhibition of NFκB activation.
While inhibition of NFκB activation augments induction of apoptosis, we asked whether it would further affect apoptosis resistance. CEM-R cells are resistant towards apoptosis mediated by doxorubicin and certain DILs. In both cell lines, apoptosis-sensitive CEM cells and apoptosis-resistant CEM-R cells, TRAIL mediates activation of NFκB comparable to activation by PMA/Ionomycin (Fig 4A). Additionally, basal NFκB activity of unstimulated cells was not consistently different between CEM and CEM-R cells (data not shown). Using LLnL, CEM-R cells were clearly sensitized towards induction of apoptosis by high doses of TRAIL (Fig 4B), anti–APO-1, and TNFα (data not shown). Most importantly, LLnL also mediated sensitivity for induction of apoptosis by doxorubicin in these otherwise completely resistant cells (Fig 4C). Similarly, LLnL restored DIL and doxorubicin sensitivity in the apoptosis-resistant cell lines Jurkat-R and BOE (data not shown). Likewise, transient overexpression of mutant IκBα strongly increased apoptosis sensitivity, predominantly for TRAIL and TNFα (Fig 4D). These data suggest that inhibition of NFκB activation enables induction of apoptosis in otherwise resistant cell lines.
Attenuation of apoptosis resistance in primary leukemia cells by inhibition of NFκB activation.
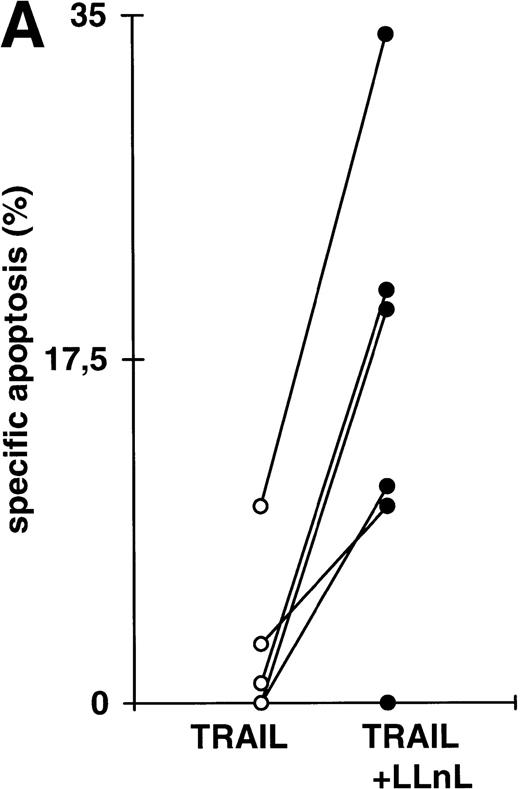
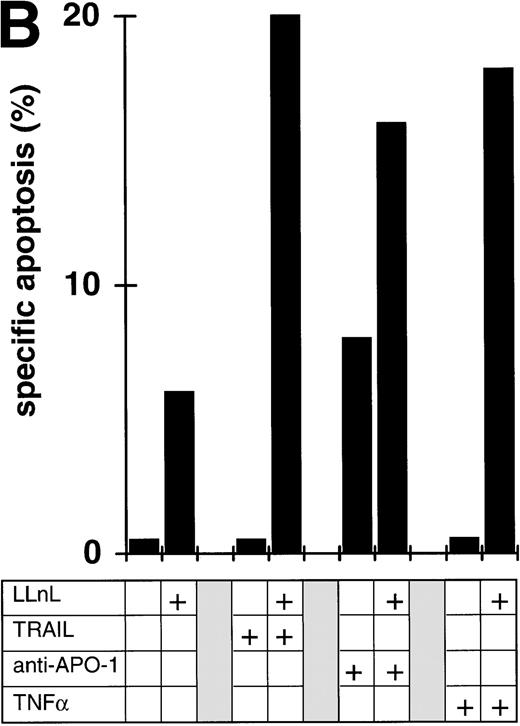
To see whether our findings obtained with cell lines would be relevant for primary tumor cells, we studied bone marrow cells from 6 patients with acute leukemias. Because of the increased spontaneous apoptosis, short-term incubations were performed. In each case, the experiment was terminated when the percentage of spontaneous apoptosis exceeded 35%. Leukemic cell from all patients displayed almost complete resistance towards induction of apoptosis by DILs ex vivo. As shown in Fig 5A, inhibition of NFκB activation by LLnL led to a strong increase in sensitivity for TRAIL induced apoptosis in 5 of 6 patient samples. In 2 patients, we further tested apoptosis induction by additional DILs. LLnL markedly increased apoptosis sensitivity triggered by CD95 or TNF-receptor I in both primary common ALL cells (Fig 5B) and primary AML cells (data not shown), suggesting that the influence of inhibition of NFκB activity on apoptosis resistance might be clinically relevant.
Attenuation of apoptosis resistance in primary leukemia cells by inhibition of NFκB activation. (A) Primary leukemia cells of 6 patients with acute leukemias were obtained from bone marrow at the time of diagnosis and separated by Ficoll gradient centrifugation. Cells were treated with TRAIL (1 μg/mL) in the presence or absence of LLnL (1 hour of preincubation, untoxic dosis, respectively, for each patient, 4 to 25 μmol/L) for 6 to 12 hours (spontaneous apoptosis <35%) and apoptosis was measured by forward side scatter analysis in FACScan flowcytometer. Data are the mean of duplicates with a standard deviation less than 10%. (B) Primary bone marrow leukemia cells of a patient with common ALL were obtained as in (A). Cells were stimulated with TRAIL (1 μg/mL), anti–APO-1 (1 μg/mL) in the presence of protein A (5 ng/mL), or TNFα (0,3 μg/mL) for 6 hours in the presence or absence of LLnL (4.25 μmol/L, 1 hour of preincubation), and apoptosis was measured as in (A). Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in two patients.
Attenuation of apoptosis resistance in primary leukemia cells by inhibition of NFκB activation. (A) Primary leukemia cells of 6 patients with acute leukemias were obtained from bone marrow at the time of diagnosis and separated by Ficoll gradient centrifugation. Cells were treated with TRAIL (1 μg/mL) in the presence or absence of LLnL (1 hour of preincubation, untoxic dosis, respectively, for each patient, 4 to 25 μmol/L) for 6 to 12 hours (spontaneous apoptosis <35%) and apoptosis was measured by forward side scatter analysis in FACScan flowcytometer. Data are the mean of duplicates with a standard deviation less than 10%. (B) Primary bone marrow leukemia cells of a patient with common ALL were obtained as in (A). Cells were stimulated with TRAIL (1 μg/mL), anti–APO-1 (1 μg/mL) in the presence of protein A (5 ng/mL), or TNFα (0,3 μg/mL) for 6 hours in the presence or absence of LLnL (4.25 μmol/L, 1 hour of preincubation), and apoptosis was measured as in (A). Data are the mean of duplicates with a standard deviation less than 10%. Similar results were obtained in two patients.
DISCUSSION
In the present study, we demonstrate that inhibition of NFκB activation augments DIL and drug-induced apoptosis and significantly decreases apoptosis resistance. DILs such as CD95-L and TNFα are known to activate NFκB independently of their cytotoxic function. We show here that TRAIL also activates NFκB in lymphoid cells. Activation of NFκB by TRAIL is independent from FADD, caspases, and induction of apoptosis, suggesting that the two intracellular signaling pathways initiated by TRAIL separate at an early stage close to the receptors and upstream from FADD.34-36 Activation of NFκB by TRAIL was not found in all lymphoid cell lines sensitive for TRAIL-induced apoptosis (data not shown). Likewise, cell type specificity of TRAIL-mediated NFκB activation has also been described in some, but not all solid tumor cell lines.25 27 However, all DILs identified so far are able to mediate at least two different signals in some cells, apoptosis and activation of NFκB.
We studied the influence of NFκB on induction of apoptosis using two different approaches to block NFκB activation. LLnL disables degradation of NFκB-inhibitory IκBα protein by the proteasome. Because inhibition of proteasome function itself may have potential side effects,37,38 we performed an extensive testing for the toxicity of LLnL. The cell-type–specific LLnL dosages used in our experiments were well beneath the cell-type specific toxicity limit of the drug (10 to 20 μmol/L, respectively). LLnL significantly increased induction of apoptosis by DILs and doxorubicin. To rule out that the increase in induction of apoptosis might be due to interference of proteasome inhibitor with the cell cycle,39a second molecular approach was performed using transfection of DNA encoding mutant IκBα protein. The mutant IκBα protein cannot be phosphorylated and subsequentially ubiquitinated, leading to inhibition of NFκB activity. Overexpression of mutant IκBα markedly increased induction of apoptosis, proving that NFκB activation interferes with DIL-induced apoptosis in lymphoid cells, eg, after stimulation with TRAIL. Similar results were obtained by transient overexpression of wild-type IκBα protein (data not shown).
Most importantly, inhibition of NFκB activation enabled apoptosis sensitivity for all DILs in otherwise apoptosis-resistant CEM-R cells. CEM-R cells were derived from the apoptosis-sensitive parental CEM cell line by continuous culture in doxorubicin for several months.40 CEM-R cells did not exhibit an MDR phenotype of drug resistance and were cross-resistant to apoptosis induction by several DILs. CEM-R cells showed a similar basal NFκB activity, similar p65 content (data not shown), and similar TRAIL-mediated NFκB activation compared with CEM cells, but a slightly increased sensitivity for LLnL (data not shown). In these cells, inhibition of NFκB activation significantly attenuated apoptosis resistance towards DILs and doxorubicin, predominantly for TRAIL and TNFα. Activation of NFκB has been found after treatment of tumor cells with cytostatic drugs.14,15 In cells cross-resistant for DIL and drug-induced apoptosis, inhibition of NFκB activation by LLnL sensitizes for both DILs and drugs. Recent evidence suggests that induction of apoptosis in target cells by cytostatic drugs may involve DILs such as the CD95 system.40-42 Drugs also increase cellular TRAIL mRNA and TNFα mRNA43 (Herr et al, manuscript submitted), suggesting a possible additional role for TRAIL and TNFα in drug-induced apoptosis. Because induction of apoptosis by anticancer drugs may involve DILs,40activation of NFκB after drug treatment may be a consequence of DIL function. Thus, LLnL might overcome drug resistance by sensitizing for DIL-induced apoptosis.
The concept that inhibition of NFκB activation increases apoptosis sensitivity was also investigated in primary tumor cells of patients with acute leukemias ex vivo. Although no specific apoptosis was induced using DIL alone, a significant increase in specific cell death could be obtained by TRAIL in the presence of LLnL in 5 of 6 patients. Recently, it was suggested that apoptosis resistance for TRAIL might be based on presence of two TRAIL receptors that contain an absent or truncated intracellular signal transducing death domain (TRAIL-R3, TRID, DcR1, LIT,26,27,30,31,44,45 and TRAIL-R4, DcR246,47). The distribution pattern of TRAIL-R3 suggested that normal, but not malignant cells were resistant towards TRAIL-induced apoptosis. In line with this, we found that normal peripheral T cells remain resistant towards TRAIL-induced apoptosis even after activation.32 Additionally, the majority of leukemic cell lines tested display a TRAIL-sensitive phenotype.32 48 However, all primary leukemia cells used here were almost completely resistant towards TRAIL-induced apoptosis. Because LLnL sensitizes patient cells for TRAIL-induced apoptosis, the molecular mechanism of apoptosis resistance seems to be more complex and might additionally involve intracellular signal proteins that may at least partially depend on NFκB activity.
The mechanism by which activation of NFκB antagonizes induction of apoptosis remains to be elucidated. Coincubation of TRAIL, CD95-L, or TNFα with an inhibitor of protein synthesis, eg, cycloheximide, also augments induction of apoptosis and attenuates apoptosis resistance (data not shown). Thus, de novo protein synthesis protects cells from DIL-induced cytotoxicity probably by induction of protective genes eventually mediated by NFκB. NFκB-dependent proteins might interfere with pathways leading to caspase activation and might participate in generating abundant cross-resistances between different apoptosis-inducing agents. Further studies are needed to detect the effectors of the NFκB activation signal as well as the therapeutic potential of interference with NFκB to overcome apoptosis resistance in clinical settings.
ACKNOWLEDGMENT
The authors thank C. Friesen for providing CEM-R and Jurkat-R cells and D. Suess, R. Zucic, and E. Musiol for excellent technical help.
Supported by Deutsche Forschungsgemeinschaft.
Address reprint requests to Prof Dr Klaus-Michael Debatin, University Childrens' Hospital, Prittwitzstr. 43, D-89075 Ulm, Germany.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal