Abstract
A lymphoma with the characteristic features of Hodgkin's disease (HD) occasionally develops in patients with B-cell chronic lymphocytic leukemia (CLL), and has been called Richter's syndrome with HD features. In such cases, large tumor cells have the morphological and immunophenotypic features of classical Hodgkin and Reed-Sternberg (H-RS) cells. However, it is not known whether the H-RS cells arise from transformation of the underlying CLL cells or from a different pathological process. We report herein a study of the clonal relationship between the CLL cells and the H-RS cells in three cases of Richter's syndrome with HD features by using a single cell assay. We isolated single CLL cells and H-RS cells from immunostained tissue sections by micromanipulation. The immunoglobulin heavy chain gene (IgH) complementarity determining region (CDR) III of each cell was amplified by the polymerase chain reaction (PCR). The products were then compared by gel electrophoresis and nucleotide sequencing. The IgH CDRIII sequences from the H-RS cells were identical to those from the CLL cells in two cases. In one case, the clonal relationship between the two types of cells could not be determined because PCR products could not be obtained from any of the H-RS cells. This study shows that the H-RS cells and the CLL cells belong to the same clonal population in some cases of Richter's syndrome with HD features. Furthermore, our findings indicate that mature B cells can undergo transformation to cells with the features of H-RS cells, in association with a cellular background typical of HD. This study also supports recent findings suggesting that the H-RS cells in classical HD are derived from transformed B cells.
B-CELL CHRONIC lymphocytic leukemia (CLL) is an indolent lymphoproliferative disease derived from mature B cells. Approximately 3% to 10% of cases of CLL develop high-grade lymphoma, a process known as Richter's syndrome.1,2 Usually, these high-grade lymphomas consist of monomorphic large cells and are classified morphologically as large-cell non-Hodgkin's lymphoma. In most cases, the high-grade component is considered to represent a blastic transformation of the CLL cells.3-5 However, in a small number of the cases, the high-grade component has characteristics of Hodgkin's disease (HD).6-17 In some of these cases, the large cells not only resemble Hodgkin and Reed-Sternberg (H-RS) cells, but also exhibit the typical H-RS cell immunophenotype (CD15+, CD30+, and lineage −). An interesting question is whether the H-RS cells in this form of Richter's syndrome represent transformation of the underlying CLL cells or a different pathological process. However, it is impossible to answer this question with the usual study methods such as Southern blotting, because of the paucity of H-RS cells, or polymerase chain reaction (PCR) using DNA extracted from whole sections, because of the presence of multiple cell populations. In this study, the clonal relationship between the CLL cells and the H-RS cells in three cases of Richter's syndrome with HD features was determined using a single cell assay. Single CLL cells and H-RS cells were isolated from immunostained sections of these cases by micromanipulation, and the immunoglobulin heavy chain gene (IgH) complementarity determining region (CDR) III of each cell was amplified by PCR. The products were then compared by gel electrophoresis and nucleotide sequencing.
MATERIALS AND METHODS
Patients.
Three cases of Richter's syndrome with HD features were selected from the database of the Lymphoma Registry at the University of Nebraska Medical Center, the Cancer Control Agency of British Columbia, and the consultation files of one of the authors (W.C.C.). All of the patients were men, ages 59 to 61 years, with a previous history of CLL before presenting with a lymphoma resembling HD. Tissue samples fixed in formalin solution and embedded in paraffin were available for study.
Immunohistochemistry.
Four-micron paraffin sections were cut and stained using a three-step immunoperoxidase method.18 The following antibodies were used as primary antibodies: L26 (anti-CD20), polyclonal anti-CD3, UCHL1 (anti-CD45RO), leukocyte common antigen (anti-CD45), and BerH2 (anti-CD30), purchased from Dako Corporation (Carpinteria, CA); Leu22 (anti-CD43) and LeuM1 (anti-CD15) from Becton Dickinson (San Jose, CA); and MB2 from Biotest (Dreieich, Germany).
Single cell isolation from tissue sections.
Four-micron sections were cut from a formalin-fixed, paraffin-embedded tissue specimen and stained with antibodies against CD20 (cases 1 and 3) or MB2 (case 2) for isolating B-CLL cells, with antibodies against CD15 (case 2) or CD30 (cases 1 and 3) for isolating H-RS cells, and with an antibody against CD3 (cases 1 and 2) for isolating T cells. Before immunostaining with anti-CD15, CD30, or CD3, the sections were digested with trypsin (0.1%, type II; Sigma, St Louis, MO) in Tris-buffer (pH 7.6) for 30 minutes at 37°C. By using an inverted microscope (Nikon, New York, NY), stained single cells were isolated with a microknife and micropipette attached to hydraulic micromanipulators (Narishige, New York, NY), and expelled into a tube containing PCR buffer.19 Before cell harvest, sections stained with anti-CD20 were digested for 20 to 30 minutes at 37°C with a mixture of collagenase (0.3%, type XI; Sigma) and trypsin (0.25%, type II; Sigma) dissolved in phosphate-buffered saline (pH 7.4). During the micromanipulation, the section was overlaid with sterile distilled water which was replaced after the isolation of each cell. The distilled water overlying the section was sampled periodically to serve as a negative supernatant control.
PCR amplification of single cells.
The IgH CDRIII of each single cell was amplified by a seminested PCR.20 In both PCR rounds, a consensus primer to the 3′ end of the third framework region of the VH genes (sequence: 5′CTGTCGACACGGCCGTGTATTACTG3′) was used.21 Two different consensus primers to the 3′ end of the antisense JH segments, a JH external primer (sequence: 5′ACCTGAGGAGACGGTGACC3′), and a JH nested primer (sequence: 5′ACCAGGGTCCCTTGGCCCCA3′) were used in the first and second rounds of the PCR, respectively. Single cells were placed into 25 μL PCR buffer (50 mmol/L KCI, 10 mmol/L Tris-HCl at pH 8.3, 2.5 mmol/L MgCl2, 0.1 mg/mL gelatin, 0.45% Nonidet P40, 0.45% Tween 20) containing 200 μg/mL proteinase K. The reaction mixture was covered with 50 μL of mineral oil and incubated at 55°C for 3 hours. Proteinase K was then inactivated at 96°C for 10 minutes. A PCR cycle consisted of annealing for 40 seconds at 55°C, extension for 40 seconds at 72°C, and denaturation for 40 seconds at 94°C. The first round of PCR was performed for 30 cycles and the second round for 35 cycles. Two microliters of a 1:200 dilution of the PCR product from the first round was used as the template for the second round of PCR. In both rounds, the PCR mixture contained 200 μmol/L of each dNTP and 0.2 μmol/L of each primer in a total volume of 50 μL of PCR buffer. Taq polymerase, 1.5 U, was added to each reaction by using a hot start procedure. The amplified products were electrophoresed on 8% polyacrylamide gels and visualized with UV light after ethidium-bromide staining.
DNA sequencing.
In cases 1 and 2, representative IgH CDRIII PCR products of the amplifiable CLL and H-RS cells were sequenced to determine their clonal relationship. For this purpose, the original IgH CDRIII PCR product was reamplified in a 50 μL volume by using VH and JH nested primers with M13 sequences attached to their 5′ end (sequence: M13 + VH, TGTAAAACGACGGCCAGTCTGTCGACACGGCCGTGTATTACTG; M13R + JH, GGAAACAGCTATGACCATGACCAGGGTCCCTTGGCCCCA). The products were gel-purified and directly sequenced with primers complementary to M13 or M13R by means of a cycle sequencing procedure with fluorescent dideoxynucleotide terminators (Applied Biosystems, Foster City, CA).22 The sequences were analyzed with the Applied Biosystem 373A sequenator (Applied Biosystems).
RESULTS
Histological and immunohistochemical findings.
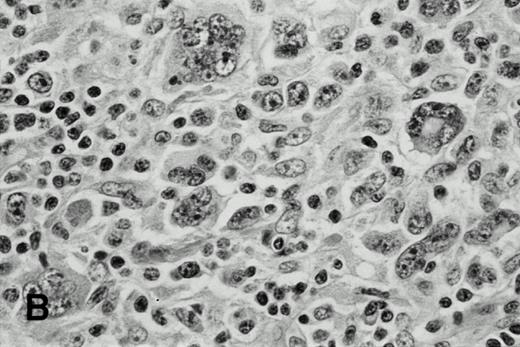
In case 1, the cervical lymph nodes showed a proliferation of small lymphocytes with round nuclei, consistent with CLL. In addition, large highly pleomorphic cells resembling H-RS cells were present in small clusters among small lymphocytes (Fig 1A).In case 2, the lymph nodes showed subtotal obliteration of the architecture by a polymorphic infiltrate of small lymphocytes and histiocytes, with scattered H-RS cells admixed. In case 3, an axillary lymph node exhibited nodules of HD, with H-RS cells present in a typical polymorphous inflammatory background (Fig 1B), which was sharply delineated from areas with CLL.
H-RS cells in case 1 (A) and case 3 (B), (hematoxylin and eosin, original magnification × 400). H-RS cells were present among small lymphocytes in case 1, whereas they were present in a polymorphous inflammatory background in case 3.
H-RS cells in case 1 (A) and case 3 (B), (hematoxylin and eosin, original magnification × 400). H-RS cells were present among small lymphocytes in case 1, whereas they were present in a polymorphous inflammatory background in case 3.
The results of the immunohistochemical studies are shown in Table1 and Fig 2.The CLL cells were reactive with B-cell markers in all three cases, and also expressed CD43 in cases 2 and 3, as is commonly observed in B-CLL.23 The H-RS cells expressed CD15 and CD30 in all three cases. In case 1, a small subset of the H-RS cells also expressed CD20. The small lymphocytes forming the inflammatory background of the HD component in cases 2 and 3 were mostly T cells. Scattered B lymphocytes were also present, more prominently in case 2 where the Hodgkin's foci were less sharply demarcated from the CLL areas as compared with case 3. In case 1, although the typical cellular background of HD was absent, most of the small lymphocytes associated with the H-RS cell areas were T cells which often formed a ring around the H-RS cells.
Immunophenotype of CLL and H-RS Cells
| Case . | Cells . | CD20 . | MB2 . | CD43 . | CD3 . | CD45RO . | CD45 . | CD15 . | CD30 . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CLL | + | ND | − | − | − | + | − | − |
| H-RS | ∓-150 | ND | − | − | − | − | + | + | |
| 2 | CLL | − | + | + | − | ND | ND | − | − |
| H-RS | − | − | − | − | ND | ND | + | + | |
| 3 | CLL | + | + | + | − | − | + | − | − |
| H-RS | − | + | − | − | − | − | + | + |
| Case . | Cells . | CD20 . | MB2 . | CD43 . | CD3 . | CD45RO . | CD45 . | CD15 . | CD30 . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CLL | + | ND | − | − | − | + | − | − |
| H-RS | ∓-150 | ND | − | − | − | − | + | + | |
| 2 | CLL | − | + | + | − | ND | ND | − | − |
| H-RS | − | − | − | − | ND | ND | + | + | |
| 3 | CLL | + | + | + | − | − | + | − | − |
| H-RS | − | + | − | − | − | − | + | + |
Abbreviation: ND, not done.
A small fraction of the cells are CD20+.
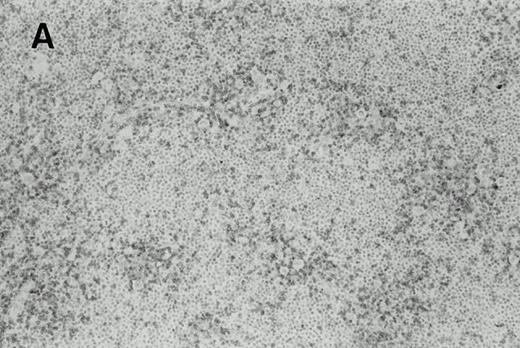
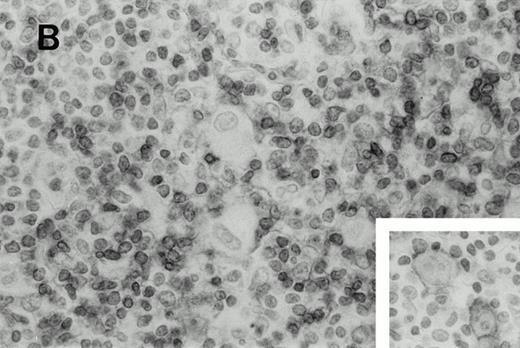
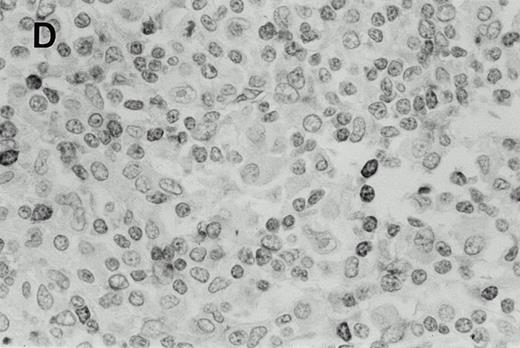
Immunohistochemistry of CD3 in case 1: (A) low (40×), (B) high (400×). CD3+ T cells are associated with clusters of H-RS cells, often forming a ring around the H-RS cells. Inset shows CD30+ H-RS cells. Immunohistochemistry of MB2 in case 2: (C) low (40×), (D) high (400×). MB2+ CLL cells are present around a nodule of HD. H-RS cells are admixed with T cells and histiocytes and only scattered MB2+ small lymphocytes are present.
Immunohistochemistry of CD3 in case 1: (A) low (40×), (B) high (400×). CD3+ T cells are associated with clusters of H-RS cells, often forming a ring around the H-RS cells. Inset shows CD30+ H-RS cells. Immunohistochemistry of MB2 in case 2: (C) low (40×), (D) high (400×). MB2+ CLL cells are present around a nodule of HD. H-RS cells are admixed with T cells and histiocytes and only scattered MB2+ small lymphocytes are present.
PCR amplification and sequence analysis of the IgH CDRIII.
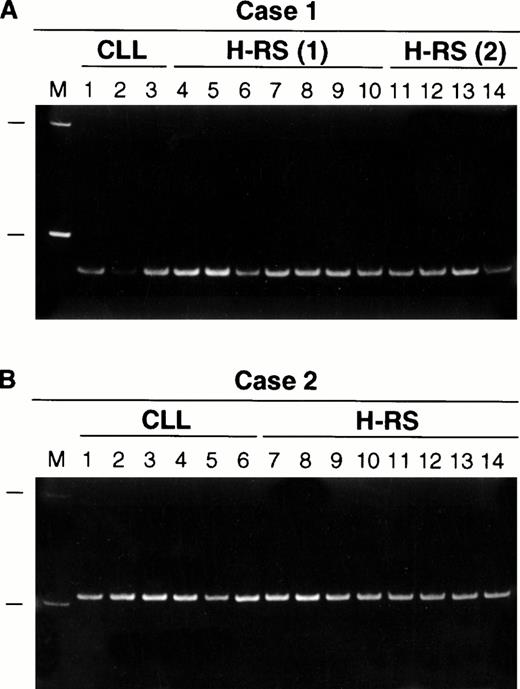
The results of IgH CDRIII PCR performed on single cells are summarized in Table 2 and illustrated in Fig3. In all three cases, PCR products were obtained from 20% to 30% of the small B cells. They showed an identical length in each case, indicating that they were monoclonal. In cases 1 and 2, PCR products which were identical in length to those from the small B cells were also obtained from 40% to 45% of the H-RS cells (Fig 3). Because a small population of H-RS cells in case 1 expressed CD20, the experiment was repeated after staining a section with CD20, and negative H-RS cells were isolated and studied. The PCR products obtained from these CD20− cells were also identical in size to those obtained from the small B cells. In case 3, IgH CDRIII products were obtained from 12/40 small B cells and they were of identical size, but PCR products could not be obtained from the H-RS cells. In cases 1 and 2, T cells were isolated as negative controls. No PCR products were obtained from the T cells in case 1, as expected. However, in case 2, a PCR product was obtained from one of 20 T cells, but it was different in length from the PCR products obtained from the B cells and the H-RS cells. A total of 54 supernatants were sampled as negative controls in the three cases, and a PCR product was amplified from only one supernatant, again different in size from those obtained from the corresponding CLL cells.
IgH CDRIII PCR Amplification of Single Cells
| Case . | B Cells . | H-RS Cells . | T Cells . |
|---|---|---|---|
| 1 | 4/20 | 9/20 | 0/20 |
| 5/16* | |||
| 2 | 6/20 | 8/20 | 1/20 |
| 3 | 5/20 | 0/20 | |
| 7/20 | 0/20 |
| Case . | B Cells . | H-RS Cells . | T Cells . |
|---|---|---|---|
| 1 | 4/20 | 9/20 | 0/20 |
| 5/16* | |||
| 2 | 6/20 | 8/20 | 1/20 |
| 3 | 5/20 | 0/20 | |
| 7/20 | 0/20 |
Results are expressed as the number of amplifiable cells/total number of cells analyzed.
CD20− H-RS cells were isolated.
Polyacrylamide gel electrophoresis of IgH CDRIII PCR products obtained from single CLL cells and H-RS cells: (A) case 1, (B) case 2. All PCR products are identical in size in each case. M, 100 basepair (bp) molecular size ladder with 100- and 200-bp markers shown. H-RS(1) indicates that the PCR products were obtained from CD30+ H-RS cells. In a separate experiment, CD20− H-RS cells were isolated for study and the PCR products are shown under H-RS(2).
Polyacrylamide gel electrophoresis of IgH CDRIII PCR products obtained from single CLL cells and H-RS cells: (A) case 1, (B) case 2. All PCR products are identical in size in each case. M, 100 basepair (bp) molecular size ladder with 100- and 200-bp markers shown. H-RS(1) indicates that the PCR products were obtained from CD30+ H-RS cells. In a separate experiment, CD20− H-RS cells were isolated for study and the PCR products are shown under H-RS(2).
Sequence analysis was performed on three PCR products from the B-CLL cells, four PCR products from the CD30+ H-RS cells, and two PCR products from the CD20− H-RS cells in case 1. Sequence analysis was also performed on three PCR products from the B-CLL cells and four PCR products from the H-RS cells in case 2. All of the PCR products were identical in each case (Table3).
Nucleotide Sequences of the IgH CDRIII of Single B Cells and H-RS Cells
| Case . | VH . | N-DH-N* . | JH . |
|---|---|---|---|
| 1 | DP-84 | DN1 | JH4 |
| TGCAAGAG | ggaAGCAGCAGCAGCTGGcccc | CTTTGACTAC | |
| 2 | DP-8 | DN1 | JH6 |
| TGCGAG | ggagaaagTAGCAGCAGCTGGTACgttcct | CTACTACTACTACGGTATGGACGTC |
| Case . | VH . | N-DH-N* . | JH . |
|---|---|---|---|
| 1 | DP-84 | DN1 | JH4 |
| TGCAAGAG | ggaAGCAGCAGCAGCTGGcccc | CTTTGACTAC | |
| 2 | DP-8 | DN1 | JH6 |
| TGCGAG | ggagaaagTAGCAGCAGCTGGTACgttcct | CTACTACTACTACGGTATGGACGTC |
Assignment of VH, D, and JH was performed by comparing the sequence with IgH germline sequences in V-BASE (http://www.mrc-cpe.cam.ac.uk/imt-doc/vbase-home-page.html) using the FASTA program.
Lower case letters represent N-region sequence.
DISCUSSION
To determine the clonal relationship between H-RS cells and CLL cells, we examined the IgH CDRIII of both types of cells from three cases of Richter's syndrome with HD features using single-cell analysis. The H-RS cells had the morphological and immunophenotypical features typical of classical HD in all three cases. We found that the IgH CDRIII sequences from the H-RS cells were identical to those from the CLL cells in two cases (cases 1 and 2). In one case (case 3), we could not determine the clonal relationship between the CLL cells and the H-RS cells because PCR products could not be obtained from any of the single H-RS cells. These findings indicate that the H-RS cells and the CLL cells belong to the same clonal population in some cases of Richter's syndrome with HD features.
HD is reported to be occasionally associated with CLL,6-17and a recent study showed that HD is one of the most common secondary neoplasms in patients with CLL.24 Two types of Richter's syndrome with HD features have been reported: one type with H-RS cells scattered in a background of CLL cells (type 1)9-12 and the other type with H-RS cells present in a typical polymorphous inflammatory background separate from the CLL (type 2).6-8,14-17 Generally, it has been assumed that the H-RS cells in type 1 disease represent histological progression of the underlying CLL cells, especially when the H-RS cells also express B-cell markers, whereas type 2 is thought to represent the occurrence of two separate diseases, based on both the histological and immunophenotypical findings. However, recent studies have suggested a lymphoid, usually B-cell, origin for the H-RS cell, not only in lymphocytic predominance HD but also in de novo classical HD.25-27 Thus, it is necessary to reconsider the assumption that the two lesions in type 2 disease are unrelated.
In case 1, the H-RS cells were present in clusters among the CLL cells. Although the typical cellular background of HD was absent, immunohistochemical stains showed that the lymphocytes associated with the H-RS cell clusters were mostly T cells, which often formed rings around the H-RS cells, as is characteristically seen in HD.28 In case 2, the H-RS cells were present in a polymorphous inflammatory background as in classical HD, but CLL cells were also scattered within the inflammatory background, especially at the edges of the HD component. Case 1 seems to fall into the above-mentioned type 1 category of disease, whereas case 2 is more similar to type 2 disease. It has been reported that some cases of type 1 disease may progress to disseminated HD with the disappearance of CLL.9 10 Our case 2 may represent such a case. In case 3, the HD component, in which the H-RS cells were present in a typical inflammatory background, was sharply delineated from the CLL as seen in type 2 disease. However, in this case we could not obtain IgH CDRIII products from any of the H-RS cells. We must be careful before concluding that the H-RS cells in case 3 are clonally unrelated to the CLL cells and the two components are separate disease processes. The possibility remains that somatic mutations on the IgH gene during the progression from the CLL cells to the H-RS cells may hamper PCR amplification of the gene. More type 2 cases need to be studied to arrive at a general conclusion concerning the clonal relationship between the CLL cells and the H-RS cells in this subtype.
In cases 1 and 2, we have shown conclusively that the H-RS cells and the corresponding CLL cells had identically rearranged IgH genes. In case 1, a small percentage of the H-RS cells expressed CD20. Therefore, we repeated the experiment by isolating CD20 negative H-RS cells and clonal identity with the CLL cells was again shown. In both cases, all the CDRIII sequences, including those from the H-RS cells, were identical, showing no intraclonal variation, as would be expected in CLL.29 The possibility that the amplified CDRIII products from the H-RS cells were caused by contamination is very unlikely because all negative controls, except for one T cell (2.5%) and one supernatant (2%), showed no amplified products. This contamination rate is far lower than the amplification rate of the H-RS cells (39%) and, in both instances, the amplicon differed in size from the corresponding clonal products. Furthermore, the H-RS cells in case 1 were mostly surrounded by T cells and, in case 2, most of the background lymphocytes were T cells. During micromanipulation, H-RS cell nuclei, instead of whole cells, were removed to avoid disturbing the surrounding cells and the sections were sufficiently thin so that individual cells could be clearly identified. The higher amplification rate of the H-RS cells compared with the CLL cells came as a surprise. It is possible that the H-RS cells were aneuploid or tetraploid and therefore more copies of IgH genes were present per cell.30
Our findings have significant theoretical implications with regard to the origin of H-RS cells. We have clearly shown that mature B cells can transform into cells with the morphology and immunophenotype of H-RS cells. This cellular transformation may also be accompanied by a change in the cellular milieu with numerous T cells and histiocytes as seen in case 2. These observations are consistent with the recent finding suggesting that H-RS cells in classical HD are derived from transformed B cells.27 Perhaps the H-RS cells occurring in CLL and classical HD share common genetic alterations that result in the morphologic and immunophenotypic features, cytokine production profile, and stromal reactions that we recognize as HD.
ACKNOWLEDGMENT
The author thanks the secretarial and technical assistance of Jannene Sass, Michael Lambert, and George Pallas.
Supported by Award No. LB595 from the Department of Health, State of Nebraska.
Address reprint requests to Wing C. Chan, MD, Department of Pathology and Microbiology, University of Nebraska Medical Center, 600 S 42nd St, Omaha, NE 68198-3135.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1784 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal