Abstract
Osteoclasts are bone resorbing cells of hematopoietic origin; however, a progenitor cell population that gives rise to mature osteoclasts remains elusive. We have characterized a unique cell surface phenotype of clonogenic osteoclast progenitors (colony-forming unit–osteoclast [CFU-O]) and obtained a marrow cell population selectively enriched for these progenitors. Whole bone marrow cells were sequentially separated based on physical and cell surface characteristics, and the presence of CFU-O and other hematopoietic progenitors was examined. CFU-O was enriched in a nonadherent, low-density, lineage-marker–negative (Lin−), Thy1.2-negative (Thy1.2−), Sca1-negative (Sca1−), and c-kit–positive (c-kit+) population, as were the progenitors that were responsive to macrophage–colony-stimulating factor(CSF; CFU-M), granulocyte-macrophage-CSF (CFU-GM), and stem cell factor (CFU-SCF). When the Lin−Thy1.2−Sca1−population was divided into c-kithigh and c-kitlow populations based on c-kit fluorescence, over 88% of CFU-M, CFU-GM, and CFU-SCF were found in the c-kithighpopulation. In relation to the above mentioned hematopoietic progenitors, CFU-O was significantly higher in the c-kitlowpopulation: 80% of progenitors present in the c-kitlowpopulation were CFU-O. The CFU-O in both c-kithigh and c-kitlow populations showed key features of the osteoclast: multinucleated tartrate-resistant acid phosphatase–positive cell formation, expressions of vitronectin receptors, c-src and calcitonin receptors, and bone resorption. We have identified a progenitor cell population in the earliest stage of the osteoclast lineage so far described and developed a method to isolate it from other hematopoietic progenitors. This should help pave the way to understand the molecular mechanisms of osteoclast differentiation.
T HE HEMATOPOIETIC ORIGIN of osteoclasts has been clearly shown by a number of in vivo1-4 and in vitro studies5,6; however, the precise identity of osteoclast progenitors and their relationship to other hematopoietic progenitors are still controversial.7-10 Therefore, questions critical to the understanding of the cellular and molecular mechanisms of differentiation from hematopoietic stem cells to the osteoclast lineage still need to be addressed.
We have previously identified a distinctive population of colony-forming cells that, after being stimulated by conditioned medium of a bone-resorbing murine mammary carcinoma, gave rise to mononuclear cells expressing several key features of osteoclasts in murine bone marrow cultures.11 The factor responsible for this osteoclast colony formation, osteoclast colony stimulating factor (O-CSF), has been isolated and is currently undergoing further molecular characterization.12 Our previous functional studies of O-CSF–responsive osteoclast progenitor cells suggested that they were distinct from macrophage progenitors: O-CSF–responsive cells were relatively resistant to 5 fluorouracil treatment,11and they could survive for many days in a growth factor-free culture condition.13 These features were not observed for macrophage progenitors. In this investigation, we attempted to further characterize clonogenic osteoclast progenitors by studying their physical properties and expressions of stem-cell–associated cell surface molecules. The main purpose of this was to distinguish osteoclast progenitors from the hematopoietic progenitors that are responsive to macrophage colony-stimulating factor (M-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and stem cell factor (SCF). We have succeeded in identifying a fraction of bone marrow cells that are predominantly enriched for clonogenic osteoclast progenitors.
MATERIALS AND METHODS
Mice.
Female C57Black6 mice obtained from Jackson Laboratory (Bar Harbor, ME) were used as bone marrow donors at 8 to 13 weeks of age. They were housed in the vivarium of the University of Washington, and both the animal care and the experiments were conducted in accordance with the institutional guidelines approved by the National Institute of Health. Femurs and humeri were aseptically removed after the mice were sacrificed, and bone marrow cells were extracted by grinding the femurs and humeri as previously described.11 Whole bone marrow cells were suspended in Medium 199 (Bio-Whittaker, Walkersville, MD) containing 2% fetal calf serum (FCS; Hyclone Laboratories, Logan, UT).
Adherent cell depletion by Sephadex G10 column.
An adherent cell-depleted bone marrow cell suspension was obtained by passage through columns of Sephadex G10 based on a technique previously described.14 A whole bone marrow cell suspension, containing 107 to 108 cells, was applied to a 10-mL syringe containing 8 mL of washed and autoclaved Sephadex G10 (Pharmacia Biotech, Piscataway, NJ). After 45 minutes of incubation at 37°C, nonadherent cells were eluted with prewarmed Medium 199.
Density gradient.
Percoll solutions with specific densities of 1.10, 1.09, 1.07, and 1.06 g/mL in 0.15 mol/L NaCl were prepared according to the manufacturer's protocol (Pharmacia Biotech), and the gradient was formed by sequentially layering 2 mL of each solution in round-bottom polypropylene tubes. On top of the gradient, 2 mL of the nonadherent cell suspension containing 107 to 108 cells was layered. The gradient was centrifuged at 1,000g at 4°C for 25 minutes.15 Cells at the interfaces 1.06/1.07 g/mL and 1.07/1.09 g/mL were pooled and washed with cold 0.15 mol/L NaCl and then with phosphate-buffered saline (PBS) containing 2% FCS and 0.1% NaN3.
Magnetic bead depletion of lineage-positive cells.
Cells obtained with the previously explained procedures were first incubated with a cocktail of rat monoclonal antibodies (MoAbs) specific for murine B lymphocytes (B220), granulocytes (Gr-1; PharMingen, San Diego, CA), macrophages (Mac-1), and erythroid cells (YW25.12.716; a gift from Dr S.M. Watt, Imperial Cancer Research Fund, London, UK). After a 30-minute incubation on ice, the cells were washed with PBS containing 0.5% bovine serum albumin (BSA) and 5 mmol/L EDTA, resuspended, and counted. Goat antirat IgG beads (Miltenyi Biotech Inc, Sunnyvale, CA) were added at the concentration of 20 μL/107 cells then incubated in the refrigerator for 20 minutes, and finally washed twice with PBS/0.5% BSA/5 mmol/L EDTA. Cells negative for the above lineage markers (Lin−) were selected using MACS magnetic cell separation technology (Miltenyi Biotech Inc) according to the manufacturer's protocol.
Immunofluorescent staining and cell sorting.
The following MoAbs were used for cell surface labeling and cell sorting: anti–c-kit, anti-Thy1.2, and anti-Sca1 (PharMingen). The MoAbs were used either biotinylated or fluoresceinated. Biotinylated MoAb was detected with streptavidin-conjugated phycoerythrin (Caltag Laboratories, South San Francisco, CA). After being labeled with the above MoAbs, Lin− cells were sorted using FACStarplus (Becton Dickinson, Mountain View, CA). The following rat immunoglobulins were used as isotype controls: biotinylated IgG2b, fluoresceinated IgG2a, and fluoresceinated IgG2b (PharMingen).
Growth factor reagents.
A clonal cell line, CESJ3, derived from a hypercalcemia and granulocytosis-inducing murine mammary carcinoma12 was cultured in serum-free HL-1 medium (Bio-Whittaker) as a source of O-CSF. CESJ3 cells also have been shown to produce G-CSF and M-CSF, but not GM-CSF or interleukin-3 (IL-3).17 The culture supernatant was concentrated approximately 500-fold by ultrafiltration, filtered (0.22 μm), and stored in aliquots at −70°C. The conditioned medium of CESJ3 cells prepared in this manner was designated as CESJ medium and was used to stimulate osteoclast progenitors at the optimal concentration. Recombinant murine (rm) SCF and M-CSF were kindly provided by Dr S. Lyman (Immunex, Seattle, WA) and Dr L. Rohrschneider (Fred Hutchinson Cancer Research Center, Seattle, WA), respectively. rm GM-CSF, IL-3, and IL-6 were purchased from Genzyme (Cambridge, MA). The following concentrations of CSFs were used throughout these experiments: CESJ medium 5% vol/vol, rmM-CSF 240 U/mL, rmGM-CSF 50 ng/mL, rmSCF 50 ng/mL, rmIL-3 20 ng/mL, and rmIL-6 20 ng/mL.
Colony-forming unit (CFU) assays.
Progenitors were analyzed by colony formation in culture medium containing agar.11 Bone marrow cells were cultured in 15 × 10 mm Linbro wells (ICN Biomedicals Inc, Costa Mesa, CA) at 103 to 105 cells/mL, depending on the degree of fractionation, in supplemented Medium 199 containing 20% FCS and 0.3% Bacto agar (Difco Laboratories, Detroit, MI) in the presence of defined CSFs. In some experiments, agar was replaced by 0.25% agarose (FMC BioProducts, Rockland, ME). Agar cultures were incubated at 37°C in a humidified atmosphere with 5% CO2 for 14 days. The colonies that developed during the culture period were stained for tartrate-resistant acid phosphatase (TRAPase), a specific enzyme marker for the murine osteoclast, and then counterstained with Hemal blue.11 Colonies derived from osteoclast progenitors were identified by their composition of cells stained for TRAPase. The cells from these colonies expressing TRAPase activity were bright red, distinctive from the blue color of negative cells. Colonies, defined as groups of 50 or more cells, were examined under a microscope and were classified into three categories based on the percentage of red stained cells in a colony. TRAPase-positive colonies contained >90% positive cells, mixed colonies had 10% to 90% positive cells, and negative contained <10% positive cells.11 All colonies appearing in an agar plate were scored, and the results were expressed as colony numbers per unit of cell numbers. For practical reasons, progenitors that formed colonies in response to CESJ medium, M-CSF, GM-CSF, and SCF (plus IL-3 and IL-6) were designated as CFU-O, CFU-M, CFU-GM, and CFU-SCF, respectively.
Chamber slide cultures of fractionated cells.
Lin−Thy1.2−Sca1−c-kithigh(c-kithigh) or Lin−Thy1.2−Sca1−c-kitlow(c-kitlow) cells were cultured in Lab-Tek 4-well chamber slides (Nunc Inc, Naperville, IL) at 500 cells/well in supplemented Medium 199 containing 20% FCS in the presence of CESJ medium, M-CSF, or GM-CSF. The cultures were fed with the above culture medium containing CSFs twice a week and incubated for 14 days. On day 14, the cells that had attached to the chamber slides were fixed and stained for TRAPase.
Immunocytochemical analysis of osteoclast markers.
c-kithigh or c-kitlow cells were cultured in the presence of CESJ medium as described previously in CFU assays at 103 cells/mL in agarose medium using 35-mm Petri dishes. On day 14, individual colonies were sterilely lifted using an Eppendorf pipet under an inverted microscope and dispersed in 100 μL minimum essential medium-α (α-MEM; GIBCO, Grand Island, NY) containing 15% FCS. Some colonies were cytospotted onto glass slides. The other colonies were cocultured with ST2 cells,18 a bone marrow–derived stromal cell line (Riken Cell Bank, Tsukuba, Japan), in 4-well chamber slides for up to 14 additional days in the presence of 10–8 mol/L 1,25(OH)2D3 (Calbiochem, San Diego, CA) and 10–6 mol/L hydrocortisone (Upjohn, Kalamazoo, MI), replacing half of the culture medium with fresh medium twice a week. The cytospot slides and chamber slides at the indicated time were fixed with 1% formaldehyde for immunoperoxidase staining. A rabbit polyclonal antibody that recognizes both the αv and β3 subunits of the vitronectin receptor complex (GIBCO) was used at a 1:100 dilution (antivitronectin receptors [anti-VTR]). A MoAb against chicken pp60src (Oncogene Science Inc, Manhasset, NY) diluted in PBS containing 0.1% BSA and 0.05% Tween 20 was used at a 1:100 dilution (anti-src). B220 at a 1:10 dilution was used as a negative control. The slides were incubated with anti-VTR, anti-src, or anti-B220 overnight at 4°C followed by a 30-minute incubation with an appropriate biotinylated secondary antibody: antirabbit IgG, antimouse IgG, or antirat IgG (Southern Biotechnology, Birmingham, AL) at a 1:200 dilution. Expressions of VTR and c-src were detected using a Vectastain ABC kit (Vector Laboratories, Burlingame, CA) and Sigma Fast DAB (Sigma Chemical Co, St Louis, MO).
Expression of calcitonin receptors.
The cells from individual CFU-O–derived colonies in c-kithigh and c-kitlow populations were cocultured with ST2 cells in chamber slides in the presence of 1,25(OH)2D3 and hydrocortisone as described previously. At the indicated time, the cells in the chamber slides were exposed to 0.2 nmol/L 125I-calcitonin (Peninsula Laboratories, Belmont, CA) in Medium 199 containing 0.1% BSA for 1 hour at 22°C following the method previously described.19 Nonspecific binding was assessed by including an excess amount (300 nmol/L) of unlabeled calcitonin in certain chambers. After labeling, the slides were washed with PBS, fixed, and stained for TRAPase. Slides were then coated with NBT-2 nuclear emulsion (Kodak, Rochester, NY), exposed in the dark for 2 weeks at 4°C, developed, and counterstained. Cells were evaluated for TRAPase and calcitonin receptor expression using a light microscope.
Formation of resorption pits on dentine slices.
Sperm whale dentine slices (150 μm thick), prepared with a low speed diamond saw, were sterilized by ultraviolet irradiation for 30 min/side and placed into Lab-Tek 8-well chamber slides in which monolayers of ST2 cells were previously established. Slices were soaked overnight in α-MEM at 37°C. c-kithigh or c-kitlowcells were cultured as described previously in the presence of CESJ medium in agarose medium. On day 14, individual colonies were sterilely lifted, and the cell suspensions representing individual colonies were overlaid onto the dentine slices in chamber slides. Cells and dentine slices were incubated for 14 days in the presence of 10–8mol/L 1,25 (OH)2D3 and 10–6 mol/L hydrocortisone, replacing half of the culture medium with fresh medium twice a week. Resorption pits were visualized using a modification of a previously described method.20 The dentine slices were soaked in 50% bleach for 10 minutes and ultrasonicated for 1 minute to release cells attached to the slices. After being washed sequentially with deionized water and methanol, the slices were stained with 2% Coomassie brilliant blue (Sigma Chemical Co) in methanol and air dried. Excess Coomassie blue was removed by moistening the dentine slices with 1 mol/L NaOH and then scrubbing them with a smooth-surfaced paper. The dentine slices were examined for resorption pits using a dissecting microscope.
RESULTS
Stepwise enrichment of osteoclast progenitors from adult mouse bone marrow.
To determine whether osteoclast progenitors can be enriched in a selected cell population, we have separated whole bone marrow into different cell populations by sequential cell fractionation and examined the incidence of osteoclast, as well as other hematopoietic progenitors, in each population. Consistent with our previous observations,11 13 over 95% of CFU-O–derived colonies formed in response to CESJ medium were intensely TRAPase-positive whereas >99% of M-CSF–stimulated colonies were TRAPase-negative macrophage colonies. Approximately, 15% to 50% of GM-CSF–stimulated colonies were of TRAPase-positive and TRAPase-mixed types; the others were TRAPase-negative macrophage and/or granulocyte colonies. The combination of SCF, IL-3, and IL-6 stimulated a variety of multilineage hematopoietic colonies including a few (<10%) TRAPase-positive colonies. However, in contrast to the strong TRAPase positivity exhibited by CFU-O–derived colonies, the TRAPase reaction of colonies formed in response to GM-CSF or SCF cocktail was weak and appeared pale red. As the sequential separation steps proceeded, the incidence of CFU-O, as well as other hematopoietic progenitors, increased in fractionated bone marrow samples (Table 1). Approximately a 26-fold enrichment of CFU-O was achieved in a nonadherent, low-density, Lin− population that represented less than 1% (0.55%) of the original bone marrow sample. CFU-M, CFU-GM, and CFU-SCF were also enriched in this population, 29-fold, 35-fold, and 42-fold, respectively.
CFU Incidence of Sequentially Fractionated BM Cell Populations
| BM Population . | % of Whole BM . | CFU-O/105Cells . | CFU-M/105 Cells . | CFU-GM/105Cells . | CFU-SCF/105 Cells . |
|---|---|---|---|---|---|
| Whole BM | 100 | 78 ± 17 | 59 ± 13 | 50 ± 14 | 115 ± 12 |
| Nonadherent (nA) | 25.4 ± 6 | 166 ± 36 | 105 ± 27 | ND | 236 ± 29 |
| Low-density (nAlowD) | 15.1 ± 1.3 | 229 ± 11 | 177 ± 43 | ND | 361 ± 21 |
| High-density (nAhighD) | 1.5 ± 0.3 | 81 ± 39 | 103 ± 23 | ND | 153 ± 54 |
| nAlowDLin+(Lin+) | 6.1 ± 0.4 | 79 ± 25 | 33 ± 7 | 35 ± 10 | 50 ± 19 |
| nAlowDLin−(Lin−) | 0.55 ± 0.16 | 1,995 ± 480 | 1,746 ± 416 | 1,742 ± 469 | 4,780 ± 1,666 |
| BM Population . | % of Whole BM . | CFU-O/105Cells . | CFU-M/105 Cells . | CFU-GM/105Cells . | CFU-SCF/105 Cells . |
|---|---|---|---|---|---|
| Whole BM | 100 | 78 ± 17 | 59 ± 13 | 50 ± 14 | 115 ± 12 |
| Nonadherent (nA) | 25.4 ± 6 | 166 ± 36 | 105 ± 27 | ND | 236 ± 29 |
| Low-density (nAlowD) | 15.1 ± 1.3 | 229 ± 11 | 177 ± 43 | ND | 361 ± 21 |
| High-density (nAhighD) | 1.5 ± 0.3 | 81 ± 39 | 103 ± 23 | ND | 153 ± 54 |
| nAlowDLin+(Lin+) | 6.1 ± 0.4 | 79 ± 25 | 33 ± 7 | 35 ± 10 | 50 ± 19 |
| nAlowDLin−(Lin−) | 0.55 ± 0.16 | 1,995 ± 480 | 1,746 ± 416 | 1,742 ± 469 | 4,780 ± 1,666 |
A total of 2 × 103 to 1 × 105 cells/mL were stimulated with CESJ medium, M-CSF, GM-CSF, and SCF + IL-3 + IL-6. Incidence was obtained from the total number of colonies formed in response to each growth stimuli. The results represent mean values ± SD from duplicate wells in at least three independent experiments.
Abbreviations: CFU, colony-forming unit; BM, bone marrow; ND, not determined.
Analysis of stem cell-associated cell-surface markers expressed by progenitors in the Lin− population.
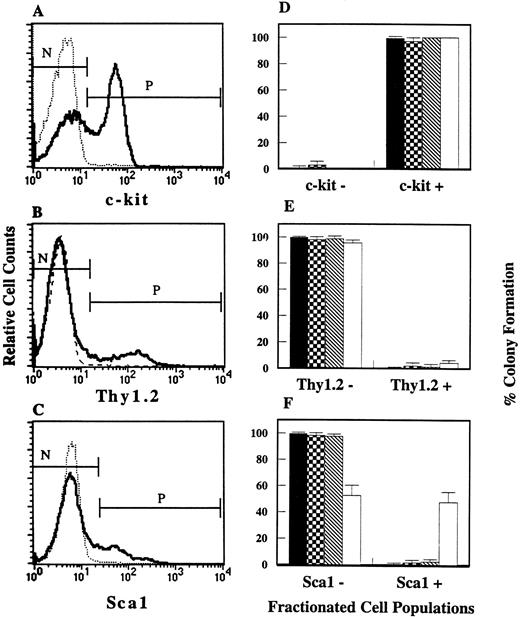
In our attempt to distinguish osteoclast progenitors from other hematopoietic progenitors, we examined the expressions of cell surface markers, c-kit, Thy1.2, and Sca1, on osteoclast progenitors. Lin− cells were stained with MoAbs to either c-kit, Thy1.2, or Sca1, and then each antigen positive or negative cell was sterilely sorted using FACStarplus. Fractionated cells were analyzed for progenitors. As shown in Fig1, over 99% of CFU-O were c-kit+, Thy1.2−, and Sca1−. CFU-M and CFU-GM showed similar cell surface phenotypes to CFU-O whereas CFU-SCF was c-kit+, Thy1.2−, but heterogeneous regarding Sca1 expression.
Incidence of CFU-O, CFU-M, CFU-GM, and CFU-SCF in FACS fractionated cell populations. Flow cytometric histograms of Lin− cells stained for (A) c-kit, (B) Thy1.2, and (C) Sca1 are shown on the left. The dotted lines indicate isotype controls. N and P indicate the negative and positive gates for cell collection. One or 2 × 103cells/mL from each cell fraction were stimulated with CESJ medium, M-CSF, GM-CSF, or SCF + IL-3 + IL-6 for progenitor analysis. Bar graphs on the right (D-F) represent colony formation from each selected cell population. Means and standard deviations (SDs) from duplicate wells of three independent experiments are shown. (▪) CFU-O; (▩) CFU-M; (▧) CFU-GM; (□) CFU-SCF.
Incidence of CFU-O, CFU-M, CFU-GM, and CFU-SCF in FACS fractionated cell populations. Flow cytometric histograms of Lin− cells stained for (A) c-kit, (B) Thy1.2, and (C) Sca1 are shown on the left. The dotted lines indicate isotype controls. N and P indicate the negative and positive gates for cell collection. One or 2 × 103cells/mL from each cell fraction were stimulated with CESJ medium, M-CSF, GM-CSF, or SCF + IL-3 + IL-6 for progenitor analysis. Bar graphs on the right (D-F) represent colony formation from each selected cell population. Means and standard deviations (SDs) from duplicate wells of three independent experiments are shown. (▪) CFU-O; (▩) CFU-M; (▧) CFU-GM; (□) CFU-SCF.
Enrichment of osteoclast progenitors based on the level of c-kit expression.
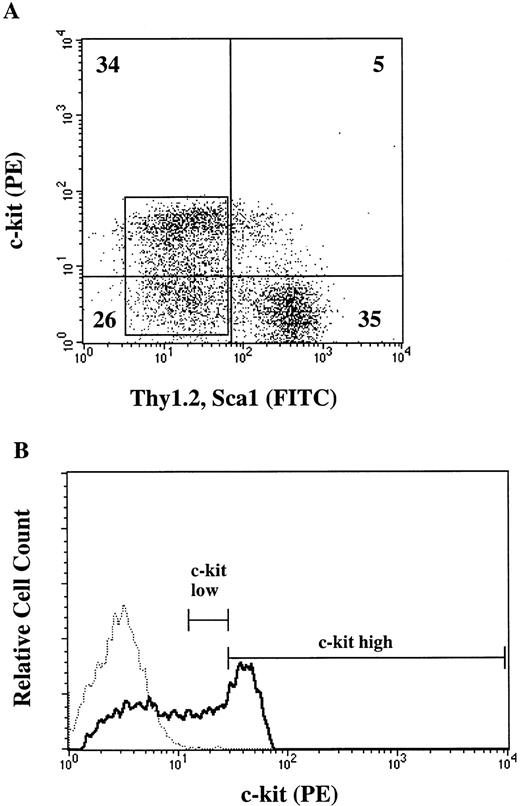
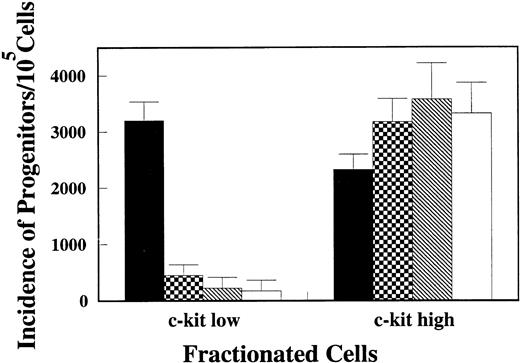
Because CFU-O appeared in the same populations as CFU-M, CFU-GM, and CFU-SCF, we attempted to isolate osteoclast progenitors by different degrees of c-kit expression. Lin− cells were again stained with c-kit, Thy1.2, and Sca-1 and analyzed by FACStarplus (Fig 2A). Thy1.2 and Sca1 negative (Lin−Thy1.2−Sca1−) cells were selected, and then the c-kit+ cells were arbitrarily divided into c-kithigh and c-kitlowgroups as shown in Fig 2B. c-kithigh cells represented 28.6% and c-kitlow cells 29.1% of the Lin−Thy1.2−Sca1−cell population. The two populations of cells were sorted and analyzed for progenitors. The majority (>88%) of CFU-M, CFU-GM, and CFU-SCF were found in the c-kithigh population. Interestingly, the c-kit expression in CFU-O was heterogeneous (Fig 3). Fifty seven percent of CFU-O was in the c-kitlow population and 43% in the c-kithigh population. As a result, CFU-O was further and selectively enriched in the c-kitlow population; 80% of the progenitors we examined in this population were osteoclast progenitors. In contrast, CFU-O constituted 23% of total progenitors in the c-kithigh population. Morphologically, colonies derived from CFU-O in the c-kitlow population were smaller than those derived from CFU-O in the c-kithigh population.
Expression of cell surface markers (c-kit, Thy1.2, and Sca1) on Lin− cells. (A) Two-color flow cytometric analysis of lin− cells stained for c-kit, Thy1.2, and Sca-1. Numbers represent percent of cells in the given areas. (B) Fluorescence histogram of Lin−Thy−Sca− cells (boxed area in A) analyzed for c-kit expression. The dotted line indicates an isotype control. Lines indicate the gate for the collection of c-kitlow and c-kithigh cells.
Expression of cell surface markers (c-kit, Thy1.2, and Sca1) on Lin− cells. (A) Two-color flow cytometric analysis of lin− cells stained for c-kit, Thy1.2, and Sca-1. Numbers represent percent of cells in the given areas. (B) Fluorescence histogram of Lin−Thy−Sca− cells (boxed area in A) analyzed for c-kit expression. The dotted line indicates an isotype control. Lines indicate the gate for the collection of c-kitlow and c-kithigh cells.
Incidence of CFU-O, CFU-M, CFU-GM, and CFU-SCF in c-kithigh and c-kitlow populations. One × 103cells/mL of c-kithigh and c-kitlow cells were plated in agar medium in the presence of CESJ medium, M-CSF, GM-CSF, and SCF + IL-3 + IL-6. Colonies developed during a 14-day culture period were evaluated. Values represent means and SDs of quadruple wells in a representative experiment. (▪) CFU-O; (▩) CFU-M; (▧) CFU-GM; (□) CFU-SCF.
Incidence of CFU-O, CFU-M, CFU-GM, and CFU-SCF in c-kithigh and c-kitlow populations. One × 103cells/mL of c-kithigh and c-kitlow cells were plated in agar medium in the presence of CESJ medium, M-CSF, GM-CSF, and SCF + IL-3 + IL-6. Colonies developed during a 14-day culture period were evaluated. Values represent means and SDs of quadruple wells in a representative experiment. (▪) CFU-O; (▩) CFU-M; (▧) CFU-GM; (□) CFU-SCF.
Liquid culture of CFU-O–enriched populations.
c-kithigh or c-kitlow cells were cultured in chamber slides in the presence of CESJ medium, M-CSF, or GM-CSF for 14 days, and the morphology of the cells attached to the slides was examined after being stained for TRAPase. TRAPase-positive multinucleated cells (more than three nuclei) were formed when either c-kithigh or c-kitlow cells were cultured in the presence of CESJ medium. However, cells from these same fractions cultured in the presence of M-CSF or GM-CSF did not yield any TRAPase-positive multinucleated cells (Table 2).
TRAPase-Positive Cell and Multinucleated Cell Formation From Fractionated Cells in the Presence of CESJ Medium, M-CSF, and GM-CSF
| . | % TRAPase-Positive Cells . | Multinucleated TRAPasePositive Cells/Well . | n . |
|---|---|---|---|
| O-CSF | 98.6 ± 3.1 | 5.8 ± 4.3 | 8 |
| M-CSF | 0 | 0 | 8 |
| GM-CSF | 3.2 ± 4.1 | 0 | 8 |
| . | % TRAPase-Positive Cells . | Multinucleated TRAPasePositive Cells/Well . | n . |
|---|---|---|---|
| O-CSF | 98.6 ± 3.1 | 5.8 ± 4.3 | 8 |
| M-CSF | 0 | 0 | 8 |
| GM-CSF | 3.2 ± 4.1 | 0 | 8 |
c-kithigh or c-kitlow cells (500 cells/well) were cultured in 4-well chamber slides in the presence of CESJ medium, M-CSF, or GM-CSF for 14 days. The cells in four random fields per chamber were enumerated as TRAPase-positive or -negative type. Whole fields were examined to identify TRAPase-positive multinucleated cells (>3 nuclei). Because the results for c-kithigh and c-kitlow cells were the same, n reflects the combined total wells.
Expression of osteoclast markers.
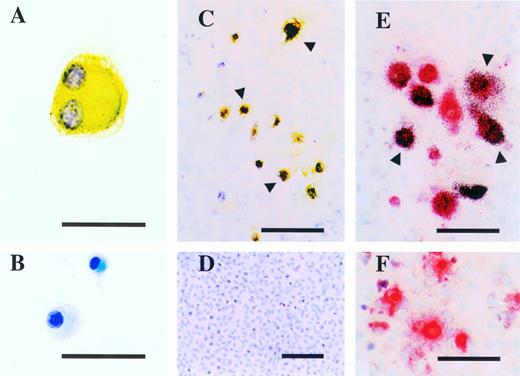
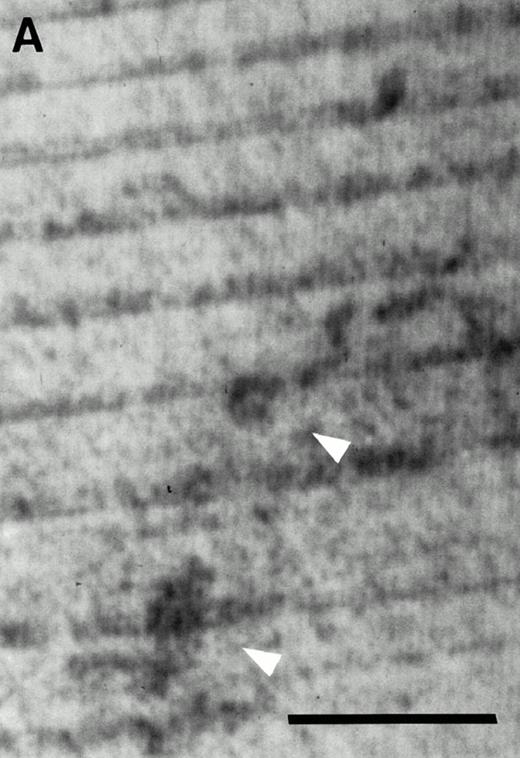
The cell surface phenotype of CFU-O–derived colonies from c-kithigh and c-kitlow populations was examined first on cytospot samples prepared on day 14 of culture in agarose. All CFU-O–derived colonies examined so far expressed αvβ3 vitronectin receptors (Fig 4A). The cells from individual CFU-O–derived colonies expressed additional osteoclast markers, c-src and calcitonin receptors, when these cells were further cultured on ST2 cells in the presence of 1,25(OH)2D3 and hydrocortisone for more than 7 days (Fig 4C and E).
Expression of osteoclast markers on CFU-O derived colony cells in the c-kitlow population. (A) Cytospot samples prepared from day-14 agarose cultures were stained for αvβ3 vitronectin receptors. Note a large multinucleated cell showing positive reaction to anti-VTR. (B) Negative control stained for B220. (C) Cells from individual CFU-O–derived colonies cocultured for 7 days with ST2 cells in the presence of 1,25(OH)2D3 and hydrocortisone and then stained with anti-src. Note a group of cells expressing c-src (arrowheads) over the monolayer of ST2 cells in the background. (D) No positive staining on ST2 cells that were reacted with anti-src. (E) Autoradiography of cells from individual CFU-O–derived colonies cocultured for 14 days with ST2 cells in the presence of 1,25(OH)2D3and hydrocortisone and then exposed to 125I-CT. Note a group of TRAPase-positive cells covered with dense silver grains (arrowheads) indicating the expression of calcitonin receptors. (F) Absence of 125I-CT binding to cells incubated with an excess amount of cold CT. Scale bars represent 50 μm (A and B), 100 μm (C, E, and F), and 150 μm (D).
Expression of osteoclast markers on CFU-O derived colony cells in the c-kitlow population. (A) Cytospot samples prepared from day-14 agarose cultures were stained for αvβ3 vitronectin receptors. Note a large multinucleated cell showing positive reaction to anti-VTR. (B) Negative control stained for B220. (C) Cells from individual CFU-O–derived colonies cocultured for 7 days with ST2 cells in the presence of 1,25(OH)2D3 and hydrocortisone and then stained with anti-src. Note a group of cells expressing c-src (arrowheads) over the monolayer of ST2 cells in the background. (D) No positive staining on ST2 cells that were reacted with anti-src. (E) Autoradiography of cells from individual CFU-O–derived colonies cocultured for 14 days with ST2 cells in the presence of 1,25(OH)2D3and hydrocortisone and then exposed to 125I-CT. Note a group of TRAPase-positive cells covered with dense silver grains (arrowheads) indicating the expression of calcitonin receptors. (F) Absence of 125I-CT binding to cells incubated with an excess amount of cold CT. Scale bars represent 50 μm (A and B), 100 μm (C, E, and F), and 150 μm (D).
Formation of resorption pits by CFU-O–enriched populations.
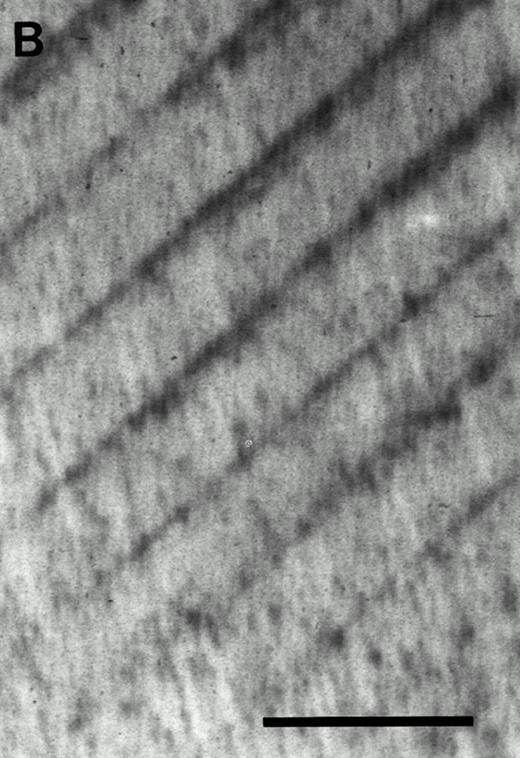
Cell suspensions from individual CFU-O–derived colonies formed in response to CESJ medium from c-kithigh or c-kitlow cells were cocultured with dentine slices in the presence of 1,25(OH)2D3 and hydrocortisone. As shown in Fig 5, resorption lacunae were observed when dentine slices were cocultured with cell suspensions from CFU-O–derived colonies in the presence of ST2 cells. Thus, it was confirmed that the isolated CFU-O could indeed give rise to functional osteoclasts.
Resorption pits formed by CFU-O–derived colonies in the c-kithigh cell population. (A) Cells from a colony developed in the presence of CESJ medium were cocultured with a dentine slice on ST2 cells for 14 days as described in the Materials and Methods section. Pits, indicated by arrowheads, were visualized by staining with Coomassie blue. (B) A control dentine slice incubated only with ST2 cells for 14 days. Multiple dark dots and lines seen throughout the specimen are dentinal tubules, whereas parallel regular lines are saw marks. Scale bars represent 100 μm (A and B).
Resorption pits formed by CFU-O–derived colonies in the c-kithigh cell population. (A) Cells from a colony developed in the presence of CESJ medium were cocultured with a dentine slice on ST2 cells for 14 days as described in the Materials and Methods section. Pits, indicated by arrowheads, were visualized by staining with Coomassie blue. (B) A control dentine slice incubated only with ST2 cells for 14 days. Multiple dark dots and lines seen throughout the specimen are dentinal tubules, whereas parallel regular lines are saw marks. Scale bars represent 100 μm (A and B).
DISCUSSION
Several studies involving bone marrow or spleen cell transplantation4,21 have indicated that osteoclasts are derived from hematopoietic cells. This concept was later confirmed by in vitro experiments in which periosteum-free bone rudiments were cocultured with a variety of hematopoietic cells as a source of osteoclasts.5,6 A study by Scheven et al22further showed the generation of osteoclasts from bone marrow fractions enriched for hematopoietic stem cells. Other investigators have supported the concept that osteoclasts are derived from immature hematopoietic cells capable of colony formation7,23-25 and from hematopoietic stem cell lines.26-28 However, the precise lineage of osteoclasts and their relationship to other hematopoietic cells are controversial. Studies have shown that CFU-G– and CFU-GM–, but not CFU-M–enriched bone marrow fractions, have given rise to osteoclasts in vivo and in vitro8,9; yet, these investigators did not distinguish whether CFU-G and CFU-GM themselves gave rise to osteoclasts or cells that were copurified with them were the progenitors of osteoclasts. Although the identification of osteoclast progenitors from granulocyte-macrophage7 or multilineage hematopoietic colony-forming cells23-25 has been attempted, only a fraction of cells from primary colonies gave rise to osteoclasts, and the identification of specific osteoclast progenitors was exceedingly difficult because the primary colonies did not exhibit any identifiable osteoclast characteristics.
Precise identification of the osteoclast progenitor has been difficult because of the lack of an osteoclast-specific growth factor. We have previously isolated a murine O-CSF, a soluble product of a bone resorbing tumor,12 and have established an in vitro colony assay system to identify clonogenic osteoclast progenitors.11 In this system, the great majority of the colonies formed in response to O-CSF are intensely TRAPase positive: a phenomenon not observed with any other hematopoietic growth factors, either alone or combined, so far examined. This osteoclast colony assay system allows us to examine previously unrevealed progenitors of the osteoclast in their early stages of development. Our recent13 and present studies have unveiled several unique characteristics of osteoclast progenitors.
In this study we have delineated osteoclast progenitors by their physical properties and expressions of cell surface molecules associated with hematopoietic stem cells. First, consistent with previous studies,22 26 osteoclast progenitors were found in a marrow cell population that had been enriched in immature hematopoietic progenitors: the nonadherent, low-density, and lineage marker-negative cell population.
Second, osteoclast progenitors expressed c-kit but did not have Thy1.2 and Sca1 on their cell surface. This is in contrast to hematopoietic stem cells that are c-kit– and Sca1-positive and do not express or express a low level of Thy1.2.29 Although c-kit was found in both stem cells and progenitors,30 cells that also expressed Sca1 are considered more primitive. In this regard our results were compatible: O-CSF–responsive osteoclast progenitors along with other hematopoietic progenitors were immature but not as primitive as Sca1-positive stem cells, which responded to SCF, IL-3, and IL-6 (Fig 1F). c-kit is a tyrosine kinase receptor for SCF, and c-kit–SCF interaction is essential for the survival and proliferation of hematopoietic stem cells and immature progenitors.31,32However, our previous study has shown that SCF is not essential for the differentiation of our osteoclast progenitors.13
Third, osteoclast progenitors were selectively enriched in the c-kitlow cell population in relation to other hematopoietic progenitors, the majority of which were found in the c-kithigh population. Previously, a dissection study of murine hematopoietic progenitors by c-kit expression has described c-kitlow cells as primitive, dormant, multipotent progenitors; c-kithigh cells as actively cycling progenitors; and c-kit very low or undetectable as most of the mature blood cells.33 Human primitive hematopoietic progenitors have also been enriched in c-kitlow cells.34 In this study, O-CSF responsive osteoclast progenitors were found in both c-kithigh and c-kitlow cell populations; however, they formed morphologically different colonies: c-kithigh CFU-O was more inclined to form large macroscopic TRAPase-positive colonies, whereas c-kitlow CFU-O formed small colonies (50 to 100 cells) and clusters (8 to 50 cells). One possible explanation for this is that c-kitlow cells are dormant and require additional factor(s) or a longer incubation period to form larger colonies. Alternatively, these cells may be more mature and may be losing their ability to proliferate. These questions need to be addressed in future studies.
Fourth, both c-kithigh and c-kitlow cell populations gave rise to TRAPase-positive multinucleated cells when cultured in the presence of CESJ medium without the addition of 1,25(OH)2D3. Cells from these same populations cultured in the presence of M-CSF or GM-CSF did not form such TRAPase-positive multinucleated cells. In a previous report,23 1,25(OH)2D3 was essential for induction of multinucleated cell formation and TRAPase activity from a population enriched for hematopoietic stem cells in the presence of IL-3 or GM-CSF. On the other hand, another study35suggested that 1,25(OH)2D3 was not required and, in fact, was inhibitory in the formation of TRAPase-positive cells from progenitors derived from murine long-term bone marrow cultures that were stimulated with M-CSF or GM-CSF. O-CSF–responsive progenitors do not require osteotropic hormones, such as 1,25(OH)2D3, for TRAPase-positive multinucleated cell formation. Furthermore, in our culture system, addition of 1,25(OH)2D3 did not have a significant effect on the formation of TRAPase-positive cells in the presence of CESJ medium, M-CSF, or GM-CSF (unpublished data). Such differences in the necessity of 1,25(OH)2D3could be explained by several factors, including culture conditions, cell sources, and/or differentiation stages of progenitors used for the studies.
Fifth, TRAPase-positive cells derived from individual CFU-O expressed other osteoclast markers, namely VTRs, c-src, and calcitonin receptors. VTR expression was observed in cells cultured in the presence of CESJ medium for 14 days; however, c-src and calcitonin receptor expression required an additional culture period on ST2 cells in the presence of 1,25(OH)2D3 and hydrocortisone. These results are consistent with current understandings that VTR expression precedes calcitonin receptor expression,36 and c-src expression is required in relatively late stages of osteoclast differentiation.37 The results also support the view that our osteoclast progenitor populations are still in an immature stage of osteoclast differentiation.
Finally, cells derived from isolated, individual CFU-O were able to form resorption pits, the definitive identification of the functional osteoclast, when they were cocultured with dentine slices in the presence of 1,25(OH)2D3, hydrocortisone, and stromal cells. These pits were relatively small and shallow, probably reflecting the immature nature of our osteoclasts. This finding confirms that our isolated CFU-O is, indeed, the osteoclast progenitor. Other studies conducted using multilineage colony-derived cells also have shown that small pits can be formed by osteoclasts.7,24 CFU-O–derived cells may require additional factors, such as osteoblastic cells or factors derived from osteoblastic cells, to fully differentiate into active functional osteoclasts as suggested by others.38 39
In conclusion, using a sequential cell separation, we have shown that osteoclast progenitors can be distinguished and isolated from other hematopoietic progenitors, thus strongly indicating the presence of previously unknown and unique progenitors of the osteoclast lineage. O-CSF–responsive progenitors appear to be distinct and belong to an earlier stage of osteoclast differentiation than those described previously. The method for osteoclast progenitor isolation established here will be useful for analyzing underlying molecular mechanisms of early stages of osteoclast differentiation.
ACKNOWLEDGMENT
We thank Kathy Allen for her excellent technical assistance with the cell sorting and Jody Lottsfeldt and Lynn Ferguson for their critical reading of this manuscript.
Supported in parts by grants from the National Institute of Health (AR-42657) and Ostex International Inc, Seattle, WA.
Address reprint requests to Minako Y. Lee, MD, Department of Biological Structure, 357420, University of Washington School of Medicine, Seattle, WA 98195-7420.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal