Abstract
AML1, a gene on chromosome 21 encoding a transcription factor, is disrupted in the (8;21)(q22;q22) and (3;21)(q26;q22) chromosomal translocations associated with myelogenous leukemias; as a result, chimeric proteins AML1/ETO(MTG8) and AML1/Evi-1 are generated, respectively. To clarify the roles of AML1/ETO(MTG8) and AML1/Evi-1 in leukemogenesis, we investigated subcellular localization of these chimeric proteins by immunofluorescence labeling and subcellular fractionation of COS-7 cells that express these chimeric proteins. AML1/ETO(MTG8) and AML1/Evi-1 are nuclear proteins, as is wild-type AML1. Polyomavirus enhancer binding protein (PEBP)2β(core binding factor [CBF]β), a heterodimerizing partner of AML1 that is located mainly in the cytoplasm, was translocated into the nucleus with dependence on the runt domain of AML1/ETO(MTG8) or AML1/Evi-1 when coexpressed with these chimeric proteins. When a comparable amount of wild-type AML1 or the chimeric proteins was coexpressed with PEBP2β(CBFβ), more of the cells expressing the chimeric proteins showed the nuclear accumulation of PEBP2β(CBFβ), as compared with the cells expressing wild-type AML1. We also showed that the chimeric proteins associate with PEBP2β(CBFβ) more effectively than wild-type AML1. These data suggest that the chimeric proteins are able to accumulate PEBP2β(CBFβ) in the nucleus more efficiently than wild-type AML1, probably because of the higher affinities of the chimeric proteins for PEBP2β(CBFβ) than that of wild-type AML1. These effects of the chimeric proteins on the cellular distribution of PEBP2β(CBFβ) possibly cause the dominant negative properties of the chimeric proteins over wild-type AML1 and account for one of the mechanisms through which these chimeric proteins contribute to leukemogenesis.
THE AML1 GENE was first identified as the gene on chromosome 21 that is disrupted in the (8;21)(q22;q22) translocation associated with acute myelogenous leukemia.1 In t(8;21)(q22;q22), the gene rearrangement results in the production of an AML1/ETO(MTG8) fusion protein.2,3 Previously we reported that the AML1gene is also disrupted and fused with the Evi-1 gene in the (3;21)(q26;q22) translocation associated with the blastic crisis of chronic myelogenous leukemia.4 Another group has also reported that the AML1 gene is rearranged in the (3;21)(q26;q22) translocation.5-7 Recently, it was reported that the AML1 gene is rearranged in acute lymphoblastic leukemia carrying t(12;21)(p12;q22).8,9PEBP2αB/CBFα2, which is a mouse homolog of AML1,was first identified as the gene encoding a member of the polyomavirus enhancer binding protein (PEBP) 2α family or a core binding factor (CBF) of Molony leukemia virus enhancer.10,11PEBP2α/CBFα and PEBP2β/CBFβ are components of the PEBP2/CBF heterodimer, which binds to the cores of polyomavirus and Molony leukemia virus enhancers.12,13 The mammalian PEBP2α/CBFα subunits are encoded by three distinct genes:AML1 (PEBP2αB/CBFα2), AML2(PEBP2αC/CBFα3), and AML3(PEBP2αA/CBFα1).1,10,11,14-16 A human homolog of PEBP2β/CBFβ is disrupted in inv(16)(p13q22) associated with acute myelogenous leukemia.17 These facts suggest critical roles of PEBP2/CBF in leukemogenesis.
AML1 has been shown to regulate the expression of several hematopietic lineage-specific genes, such as those for myeloperoxidase, leukocyte elastase,18 macrophage colony-stimulating factor (colony-stimulating factor 1) receptor,19,20granulocyte-macrophage colony-stimulating factor,21 and T-cell receptors.22-26 We have shown that AML1 regulates myeloid cell differentiation and transcriptional activation antagonistically by two alternative spliced forms, suggesting that a transactivation property of AML1 is necessary for myeloid cell differentiation.27 We also reported that the expression of AML1 increases before morphological and functional differentiation of U937 cells treated with all-trans retinoic acid.28It was recently shown that mice lacking AML1 die during midembryonic development because of extensive hemorrhaging and show the complete absence of definitive hematopoiesis.29 30 These findings suggest that AML1 contributes, by regulating the expression of target genes, to hematopoietic cell differentiation and proliferation.
Within the AML1 protein, there are two functional domains that have been identified. The runt domain, a 128-amino acids region of homology with the Drosophila runt protein,31 is known to be essential for DNA binding and heterodimerization with PEBP2β/CBFβ.16,32,33 AML1 specifically recognizes a consensus sequence, designated as a PEBP2 site (R/TACCRCA),33,34 whereas PEBP2β/CBFβ binds to AML1 and increases its affinity for DNA without interacting with DNA by itself.12 In our recent study, we showed that a conserved cysteine residue in the runt domain of AML1 is important for the DNA binding ability and the transforming ability.35 The proline-, serine-, threonine-rich (PST) region is essential for transcriptional activation,27,36 and this region is missing in the chimeric proteins AML1/ETO(MTG8) and AML1/Evi-1. Recently, we showed that AML1 is phosphorylated in vivo on two serine residues within the PST region with dependence on extracellular signal-regulated kinase (ERK) activation.37
Chimeric proteins generated as a result of chromosomal translocations should play causative roles in leukemogenesis. However, little is known about the mechanism for leukemic transformation in t(8;21) and t(3;21) leukemias. We and other groups have shown that AML1/ETO(MTG8) and AML1/Evi-1 dominantly suppress the functions of intact AML1 and inhibit myeloid cell differentiation.38-41 It is a useful approach for elucidation of the function of the chimeric proteins to study subcellular localization of leukemia-associated chimeric proteins and compare it with those of original wild-type proteins. For example, in t(15;17) acute promyelocytic leukemia, the alteration of subcellular localization of the wild-type protein (PML) by the chimeric protein (PML/retinoic acid receptor α [RARα]) plays an important role in leukemic transformation.42-44 In the present study, we investigated subcellular localization of AML1/ETO(MTG8) and AML1/Evi-1. Lu et al reported that PEBP2αA and PEBP2αB are nuclear proteins, whereas PEBP2β(CBFβ) is present mainly in the cytoplasm.45 Interestingly, they also reported that the N- or C-terminally truncated PEBP2αA colocalizes with PEBP2β(CBFβ) in the nucleus, in contrast to the full-size PEBP2αA, which does not colocalize with PEBP2β(CBFβ). To clarify the mechanism of leukemogenesis, it should be important to investigate how AML1/ETO(MTG8) and AML1/Evi-1 change the subcellular localization of PEBP2β(CBFβ). Using immunofluorescence and subcellular fractionation, we showed that these chimeric proteins are located in the nucleus. It was suggested that both the runt domain and the AML1/ETO(MTG8) or Evi-1 portion of the chimeric proteins are responsible for their nuclear localization. We also found that these chimeric proteins accumulate PEBP2β(CBFβ) in the nucleus with dependence on the runt domain. Then we showed that the chimeric proteins show the higher abilities to accumulate PEBP2β(CBFβ) in the nucleus than wild-type AML1. It was also shown that the chimeric proteins associate with PEBP2β(CBFβ) more effectively than wild-type AML1, implying the relationship between the binding affinity for PEBP2β(CBFβ) and the ability to accumulate it in the nucleus. These data suggest that AML1/ETO(MTG8) and AML1/Evi-1 exhibit dominant effects over wild-type AML1 owing to their efficient ability of nuclear accumulation of PEBP2β(CBFβ) and give some important clues for elucidation of transformation mechanism in leukemic cells expressing such chimeric proteins.
MATERIALS AND METHODS
Cell culture and DNA transfection.
COS-7 cells were grown in a 5% CO2 environment in Dulbecco's modified Eagle's medium (DMEM) supplemented with penicillin/streptomycin and 5% fetal calf serum. Plasmids were transfected by the DEAE-dextran method as described previously.41
Plasmid constructions.
The human AML1(AML1b)46 and AML1/Evi-1 cDNAs4were inserted into the EcoRI site of pME18S, an SRα promoter-driven expression plasmid,47 as described previously.27,41 The AML1/ETO(MTG8) cDNA was kindly provided by S.W. Hiebert (St Jude Children's Research Hospital, Memphis, TN).39 The mouse PEBP2β cDNA was kindly provided by Y. Ito (Kyoto University, Tokyo, Japan).12 TheEcoRI site was created by site-directed mutagenesis48 at positions 65 or 6 bp upstream from the AML1/ETO(MTG8) or PEBP2β translation initiation site, respectively, and these cDNAs were inserted into the EcoRI site of pME18S. The runt domain deletion mutants of AML1 (AML1ΔRD) and AML1/Evi-1 (AML1/Evi-1ΔRD) were constructed as described previously.41,49 For construction of the runt domain deletion mutant of AML1/ETO(MTG8) (AML1/ETOΔRD), theEcoRI-Apa I [519] (numbers in parentheses indicate nucleotide numbers from the start site of translation to the cutting site of the enzyme) fragment of AML1/ETO(MTG8) was replaced by the fragment from EcoRI to mutagenic Apa I [141] derived from AML1/Evi-1ΔRD. For tagging AML1, AML1/ETO(MTG8), or AML1/Evi-1 at the NH2-terminus, the influenza virus hemagglutinin (HA) epitope (YPYDVPDYA) was inserted after the first methionine codon by polymerase chain reaction (PCR).37 50 These HA-tagged cDNAs were inserted into pME18S.
Antibodies.
A rabbit polyclonal antibody to AML1 (anti-AML1 serum) was raised against maltose-binding protein fusion of AML1 as described previously.27 Polyclonal antibodies to PEBP2β (anti-PEBP2β serum) were obtained as follows. A glutathioneS-transferase (GST) fusion of PEBP2β was constructed by ligating a mutagenic EcoRI fragment of PEBP2β into theEcoRI site of the pGEX-2T vector (Pharmacia, Uppsala, Sweden). This construct was expressed in Escherichia coli BL21 cells, purified, and used to immunize a rabbit and hamsters as described previously.27
Immunofluorescent cell staining.
A total of 3 × 104 COS-7 cells per 24 mm × 24 mm coverslip were plated 12 hours before plasmid DNA transfection. The cells were incubated for 40 hours after transfection and then fixed and blocked as described.45 Briefly, the cells were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 20 minutes, permeabilized with 0.1% Nonidet P-40 in PBS for 10 minutes, and blocked in PBS containing 5% normal goat serum (GIBCO-BRL, Gaithersburg, MD) and 1 mg of bovine serum albumin fraction V (Sigma Chemical Co, St Louis, MO) per mL for 1 hour. Then the cells were incubated with anti-AML1 serum (1:1,000 in dilution) or hamster anti-PEBP2β serum (1:1,000 in dilution) for 1 hour, followed by incubation with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Zymed Laboratories, San Francisco, CA; 1:50 in dilution) or Texas Red-conjugated goat anti-hamster IgG (Cedarlane Laboratories, Ontario, Canada; 1:50 in dilution) for 1 hour. For double labeling, the mixture of anti-AML1 serum and hamster anti-PEBP2β serum was used as the primary antiserum and the mixture of FITC-conjugated goat anti-rabbit IgG, and Texas Red-conjugated goat anti-hamster IgG was used as the secondary antiserum. The cells were visualized under a Bio-Rad MRC 1024 confocal microscope (Bio-Rad Laboratories, Richmond, CA).
Subcellular fractionation and Western blotting.
Cells were homogenized in 500 μL of hypotonic suspension buffer (10 mmol/L sodium phosphate buffer [pH 7.0], 5 mmol/L EDTA, 1 mmol/L dithiothreitol [DTT], and 1 mmol/L phenylmethylsulfonyl fluoride [PMSF]) using Dounce homogenizer and separated into the nuclear (pellet) and cytoplasmic (supernatant) fractions by the centrifugation at 1,000g. One tenth of each fraction was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred onto polyvinylidene difluoride filters (Millipore Corp, Bedford, MA), then reacted with anti-AML1 serum (1:500 in dilution), HA. 11 anti-HA serum (BAbCO, Richmond, CA; 1:1,000 in dilution), or rabbit anti-PEBP2β serum (1:500 in dilution). To verify the purity of subcellular fractionation, the membranes were also reacted with the anti-actin monoclonal antibody (Boehringer Mannheim, Mannheim, Germany; 1:100 in dilution) or the anti-Rb monoclonal antibody (Pharmingen, San Diego, CA; 1:500 in dilution). The blots were visualized by Protoblot system (Promega, Madison, WI) or ECL blotting system (Amersham, Arlington Heights, IL).
In vitro binding assays.
The GST or GST-PEBP2β protein was expressed in E coli BL21 cells and purified as described previously.27 Approximately 5 μg of the GST or GST-PEBP2β protein immobilized on Glutathione Sepharose 4B (Pharmacia) was incubated with the lysates from COS-7 cells overexpressing HA-AML1, HA-AML1/ETO(MTG8), or HA-AML1/Evi-1 in lysis buffer (50 mmol/L Tris-HCl [pH 8.0], 150 mmol/L NaCl, 0.02% sodium azide, 1% Triton X-100, 100 μg of PMSF per mL, and 1 μg of aprotinin per mL) for 1 hour at 4°C with gentle rotation. The protein-GST beads were washed four times with the lysis buffer and subjected to SDS-PAGE.
Immunoprecipitation and metabolic labeling.
COS-7 cells cultured for 40 hours after transfection were obtained in 250 μL of the lysis buffer (the same buffer as we used for in vitro binding assay) per 100-mm-diameter culture dish. For immunoprecipitations, 1 mg of cell lysates per lane were mixed with 20 μL of anti-AML1 serum and rotated for 1 hour at 4°C. Then samples were incubated with protein-A-Sepharose (Sigma) for 1 hour at 4°C, followed by washing three times with lysis buffer. Immunoprecipitates were subjected to SDS-PAGE and Western-blotting with anti-AML1 serum or rabbit anti-PEBP2β serum. For metabolic labeling, COS-7 cells were cultured for 35 hours after transfection, transferred to and cultured for 4 hours in methionine-free DMEM plus 50 μCi of [35S]methionine (Tran-35S label; ICN Pharmaceuticals Inc, Costa Mesa, CA) per mL, lysed, and immunoprecipated with 5 μL (0.4 mg/mL) of 12CA5 anti-HA monoclonal antibody (Boehringer Mannheim) or 5 μL of rabbit anti-PEBP2β serum. Immunoprecipitates were analyzed by SDS-PAGE and autoradiography. All transfection experiments were performed at least twice and similar results were obtained.
RESULTS
Subcellular localization of AML1/ETO(MTG8) and AML1/Evi-1.
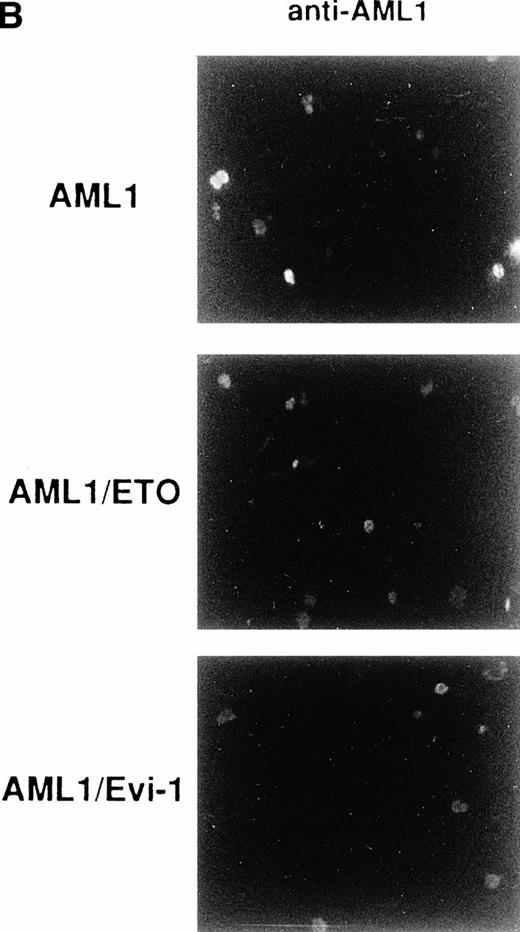
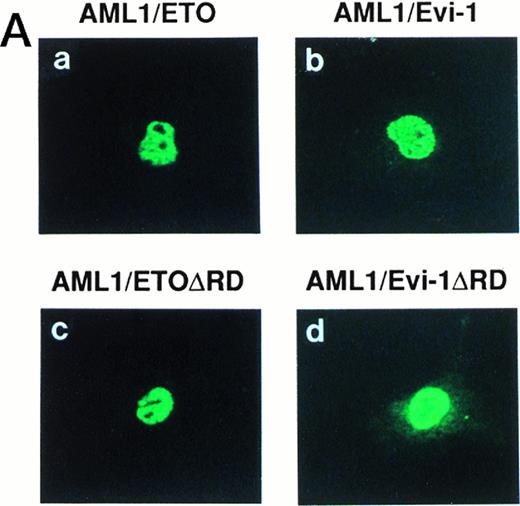
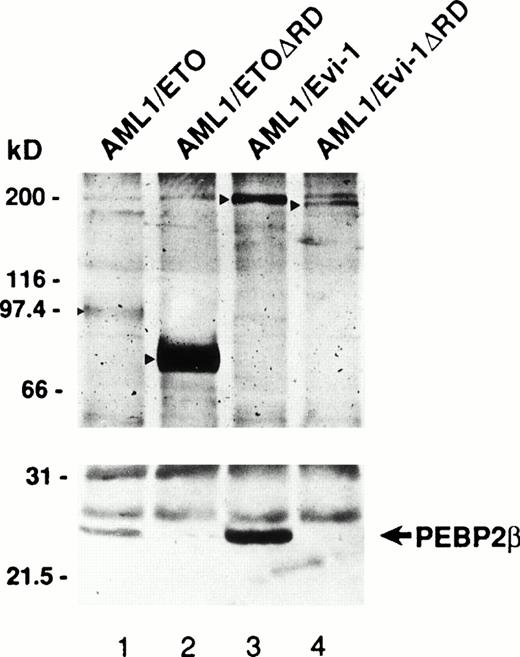
Generation of the chimeric proteins AML1/ETO(MTG8) and AML1/Evi-1 is supposed to be essential for leukemic transformation in t(8;21) and t(3;21) leukemias, respectively, and the investigation of their cellular distributions should give an important clue to elucidate the mechanism of leukemogenesis. We thus studied the subcellular localization of these chimeric proteins by indirect immunofluorescence using a rabbit polyclonal antibody to AML1 (anti-AML1) serum and FITC-conjugated anti-rabbit IgG. By Western blotting with anti-AML1 serum, the 55-kD protein was specifically detected in COS-7 cells transfected with the expression plasmid for AML1 (Fig 1A, lanes 1 to 4), as described in our other reports.25,35,37,49 The endogenous AML1 was not detected. There were two bands corresponding to AML1, and the upper band was considered to represent the phosphorylated form, as we described previously.37 In Kasumi-1 and SKH1 cell lines endogenously expressing AML1/ETO(MTG8) and AML1/Evi-1, respectively, we could not detect these chimeric proteins by immunofluorescence probably because their expression levels are too low to be detected using this antiserum (data not shown). Therefore, we overexpressed these chimeric proteins in COS-7 cells to study their subcellular localization. The endogenous AML1 proteins in COS-7 cells were below the detectable level, because COS-7 cells transfected with the empty pME18S vector were not stained with anti-AML1 serum (Fig 1B[c], see page 1690). COS-7 cells were transfected with the expression vectors for the proteins listed in Fig2 and investigated by immunofluorescence. As reported previously, wild-type AML1 was present exclusively in the nucleus (Fig 1B [d]).45 The specificity of the staining was confirmed by using preimmune serum (Fig 1B[a and b]). Two chimeric proteins, AML1/ETO(MTG8) and AML1/Evi-1, were also located in the nucleus (Fig 3A [a and b]). When observed in lower magnification, wild-type AML1 and the chimeric proteins were located in the nucleus in more than 95% of the cells successfully transfected with the plasmids (Fig 3B). The rest of the cells were slightly stained diffusely throughout the cell. In accordance with these results, Sacchi et al reported that AML1/ETO(MTG8) is detectable in the nucleus of Kasumi-1 cells by immunofluorescence labeling using antiserum that recognizes ETO(MTG8).51 Because the runt domain is known to be responsible for the nuclear localization of PEBP2αA,45 we examined whether the nuclear localization of these chimeric proteins is dependent on the runt domain. The deletion mutant of AML1 lacking the runt domain showed a perinuclear accumulation with a weak nuclear fluorescence, which is compatible with the previous report (data not shown).45 On the other hand, the mutant chimeric proteins lacking the runt domain still remained mainly in the nucleus (Fig 3A [c and d]). These data suggest that the ETO(MTG8) and Evi-1 portions of the chimeric proteins play some roles in the nuclear localization of the chimeric proteins.
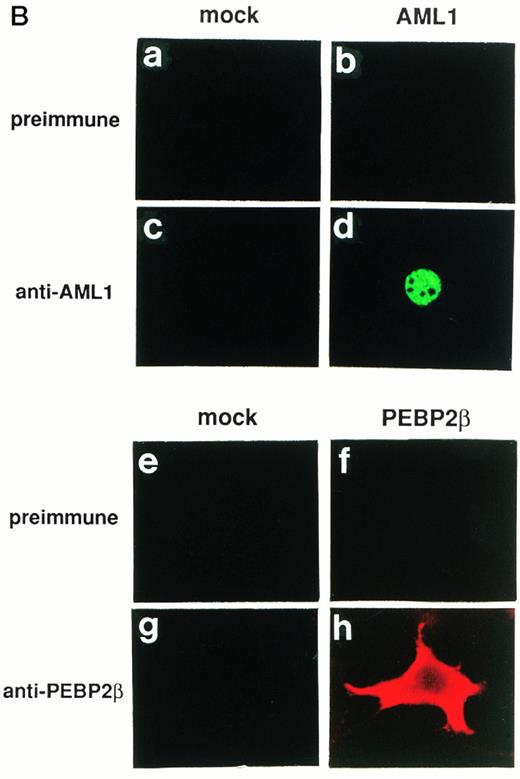
(A) Specificities of antibodies as revealed by Western blotting. A total of 3 × 104 COS-7 cells were transfected with 4 μg of pME18S (lanes 1, 3, 5, 7, 9, and 11), expression plasmid for AML1 (lanes 2 and 4), or that for PEBP2β (lanes 6, 8, 10, and 12), lysed, and subjected to Western blotting. The blots were probed with anti-AML1 serum (lanes 3 and 4), rabbit anti-PEBP2β serum (lanes 7 and 8), hamster anti-PEBP2β serum (lanes 11 and 12), as well as with the respective preimmune sera (lanes 1 and 2, 5 and 6, and 9 and 10, respectively). The AML1 or PEBP2β protein is indicated by the arrowhead. Molecular weight standards (in kilodaltons) are indicated. (B) (see page 1690) Specificities of antibodies as revealed by immunofluorescence. A total of 3 × 104 COS-7 cells were transfected with 4 μg of pME18S (a, c, e, and g), expression plasmid for AML1 (b and d), or that for PEBP2β (f and h). The cells were analyzed by immunofluorescence labeling with anti-AML1 serum (c and d) or hamster anti-PEBP2β serum (g and h) as well as with the respective preimmune sera (a and b, and e and f, respectively). Original magnification × 600.
(A) Specificities of antibodies as revealed by Western blotting. A total of 3 × 104 COS-7 cells were transfected with 4 μg of pME18S (lanes 1, 3, 5, 7, 9, and 11), expression plasmid for AML1 (lanes 2 and 4), or that for PEBP2β (lanes 6, 8, 10, and 12), lysed, and subjected to Western blotting. The blots were probed with anti-AML1 serum (lanes 3 and 4), rabbit anti-PEBP2β serum (lanes 7 and 8), hamster anti-PEBP2β serum (lanes 11 and 12), as well as with the respective preimmune sera (lanes 1 and 2, 5 and 6, and 9 and 10, respectively). The AML1 or PEBP2β protein is indicated by the arrowhead. Molecular weight standards (in kilodaltons) are indicated. (B) (see page 1690) Specificities of antibodies as revealed by immunofluorescence. A total of 3 × 104 COS-7 cells were transfected with 4 μg of pME18S (a, c, e, and g), expression plasmid for AML1 (b and d), or that for PEBP2β (f and h). The cells were analyzed by immunofluorescence labeling with anti-AML1 serum (c and d) or hamster anti-PEBP2β serum (g and h) as well as with the respective preimmune sera (a and b, and e and f, respectively). Original magnification × 600.
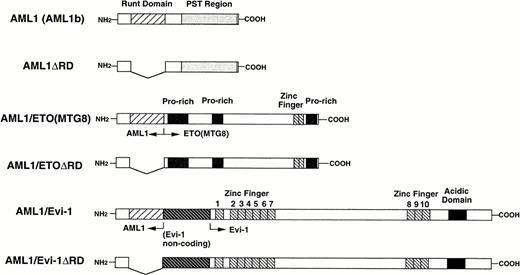
A schematic representation of full-length or mutant AML1, AML1/ETO(MTG8), and AML1/Evi-1. The runt domain and the PST region (see text) of AML1 (also called AML1b) are shown by striped and dotted boxes, respectively. Other regions of AML1/ETO(MTG8) (proline-rich regions and the zinc finger domain) and AML1/Evi-1 (the noncoding exon of Evi-1, zinc finger domains and the acidic domain) are indicated.
A schematic representation of full-length or mutant AML1, AML1/ETO(MTG8), and AML1/Evi-1. The runt domain and the PST region (see text) of AML1 (also called AML1b) are shown by striped and dotted boxes, respectively. Other regions of AML1/ETO(MTG8) (proline-rich regions and the zinc finger domain) and AML1/Evi-1 (the noncoding exon of Evi-1, zinc finger domains and the acidic domain) are indicated.
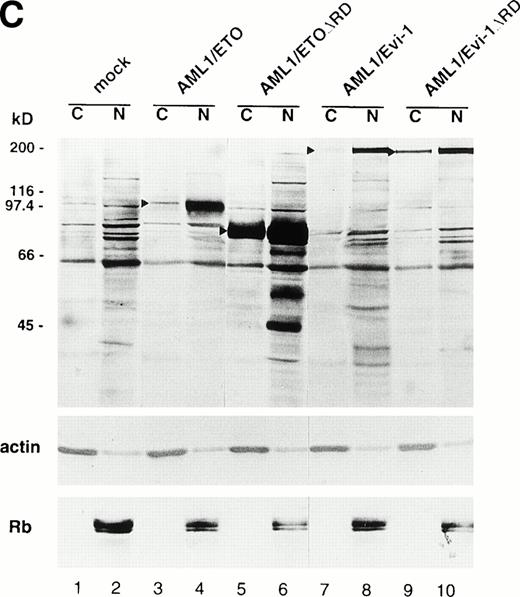
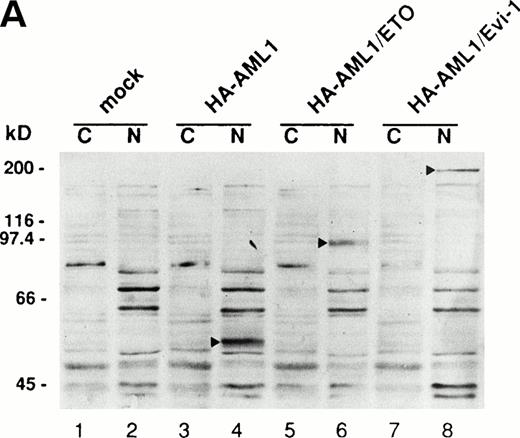
(A) (see page 1690) Subcellular localization of full-length or mutant AML1/ETO(MTG8) and AML1/Evi-1. A total of 3 × 104 COS-7 cells were transfected with 4 μg of each construct as indicated and analyzed by immunofluorescence labeling with anti-AML1 serum. Original magnification × 600. (B) The staining pattern of the cells overexpressing AML1 (upper panel), AML1/ETO(MTG8) (middle panel), or AML1/Evi-1 (lower panel) in lower magnification (× 100). (C) Identification of full-length or mutant AML1/ETO(MTG8) and AML1/Evi-1 in cytoplasmic (lanes C) and nuclear (lanes N) fractions of COS-7 cells. COS-7 cells were transfected with the same amount of each construct as indicated in (A), lysed, fractionated, and subjected to SDS-PAGE and immunoblotting using anti-AML1 serum. Each protein was expressed at the anticipated size, as marked by the arrowhead. The cell lysate of untransfected COS-7 cells was also analyzed as a control (mock). Western blotting of the actin and Rb proteins are shown as known cytoplasmic and nuclear proteins, respectively. Molecular weight standards (in kilodaltons) are indicated.
(A) (see page 1690) Subcellular localization of full-length or mutant AML1/ETO(MTG8) and AML1/Evi-1. A total of 3 × 104 COS-7 cells were transfected with 4 μg of each construct as indicated and analyzed by immunofluorescence labeling with anti-AML1 serum. Original magnification × 600. (B) The staining pattern of the cells overexpressing AML1 (upper panel), AML1/ETO(MTG8) (middle panel), or AML1/Evi-1 (lower panel) in lower magnification (× 100). (C) Identification of full-length or mutant AML1/ETO(MTG8) and AML1/Evi-1 in cytoplasmic (lanes C) and nuclear (lanes N) fractions of COS-7 cells. COS-7 cells were transfected with the same amount of each construct as indicated in (A), lysed, fractionated, and subjected to SDS-PAGE and immunoblotting using anti-AML1 serum. Each protein was expressed at the anticipated size, as marked by the arrowhead. The cell lysate of untransfected COS-7 cells was also analyzed as a control (mock). Western blotting of the actin and Rb proteins are shown as known cytoplasmic and nuclear proteins, respectively. Molecular weight standards (in kilodaltons) are indicated.
To confirm these results obtained by immunofluorescence, we also investigated subcellular localization of AML1/ETO(MTG8) and AML1/Evi-1 by a combination of subcellular fractionation and Western blotting. Cell lysates were fractionated into cytoplasmic and nuclear fractions, and the chimeric proteins were detected by anti-AML1 serum. The majority of the control proteins, actin and Rb, were detected in the cytoplasmic and nuclear fractions, respectively (Fig 3C). AML1/ETO(MTG8) was found predominantly in the nucleus as previously shown (Fig 3C, lanes 3 and 4).39 AML1/Evi-1 was also present predominantly in the nucleus (Fig 3C, lanes 7 and 8). On the other hand, a certain fraction of each deletion mutant lacking the runt domain was present in the cytoplasm (Fig 3C, lanes 5, 6, 9, and 10), indicating that the runt domain is partially required for the nuclear localization of the chimeric proteins. The difference of subcellular localization between AML1/ETO(MTG8) or AML1/Evi-1 and each deletion mutant was not clear in immunofluorescence, probably because of the sensitivity of the immunostaining.
AML1/ETO(MTG8) and AML1/Evi-1 accumulate PEBP2β in the nucleus with dependence on the runt domain.
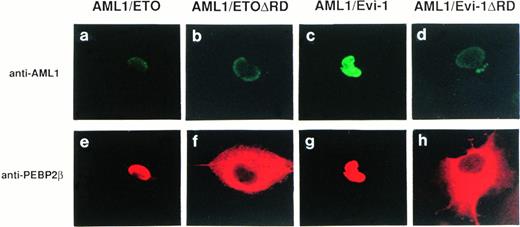
PEBP2β, a heterodimerizing partner of PEBP2α, intensifies the DNA binding ability of PEBP2α.12 However, PEBP2β is present mainly in the cytoplasm when it is overexpressed solely, and does not colocalize with PEBP2αA.45 Interestingly, the truncated PEBP2αA protein, devoid of the region either upstream or downstream from the runt domain, colocalizes with PEBP2β in the nucleus.45 The study on the cellular distribution of PEBP2β in association with that of AML1/ETO(MTG8) or AML1/Evi-1 will give a good clue to reveal the role of the chimeric proteins in leukemogenesis. Therefore, we examined subcellular localization of PEBP2β and the effects of AML1/ETO(MTG8) and AML1/Evi-1 on it. The antibodies against PEBP2β (anti-PEBP2β serum) were raised in a rabbit and hamsters. The reactivities of the antibodies were examined by Western blotting, and they specifically detected the 23-kD protein in COS-7 cells transfected with the expression plasmid for PEBP2β (Fig 1A, lanes 5 to 12). The endogenous PEBP2β in COS-7 was below the level of detection. AML1/ETO(MTG8) or AML1/Evi-1 was coexpressed with PEBP2β in COS-7 cells, and subcellular localization of each protein was determined by double staining. The chimeric proteins were detected by anti-AML1 serum and FITC-conjugated anti-rabbit IgG. PEBP2β was detected by hamster anti-PEBP2β serum and Texas Red-conjugated anti-hamster IgG. We could not evidently detect an endogenous PEBP2β protein in COS-7 cells transfected with the empty pME18S vector by immunofluorescence probably because of its low expression level (Fig 1B [g]). In 100% of the cells overexpressing PEBP2β alone, it was mainly located in the cytoplasm (Fig 1B [h]), in accordance with the previous reports.45 52 The specificity of the staining was confirmed by using preimmune serum (Fig 1B [e and f]). When AML1/ETO(MTG8) or AML1/Evi-1 was coexpressed with PEBP2β, PEBP2β colocalized with the chimeric proteins in the nucleus (Fig 4 [e and g], see page 1690). We also examined the effects of the deletion mutants of the chimeric proteins lacking the runt domain on the localization of PEBP2β. Although such deletion mutants of the chimeric proteins are located mainly in the nucleus, the deletion of the runt domain almost completely abolished their abilities to translocate PEBP2β into the nucleus (Fig 4 [f and h]). As mentioned above, the deletion mutant of AML1 devoid of the runt domain is distributed throughout the cell. When coexpressed with this mutant, PEBP2β remained in the cytoplasm (data not shown). These data indicate that AML1/ETO(MTG8) and AML1/Evi-1 have the abilities to accumulate PEBP2β in the nucleus, and these abilities are dependent on the runt domain.
Double fluorescence labeling of COS-7 cells transfected with the constructs of full-length or mutant chimeric proteins and PEBP2β. A total of 3 × 104 COS-7 cells were transfected with 4 μg of each construct together with 0.4 μg of the expression plasmid for PEBP2β. (a, b, c, and d) The chimeric proteins were detected with anti-AML1 serum. (e, f, g, and h) PEBP2β was detected with hamster anti-PEBP2β serum. Original magnification × 600.
Double fluorescence labeling of COS-7 cells transfected with the constructs of full-length or mutant chimeric proteins and PEBP2β. A total of 3 × 104 COS-7 cells were transfected with 4 μg of each construct together with 0.4 μg of the expression plasmid for PEBP2β. (a, b, c, and d) The chimeric proteins were detected with anti-AML1 serum. (e, f, g, and h) PEBP2β was detected with hamster anti-PEBP2β serum. Original magnification × 600.
AML1/ETO(MTG8) and AML1/Evi-1 associate with PEBP2β with dependence on the runt domain.
The runt domain of AML1 or PEBP2α was reported to be responsible for the heterodimerization with PEBP2β.16,32 33 As both AML1/ETO(MTG8) and AML1/Evi-1 also contain the runt domain, it is likely that they are able to heterodimerize with PEBP2β. To confirm this, we examined the association of the chimeric proteins with PEBP2β in COS-7 cells expressing AML1/ETO(MTG8) or AML1/Evi-1 together with PEBP2β. COS-7 cells were transfected with expression plasmids for each chimeric protein and PEBP2β and lysed. After immunoprecipitation with anti-AML1 serum, these immunoprecipitates were analyzed by immunoblotting using anti-AML1 serum or anti-PEBP2β serum. When the cells were transfected only with the expression plasmid for PEBP2β and immunoprecipitated with anti-AML1 serum, no PEBP2β was detected (data not shown). PEBP2β was coimmunoprecipitated with both AML1/ETO(MTG8) and AML1/Evi-1, showing the association between each chimeric protein and PEBP2β (Fig 5,lanes 1 and 3). We also examined the association of the mutant chimeric proteins devoid of the runt domain with PEBP2β. As expected, PEBP2β was not coimmunoprecipitated with these deletion mutants (Fig 5, lanes 2 and 4), indicating that the chimeric proteins associate with PEBP2β through the runt domain. Together with the finding that the runt domain is required for the accumulation of PEBP2β in the nucleus by AML1/ETO(MTG8) and AML1/Evi-1, these data suggest that the chimeric proteins accumulate PEBP2β in the nucleus by heterodimerization with PEBP2β.
Association of AML1/ETO(MTG8) and AML1/Evi-1 with PEBP2β. COS-7 cells (1 × 106) were transfected with 10 μg of the expression plasmids for full-length or mutant chimeric protein together with 1 μg of that for PEBP2β. Cells were lysed, immunoprecipitated with anti-AML1 serum, and subjected to Western blotting with anti-AML1 serum or rabbit anti-PEBP2β serum. Arrowheads show the positions of full-size and mutant chimeric proteins. The position of PEBP2β is marked by the arrow. Molecular weight standards (in kilodaltons) are indicated.
Association of AML1/ETO(MTG8) and AML1/Evi-1 with PEBP2β. COS-7 cells (1 × 106) were transfected with 10 μg of the expression plasmids for full-length or mutant chimeric protein together with 1 μg of that for PEBP2β. Cells were lysed, immunoprecipitated with anti-AML1 serum, and subjected to Western blotting with anti-AML1 serum or rabbit anti-PEBP2β serum. Arrowheads show the positions of full-size and mutant chimeric proteins. The position of PEBP2β is marked by the arrow. Molecular weight standards (in kilodaltons) are indicated.
AML1/ETO(MTG8) and AML1/Evi-1 accumulate PEBP2β in the nucleus more efficiently than wild-type AML1.
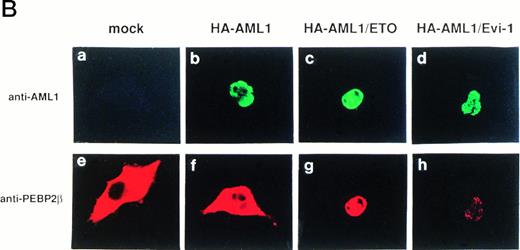
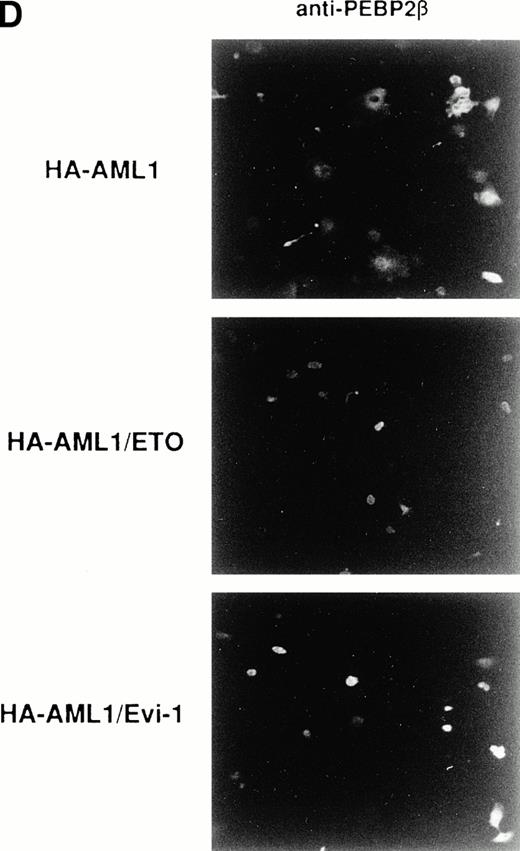
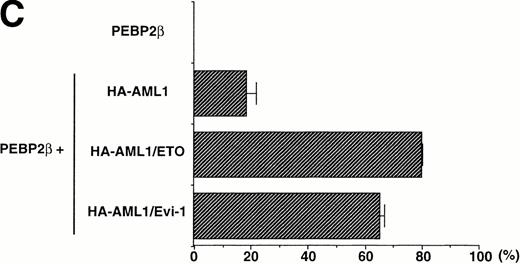
The accumulation of PEBP2β in the nucleus by AML1/ETO(MTG8) and AML1/Evi-1 may play a crucial role in leukemogenesis. To elucidate such a role, it should be important to compare the abilities to accumulate PEBP2β in the nucleus between wild-type AML1 and the chimeric proteins. For this purpose, we tried to make the expression levels of wild-type AML1 and the chimeric proteins approximately equal. We constructed expression vectors in which the expressed wild-type AML1 and chimeric proteins contain HA epitope in their NH2-terminus and detected their expression by anti-HA serum in Western blotting. The HA-tagged AML1, AML1/ETO(MTG8), or AML1/Evi-1 showed a subcellular localization similar to that of the original proteins (Fig 6B [b to d], see page 1690). In addition, HA-tagged AML1 showed a similar transactivation ability compared with the original AML1 (M. Kurokawa and H. Hirai, unpublished observation, February 1995). COS-7 cells were transfected with various amounts of the expression plasmids for the HA-tagged proteins, and the expression levels of the proteins were compared. In conditions shown in Fig 6A, the AML1, AML1/ETO(MTG8), and AML1/Evi-1 proteins were expressed to the approximately same levels. Using these conditions, each of HA-tagged proteins was coexpressed with PEBP2β in COS-7 cells, and subcellular localization of both proteins was determined by double staining. The expression levels of coexpressed PEBP2β were also comparable when assessed by Western blotting (data not shown). We observed nonspecific background signal in the nucleus when HA. 11 anti-HA serum was used for immunofluorescent labeling. Therefore, HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were detected by anti-AML1 serum and FITC-conjugated anti-rabbit IgG. PEBP2β was detected by hamster anti-PEBP2β serum and Texas Red-conjugated anti-hamster IgG. In these sets of experiments, COS-7 cells were transiently transfected and the expression levels of wild-type AML1 or the chimeric proteins, and PEBP2β in each cell may be different. Therefore, a statistical analysis is needed to estimate the ability to accumulate PEBP2β in the nucleus. It is probable that the higher amounts of wild-type AML1 or the chimeric proteins relative to that of PEBP2β accumulate PEBP2β in the nucleus more efficiently than the lower amounts, although we could not quantitatively compare the expression levels of these proteins in individual cells. As we set up conditions so that the average expression levels of wild-type AML1 and the chimeric proteins are comparable, the proportion of the cells showing the nuclear accumulation of PEBP2β is supposed to reflect the ability of these proteins to translocate PEBP2β into the nucleus. So we examined the percentage of the cells showing the nuclear accumulation of PEBP2β among the cells expressing both each HA-tagged protein and PEBP2β as a scale to estimate this ability. Some of the cells with a strong fluorescence in the nucleus also showed a weak cytoplasmic fluorescence, and they were counted as the cells showing the nuclear accumulation of PEBP2β. Less than 5% of the cells showed faint and diffuse fluorescence throughout the cell, and they were excluded from the counts. When coexpressed with HA-AML1, PEBP2β showed a nuclear accumulation in only 19% of the cells and remained in the cytoplasm in the rest of the cells (Fig 6B [f] and C). On the other hand, in 80% of the cells expressing both HA-AML1/ETO(MTG8) and PEBP2β, PEBP2β was located mainly into the nucleus (Fig 6B [g] and C). HA-AML1/Evi-1 colocalized with PEBP2β in the nucleus in 66% of the cells (Fig 6B [h] and C). The staining patterns of the cells with anti-PEBP2β serum in lower magnification shown in Fig 6D clearly show the difference in the effect on subcellular localization of PEBP2β. Most of the cells overexpressing PEBP2β also expressed the HA-tagged AML1 or chimeric proteins (data not shown). The transfection efficiencies were comparable (15% to 20% of all the cells) among the cells transfected with the HA-tagged AML1 and chimeric proteins. In the majority of the cells expressing wild-type AML1, PEBP2β was located mainly in the cytoplasm, especially in the perinuclear region (Fig 6D, upper panel). In contrast, when coexpressed with HA-AML1/ETO(MTG8) or HA-AML1/Evi-1, PEBP2β showed a nuclear pattern in most of the cells (Fig 6D, middle and lower panels). These findings indicate that AML1/ETO(MTG8) and AML1/Evi-1 have higher abilities to translocate PEBP2β into the nucleus than wild-type AML1.
(A) Expressions of the HA-tagged wild-type AML1 and chimeric proteins in COS-7 cells. A total of 3 × 104COS-7 cells were transfected with the expression plasmid for PEBP2β (0.4 μg) together with that for each HA-tagged protein. To obtain the same expression level of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1, based on several preparative experiments, we determined the amounts of the transfected plasmids as follows; HA-AML1 (0.5 μg), HA-AML1/ETO(MTG8) (1.2 μg), and HA-AML1/Evi-1 (4.0 μg). Cell lysates were fractionated into cytoplasmic (lanes C) and nuclear (lanes N) fractions and analyzed by immunoblotting using anti-HA serum. Wild-type AML1 and the chimeric proteins were expressed at the anticipated sizes, as marked by the arrowheads. The complete transfer of these three proteins was verified by confirming that all molecular weight standards were equally stained on the membrane and barely detectable on the gel by Coomassie staining after blotting (data not shown). Cell lysates of COS-7 cells transfected with only PEBP2β construct were also analyzed (mock). Molecular weight standards (in kilodaltons) are indicated. (B) (see page 1690) Double fluorescence labeling of COS-7 cells transfected with the constructs of each HA-tagged protein and PEBP2β. The amounts of the transfected expression plasmids were the same as indicated in (A). (a, b, c, and d) The HA-tagged proteins were detected with anti-AML1 serum. (e, f, g, and h) PEBP2β was detected with hamster anti-PEBP2β serum. Original magnification × 600. (C) The ability of each HA-tagged protein to accumulate PEBP2β in the nucleus. COS-7 cells were transfected with the expression plasmids as shown in (A). Two hundred cells expressing both each HA-tagged protein and PEBP2β were counted (see text). Bars show the percentages of the cells showing stronger fluorescence detected by anti-PEBP2β serum in the nucleus as compared with the fluorescence in the cytoplasm (obtained in three independent experiments). Error bars indicate one standard deviation. (D) The staining pattern of the cells with anti-PEBP2β serum overexpressing HA-AML1 (upper panel), HA-AML1/ETO(MTG8) (middle panel), or HA-AML1/Evi-1 (lower panel) in lower magnification (× 100).
(A) Expressions of the HA-tagged wild-type AML1 and chimeric proteins in COS-7 cells. A total of 3 × 104COS-7 cells were transfected with the expression plasmid for PEBP2β (0.4 μg) together with that for each HA-tagged protein. To obtain the same expression level of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1, based on several preparative experiments, we determined the amounts of the transfected plasmids as follows; HA-AML1 (0.5 μg), HA-AML1/ETO(MTG8) (1.2 μg), and HA-AML1/Evi-1 (4.0 μg). Cell lysates were fractionated into cytoplasmic (lanes C) and nuclear (lanes N) fractions and analyzed by immunoblotting using anti-HA serum. Wild-type AML1 and the chimeric proteins were expressed at the anticipated sizes, as marked by the arrowheads. The complete transfer of these three proteins was verified by confirming that all molecular weight standards were equally stained on the membrane and barely detectable on the gel by Coomassie staining after blotting (data not shown). Cell lysates of COS-7 cells transfected with only PEBP2β construct were also analyzed (mock). Molecular weight standards (in kilodaltons) are indicated. (B) (see page 1690) Double fluorescence labeling of COS-7 cells transfected with the constructs of each HA-tagged protein and PEBP2β. The amounts of the transfected expression plasmids were the same as indicated in (A). (a, b, c, and d) The HA-tagged proteins were detected with anti-AML1 serum. (e, f, g, and h) PEBP2β was detected with hamster anti-PEBP2β serum. Original magnification × 600. (C) The ability of each HA-tagged protein to accumulate PEBP2β in the nucleus. COS-7 cells were transfected with the expression plasmids as shown in (A). Two hundred cells expressing both each HA-tagged protein and PEBP2β were counted (see text). Bars show the percentages of the cells showing stronger fluorescence detected by anti-PEBP2β serum in the nucleus as compared with the fluorescence in the cytoplasm (obtained in three independent experiments). Error bars indicate one standard deviation. (D) The staining pattern of the cells with anti-PEBP2β serum overexpressing HA-AML1 (upper panel), HA-AML1/ETO(MTG8) (middle panel), or HA-AML1/Evi-1 (lower panel) in lower magnification (× 100).
AML1/ETO(MTG8) and AML1/Evi-1 show the higher affinities for PEBP2β than wild-type AML1.
If the accumulation of PEBP2β in the nucleus is dependent on its binding to the chimeric proteins, it is anticipated that the chimeric proteins associate with PEBP2β more effectively than wild-type AML1. We performed in vitro binding experiments in which E coli–expressed GST-PEBP2β immobilized on glutathione sepharose beads were incubated with the lysates from COS-7 cells overexpressing wild-type AML1 or the chimeric proteins. As shown in Fig7A, the chimeric proteins bound to GST-PEBP2β more strongly than wild-type AML1. To confirm this result in vivo, we investigated whether the chimeric proteins show the higher affinities for PEBP2β than wild-type AML1 when both proteins are coexpressed. COS-7 cells were transfected with expression plasmids for HA-AML1, one of the HA-tagged chimeric proteins, and PEBP2β. The cells were lysed and immunoprecipitated with anti-PEBP2β serum, and we tried to compare the amounts of HA-AML1 and the HA-tagged chimeric proteins coimmunoprecipitated with PEBP2β. As the molecular weight of HA-AML1 is close to that of the immunoglobulin heavy chains, it was difficult to estimate the amount of immunoprecipitated HA-AML1 by Western blotting in which anti-immunoglobulin antibody is used as a secondary antibody for the protein detection. Therefore COS-7 cells were metabolically labeled with [35S]methionine, and immunoprecipitates were detected by autoradiography. In the first place, the expression levels of wild-type AML1 and the chimeric proteins were examined by Western blotting of total cell lysates with anti-HA serum (Fig 7B [a]). The amounts of the HA-tagged chimeric proteins were less than those of HA-AML1. To estimate quantitatively the affinities for PEBP2β, [35S]methionine-labeled cell lysates were divided into two and subjected to the immunoprecipitation using 12CA5 anti-HA monoclonal antibody and to the coimmunoprecipitation with PEBP2β using anti-PEBP2β serum. Then the ratios of the amount of each HA-tagged chimeric protein to that of HA-AML1 were compared between the immunoprecipitates with anti-HA monoclonal antibody and those with anti-PEBP2β serum. In the immunoprecipitates with anti-HA monoclonal antibody, the ratios of the amounts of HA-AML1/ETO(MTG8) and HA-AML1/Evi-1 to those of HA-AML1 were 0.53 and 0.72, respectively (Fig 7B [b]). These ratios are properly assumed to reflect the ratios of the expression levels of these proteins and [35S]methionine incorporation of the proteins, because they are in accordance with the results of the Western blotting of the total cell lysates with anti-HA serum (Fig 7B [a]). On the other hand, in the immunoprecipitates with anti-PEBP2β serum, the ratios of the amounts of HA-AML1/ETO(MTG8) and HA-AML1/Evi-1 to those of HA-AML1 went up to 1.18 and 1.61, respectively (Fig 7B [c]). These data suggest that these chimeric proteins associate with PEBP2β more efficiently than wild-type AML1, and this is probably related to the higher abilities of the chimeric proteins to accumulate PEBP2β in the nucleus as compared with wild-type AML1.
Comparison of the affinities for PEBP2β between the chimeric proteins and wild-type AML1. (A) COS-7 cells (1 × 106) were transfected with the expression plasmid for HA-AML1, HA-AML1/ETO(MTG8), or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. The cells were lysed and incubated with GST (lanes 4 to 6) or GST-PEBP2β (lanes 7 to 9) linked to glutathione sepharose beads and subjected to Western blotting with anti-HA serum. Ten percent of the input was also run on the same gel (lanes 1 to 3). The positions of wild-type AML1 and the chimeric proteins are indicated by the arrowheads. Molecular weight standards (in kilodaltons) are shown. (B) COS-7 cells (1 × 106) were transfected with the expression plasmid for PEBP2β together with the construct for HA-AML1 and that for HA-AML1/ETO(MTG8) or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. COS-7 cells transfected with only PEBP2β construct were also analyzed (mock). Cells were subjected to [35S]methionine labeling and lysed. (a) Expressions of the HA-tagged chimeric proteins and wild-type AML1. Total cell lysates, including 50 μg of protein, were subjected to SDS-PAGE and Western blotting with anti-HA serum. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). Molecular weight standards (in kilodaltons) are indicated. (b) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-HA monoclonal antibody. One milligram of each cell lysate was immunoprecipitated with anti-HA monoclonal antibody and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film Corp, Kanagawa, Japan), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown. (c) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-PEBP2β serum. One milligram of each cell lysate was immunoprecipitated with rabbit anti-PEBP2β serum and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The position of PEBP2β is marked by the arrow. The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown.
Comparison of the affinities for PEBP2β between the chimeric proteins and wild-type AML1. (A) COS-7 cells (1 × 106) were transfected with the expression plasmid for HA-AML1, HA-AML1/ETO(MTG8), or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. The cells were lysed and incubated with GST (lanes 4 to 6) or GST-PEBP2β (lanes 7 to 9) linked to glutathione sepharose beads and subjected to Western blotting with anti-HA serum. Ten percent of the input was also run on the same gel (lanes 1 to 3). The positions of wild-type AML1 and the chimeric proteins are indicated by the arrowheads. Molecular weight standards (in kilodaltons) are shown. (B) COS-7 cells (1 × 106) were transfected with the expression plasmid for PEBP2β together with the construct for HA-AML1 and that for HA-AML1/ETO(MTG8) or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. COS-7 cells transfected with only PEBP2β construct were also analyzed (mock). Cells were subjected to [35S]methionine labeling and lysed. (a) Expressions of the HA-tagged chimeric proteins and wild-type AML1. Total cell lysates, including 50 μg of protein, were subjected to SDS-PAGE and Western blotting with anti-HA serum. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). Molecular weight standards (in kilodaltons) are indicated. (b) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-HA monoclonal antibody. One milligram of each cell lysate was immunoprecipitated with anti-HA monoclonal antibody and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film Corp, Kanagawa, Japan), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown. (c) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-PEBP2β serum. One milligram of each cell lysate was immunoprecipitated with rabbit anti-PEBP2β serum and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The position of PEBP2β is marked by the arrow. The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown.
DISCUSSION
In this study, we showed that the two leukemia-associated chimeric proteins, AML1/ETO(MTG8) and AML1/Evi-1, are proteins that are located in the nucleus. The runt domain is partially responsible for their nuclear localization, and it was suggested that the ETO(MTG8) and Evi-1 portions of the chimeric proteins also play some roles in it. We also found that these chimeric proteins accumulate PEBP2β in the nucleus with dependence on the runt domain, which is known to be responsible for heterodimerization between AML1 and PEBP2β. Furthermore, we showed that the chimeric proteins accumulate PEBP2β in the nucleus more efficiently than wild-type AML1. This phenomenon is probably attributed to the affinities for PEBP2β, which is higher in the chimeric proteins than in wild-type AML1.
What domains are responsible for the nuclear localization of AML1/ETO(MTG8) and AML1/Evi-1? The PST region, which is partially responsible for the nuclear translocation activity of PEBP2αA,45 is missing in the chimeric proteins, AML1/ETO(MTG8) and AML1/Evi-1 (Fig 2). The nuclear localization of these chimeric proteins and the retention of their deletion mutants lacking the runt domain in the nucleus suggest that ETO(MTG8) and Evi-1 portions of the chimeric proteins are responsible for their nuclear localization. Indeed, Evi-1 is a nuclear-localized DNA-binding protein with two zinc finger domains (Fig 2) and works as a transcription factor.53-55 We confirmed that Evi-1 is present in the nucleus in COS-7 cells that are expressing Evi-1 by immunofluorescence-labeling using anti–Evi-1 serum (K. Tanaka and H. Hirai, unpublished observation, July 1995). ETO(MTG8) has also putative functional domains as a transcription factor (Fig 2), and it was reported that ETO(MTG8) is located in the nucleus in a hematopoietic cell.56 The regions in ETO(MTG8) and Evi-1 that are required for their nuclear localization remain further to be determined.
We showed that more of the cells showed the nuclear localization of PEBP2β when AML1/ETO(MTG8) or AML1/Evi-1 was expressed together with PEBP2β, as compared with wild-type AML1. These data imply that AML1/ETO(MTG8) and AML1/Evi-1 have higher abilities to accumulate PEBP2β in the nucleus than wild-type AML1. What makes such difference of the ability between wild-type AML1 and the chimeric proteins? The chimeric proteins are able to heterodimerize with PEBP2β, and it is probable that PEBP2β is translocated into the nucleus with dependence on the binding to the chimeric proteins. Our findings suggest that the chimeric proteins heterodimerize with PEBP2β more effectively than wild-type AML1. Therefore, it is plausible that the difference in the ability to accumulate PEBP2β in the nucleus between wild-type AML1 and the chimeric proteins reflects the difference in the affinity for PEBP2β between them. If this is the case, how are the chimeric proteins able to show the higher affinities for PEBP2β than wild-type AML1? As it has been suggested that the PST region of PEBP2αA is inhibitory to its binding to PEBP2β,45 the PST region of AML1 may prevent PEBP2β from associating efficiently with AML1. In this context, it is convincing that deletion of the PST region has a significant role in the higher affinity of PEBP2β for the chimeric proteins. In addition, it is also possible that the ETO(MTG8) and Evi-1 portions of the chimeric proteins promote the binding of PEBP2β to the chimeric protein.
The difference in the ability to accumulate PEBP2β in the nucleus between wild-type AML1 and the chimeric proteins is possibly one of the mechanisms of leukemic transformation by the chimeric proteins. As AML1 is considered to play essential roles in hematopoietic cell differentiation and proliferation, the repression of the function of wild-type AML1 by the chimeric protein is a convincing mechanism of leukemogenesis in t(8;21) and t(3;21) leukemias. In t(15;17) leukemias, the PML/RARα chimeric protein is shown to act as a dominant negative oncoprotein by inhibiting the localization of PML in large nuclear bodies.42-44 We and other groups have shown that AML1/ETO(MTG8) and AML1/Evi-1 also act as dominant negative oncoproteins over intact AML1 and inhibit myeloid cell differentiation.38-41,57 These dominant negative regulations are probably achieved by the higher affinities of the chimeric proteins for DNA or for PEBP2β as compared with that of wild-type AML1. We have already shown that AML1/Evi-1 shows higher affinity for DNA than wild-type AML1.41 The higher affinities of the chimeric proteins for PEBP2β are also assumed to result in their efficient binding to DNA as compared with wild-type AML1, because the heterodimerization with PEBP2β is probably required for the efficient binding to DNA. Therefore, it is suggested that the chimeric proteins act as dominant negative proteins by decreasing the binding of wild-type AML1 to DNA.
The analyses of the effects of AML1/ETO(MTG8) and AML1/Evi-1 on subcellular localization of PEBP2β have provided new insights into the mechanisms for leukemogenesis mediated by these chimeric proteins. Further study is necessary to clarify the precise association between the localization of these proteins and the mechanisms of leukemogenesis.
ACKNOWLEDGMENT
We thank Dr Y. Ito (Kyoto University, Kyoto, Japan) for a present of a mouse PEBP2β cDNA and Dr S.W. Hiebert (St Jude Children's Research Hospital, Memphis, TN) for a gift of AML1/ETO cDNA.
Supported in part by Grants-in-Aid for Cancer Research from the Ministry of Health and Welfare and from the Ministry of Education, Science and Culture of Japan.
Address reprint requests to Hisamaru Hirai, MD, PhD, The Third Department of Internal Medicine, Faculty of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be here-by marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


![Fig. 7. Comparison of the affinities for PEBP2β between the chimeric proteins and wild-type AML1. (A) COS-7 cells (1 × 106) were transfected with the expression plasmid for HA-AML1, HA-AML1/ETO(MTG8), or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. The cells were lysed and incubated with GST (lanes 4 to 6) or GST-PEBP2β (lanes 7 to 9) linked to glutathione sepharose beads and subjected to Western blotting with anti-HA serum. Ten percent of the input was also run on the same gel (lanes 1 to 3). The positions of wild-type AML1 and the chimeric proteins are indicated by the arrowheads. Molecular weight standards (in kilodaltons) are shown. (B) COS-7 cells (1 × 106) were transfected with the expression plasmid for PEBP2β together with the construct for HA-AML1 and that for HA-AML1/ETO(MTG8) or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. COS-7 cells transfected with only PEBP2β construct were also analyzed (mock). Cells were subjected to [35S]methionine labeling and lysed. (a) Expressions of the HA-tagged chimeric proteins and wild-type AML1. Total cell lysates, including 50 μg of protein, were subjected to SDS-PAGE and Western blotting with anti-HA serum. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). Molecular weight standards (in kilodaltons) are indicated. (b) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-HA monoclonal antibody. One milligram of each cell lysate was immunoprecipitated with anti-HA monoclonal antibody and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film Corp, Kanagawa, Japan), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown. (c) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-PEBP2β serum. One milligram of each cell lysate was immunoprecipitated with rabbit anti-PEBP2β serum and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The position of PEBP2β is marked by the arrow. The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/5/10.1182_blood.v91.5.1688/3/m_blod4052907a.jpeg?Expires=1769299543&Signature=Pkm7PgsFaNirdqCqCJvEU5ncOeOj7ktUzignIQDo2MxxNgUrdLJ2ymu0GXcEe~n7QQgvdNrSiwgaHY4-Xb17c8zRLeQ7Q1l4zVnAhkf2ZweJvTIAk87uPsgjGV1Zf7DFerRYXC8y0MuN5i3wH5y8htyob8-Sxk4f3VRCtULwmB9vBwVt2t0hfPnFFV5de6okc~35I8xByKVhWzWWgJXn6xaPkRgEYwcG8uGTaPv~J5NNClUoN20wF5bHgEBhceJ0-s0~eD9zm2L3n9g2Mee1haLIjIxBX17GXD3TWC4Ea6Yyd3hp9OuZ~CwzjNbynhcVhvo8DCWom-hZl365mAp5Lw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Comparison of the affinities for PEBP2β between the chimeric proteins and wild-type AML1. (A) COS-7 cells (1 × 106) were transfected with the expression plasmid for HA-AML1, HA-AML1/ETO(MTG8), or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. The cells were lysed and incubated with GST (lanes 4 to 6) or GST-PEBP2β (lanes 7 to 9) linked to glutathione sepharose beads and subjected to Western blotting with anti-HA serum. Ten percent of the input was also run on the same gel (lanes 1 to 3). The positions of wild-type AML1 and the chimeric proteins are indicated by the arrowheads. Molecular weight standards (in kilodaltons) are shown. (B) COS-7 cells (1 × 106) were transfected with the expression plasmid for PEBP2β together with the construct for HA-AML1 and that for HA-AML1/ETO(MTG8) or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. COS-7 cells transfected with only PEBP2β construct were also analyzed (mock). Cells were subjected to [35S]methionine labeling and lysed. (a) Expressions of the HA-tagged chimeric proteins and wild-type AML1. Total cell lysates, including 50 μg of protein, were subjected to SDS-PAGE and Western blotting with anti-HA serum. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). Molecular weight standards (in kilodaltons) are indicated. (b) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-HA monoclonal antibody. One milligram of each cell lysate was immunoprecipitated with anti-HA monoclonal antibody and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film Corp, Kanagawa, Japan), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown. (c) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-PEBP2β serum. One milligram of each cell lysate was immunoprecipitated with rabbit anti-PEBP2β serum and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The position of PEBP2β is marked by the arrow. The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/5/10.1182_blood.v91.5.1688/3/m_blod4052907ba.jpeg?Expires=1769299543&Signature=P7oqYr~8LMaeZ0WmJlMhZcYjiFx~uTH0EzQ3YN5h0gVe7WuSMVthoKT4lt7y2ZrJNtA4xL-CcOIzpdBqITd26LRCcWmNM9WIj0rKhT8WfreAA3XzlrtynnzMxguzNcV~mhMWzVleX5OSDsnBzoF9vqn9sLsOr71K6wXds99zkYwETazvlSVCM-zghftpZHDqGBoMzW3rx8G4tirGA-S4aVWWMZ8UKRv2FuEfOZlHgJosnMdGwNyjNEUi-Hz04b0liNU2bjYAg3O8AEXHLDc8gBeKk-tWCZbHSo~grnZe48Thvob7MdLh8e008jkbSi83H~t~OzemLHub7AlXmr~eyw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Comparison of the affinities for PEBP2β between the chimeric proteins and wild-type AML1. (A) COS-7 cells (1 × 106) were transfected with the expression plasmid for HA-AML1, HA-AML1/ETO(MTG8), or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. The cells were lysed and incubated with GST (lanes 4 to 6) or GST-PEBP2β (lanes 7 to 9) linked to glutathione sepharose beads and subjected to Western blotting with anti-HA serum. Ten percent of the input was also run on the same gel (lanes 1 to 3). The positions of wild-type AML1 and the chimeric proteins are indicated by the arrowheads. Molecular weight standards (in kilodaltons) are shown. (B) COS-7 cells (1 × 106) were transfected with the expression plasmid for PEBP2β together with the construct for HA-AML1 and that for HA-AML1/ETO(MTG8) or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. COS-7 cells transfected with only PEBP2β construct were also analyzed (mock). Cells were subjected to [35S]methionine labeling and lysed. (a) Expressions of the HA-tagged chimeric proteins and wild-type AML1. Total cell lysates, including 50 μg of protein, were subjected to SDS-PAGE and Western blotting with anti-HA serum. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). Molecular weight standards (in kilodaltons) are indicated. (b) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-HA monoclonal antibody. One milligram of each cell lysate was immunoprecipitated with anti-HA monoclonal antibody and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film Corp, Kanagawa, Japan), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown. (c) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-PEBP2β serum. One milligram of each cell lysate was immunoprecipitated with rabbit anti-PEBP2β serum and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The position of PEBP2β is marked by the arrow. The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/5/10.1182_blood.v91.5.1688/3/m_blod4052907bb.jpeg?Expires=1769299543&Signature=lQTO~kO2nuBK5k3c9sMji-XVDiSJNmegaaAnt2spxqIZMa1YVeYwbLcFy4-meKde9Oi3zM-vKYfj5xjIzddkdW9c3BDGiqpgIo88EjU6LXEF1X4vVFhR6p9fqYDiqLP1eiai~~mXjF4r7EhvwRnKtlfOGQGE7k3BbkDIK4yfBbDyjJkT3zGjg5yrpdnWDzRuzAX6VILiRv3f9HfRY8aqMfGtiUoHuWG8xzozBGHVee-KrG-2jBAapsbWcd5ilLhjTIM9n1Sx~ly8ys9gbRw-asrgzpxbUizkU-iAVszzCdL2F22EWfEg~JxoNA6dKiJiCskrqHhjXHBo0TLz2xKw2w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Comparison of the affinities for PEBP2β between the chimeric proteins and wild-type AML1. (A) COS-7 cells (1 × 106) were transfected with the expression plasmid for HA-AML1, HA-AML1/ETO(MTG8), or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. The cells were lysed and incubated with GST (lanes 4 to 6) or GST-PEBP2β (lanes 7 to 9) linked to glutathione sepharose beads and subjected to Western blotting with anti-HA serum. Ten percent of the input was also run on the same gel (lanes 1 to 3). The positions of wild-type AML1 and the chimeric proteins are indicated by the arrowheads. Molecular weight standards (in kilodaltons) are shown. (B) COS-7 cells (1 × 106) were transfected with the expression plasmid for PEBP2β together with the construct for HA-AML1 and that for HA-AML1/ETO(MTG8) or HA-AML1/Evi-1. The amounts of the transfected expression plasmids were the same as described in the legend to Fig 6. COS-7 cells transfected with only PEBP2β construct were also analyzed (mock). Cells were subjected to [35S]methionine labeling and lysed. (a) Expressions of the HA-tagged chimeric proteins and wild-type AML1. Total cell lysates, including 50 μg of protein, were subjected to SDS-PAGE and Western blotting with anti-HA serum. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). Molecular weight standards (in kilodaltons) are indicated. (b) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-HA monoclonal antibody. One milligram of each cell lysate was immunoprecipitated with anti-HA monoclonal antibody and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film Corp, Kanagawa, Japan), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown. (c) Comparison of the amounts of the HA-tagged chimeric proteins to that of HA-AML1 immunoprecipitated with anti-PEBP2β serum. One milligram of each cell lysate was immunoprecipitated with rabbit anti-PEBP2β serum and subjected to SDS-PAGE and autoradiography. Closed arrowheads show the position of HA-AML1. Open arrowheads show the positions of HA-AML1/ETO(MTG8) (lane 2) and HA-AML1/Evi-1 (lane 3). The position of PEBP2β is marked by the arrow. The radioactivities of the bands of HA-AML1, HA-AML1/ETO(MTG8), and HA-AML1/Evi-1 were quantified by Fujix BAS 2000 (Fuji Film), and the ratios of the radioactivities of the HA-tagged chimeric proteins to those of HA-AML1 are indicated. Molecular weight standards (in kilodaltons) are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/5/10.1182_blood.v91.5.1688/3/m_blod4052907bc.jpeg?Expires=1769299543&Signature=xqBR3X4Kazm1DclpqKpeS4uc35RqiE5hK0hOFTfejGjFmhyx5wG3ZlDplkkvCUJBW6HsXR7nikKivmSWkoK~aobvehjGHNv5~ev7Q592F8bF95s4FUi6WjwSmRhu7uY-f~yQ6~svYhrPlRYqoTBxuqfsC~Uf~SW27hNtG1Yr6bmOYVqHUQ-JpsfJTGHuJq3Z3LlxkqBn9j2Ib3J9-fYKHHKdtbvTMshN1yU9hEC-MtznvS-JSnNgEND9EulYSb941Fl7x9b0BnXVygz3SkLqeCsb8gdSzMDGMzqSZ--8Pfm8dRYSHJMB7ae1Tej3d1Rn0JBjTyYdhhdfpCmZcXlQ0w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal