Abstract
The inv(16)(p13q22) and t(16;16)(p13;q22) cytogenetic abnormalities occur commonly in acute myeloid leukemia (AML), typically associated with French-American-British (FAB) AML-M4Eo subtype. Reverse transcriptase-polymerase chain reaction (RT-PCR) techniques have been recently developed to detect the presence of several variants of the resultant CBFB-MYH11 fusion gene that encodes a CBFβ-smooth muscle myosin heavy chain (SMMHC) fusion protein. We have now determined the clinical use of a polyclonal antibody [anti-inv(16) Ab] directed against a junctional epitope of the most common type of CBFβ-SMMHC fusion protein (type A), which is present in 90% of inv(16)/t(16;16) AML cases. Using flow cytometry, reproducible methods were developed for detection of CBFβ-SMMHC proteins in permeabilized cells; flow cytometric results were then correlated with cytogenetics and RT-PCR detection methods. In an analysis of 42 leukemia cases with various cytogenetic abnormalities and several normal controls, the anti-inv(16) Ab specifically detected all 23 cases that were cytogenetically positive for inv(16) or t(16;16), including a single AML case that was RT-PCR–negative. In addition to detecting all type A fusions, the anti-inv(16) Ab also unexpectedly identified the type C and type D CBFβ-SMMHC fusion proteins. Molecular characterization of one RT-PCR–positive and Ab-positive t(16;16) case with a non-type A product showed a novel previously unreported CBFB-MYH11 fusion (CBFB nt 455-MYH11 nt 1893). Flow cytometric results were analyzed using the Kolmogorov-Smirnov statistic D-value and the median value for positive samples was 0.65 (range, 0.35 to 0.77) versus 0.07 (range, −0.21 to 0.18) in the negative group (P < .0001). The overall concordance between cytogenetics and RT-PCR was 97%, whereas the concordance between flow cytometry and cytogenetics was 100%. Thus, using the anti-inv(16) Ab, all cytogenetically positive and RT-PCR–positive AML cases with inv(16) or t(16;16) could be rapidly identified. This study demonstrates the use of this antibody as an investigational tool in inv(16)/t(16;16) AML and suggests that the development of such reagents may have potential clinical diagnostic use.
THE CHARACTERIZATION and identification of specific genetic abnormalities is critical for the diagnosis and investigation of the acute leukemias. An association between chromosome 16 rearrangements and acute myelomonocytic leukemia with abnormal, dysplastic marrow eosinophils was reported nearly 15 years ago1,2 and subsequently confirmed in several reports.3-6 The pericentric inv(16)(p13q22) and related t(16;16)(p13;q22) cytogenetic abnormalities are observed in approximately 10% of de novo acute myeloid leukemia (AML) cases; such neoplasms are typically characterized by a strong association with French-American-British (FAB) AML-M4Eo morphology7 and relatively favorable therapeutic outcome.2,8-12 Both of these cytogenetic events result in the molecular fusion between theCBFB gene on 16q22 (encoding core binding factor β-subunit [CBFβ]) and the MYH11 gene on 16p13 (encoding a type II smooth muscle myosin heavy chain [SMMHC]).13 The resultant chimeric CBFβ-SMMHC protein is thought to disrupt normal myelomonopoiesis by acting as a dominant negative inhibitor of the function of endogenous CBFβ and its heterodimerizing AML1 (CBFα) partners, thereby altering the expression patterns of critical target genes.14 A murine CBFB-MYH11 knock-in model has shown that heterozygous germline introduction of this chimeric gene abrogates definitive and possibly also primitive embryonic hematopoiesis and is lethal in utero.15
In cases of CBFB-MYH11 AML studied to date, the CBFBfusion site is nearly constant at the mRNA level, with few reported exceptions.14,16,17 However, several differentCBFB-MYH11 transcripts have been described, principally as a result of heterogeneity in MYH11 genomic breakpoints,14,16,18,19 or by alternative splicing inCBFB.20 The large majority (>85%) of cases of inv(16)/t(16;16) AML are associated with a type A fusion transcript, corresponding to an in-frame CBFB nt 495-MYH11 nt 1921 junction.14 Given the specificity of fusion genes as molecular markers in a subset of acute leukemias, appropriate and timely therapy increasingly depends on the rapid identification of these entities. Reverse transcriptase-polymerase chain reaction (RT-PCR) has been widely used for this purpose, including the diagnosis of CBFB-MYH11 fusions in inv(16)/t(16;16) AML.13,16-22 Although this is an effective and sensitive technique, RT-PCR requires sufficient cells for adequate RNA isolation and careful attention to contamination control. In addition, RT-PCR may not detect all CBFB-MYH11 fusions with primer combinations that have been previously described.16 Identification of chimeric protein products arising from specific leukemia-associated genetic fusions is a potentially rapid and attractive alternative to nucleic acid-based methodologies. This approach has been recently reported for detection of the E2A-PBX1 fusion product occurring in acute lymphoblastic leukemia with the t(1;19) abnormality.23 Immunocytochemical analysis of altered macromolecular nuclear structures has also been described for recognition of the t(15;17)-associated PML-RARα fusion protein.24 Western blot analysis using CBFβ antisera has been used to distinguish the 70-kD type A and 95-kD type D CBFβ-SMMHC fusion proteins in inv(16) AML from the normal constituent 21-kD CBFβ protein.25 Previously, a novel antibody recognizing the type A CBFβ-SMMHC fusion protein was described by Liu et al.26 This antibody, named anti-inv16, was shown to detect CBFβ-SMMHC proteins in a small number of AML cases and transfected cells using both Western blotting and immunofluorescence techniques.26 We now demonstrate the successful application of this antibody using flow cytometric methodology in permeabilized cells to specifically recognize the inv(16)/t(16;16)-associated CBFβ-SMMHC in a large series of AML cases. During the course of this study, a novel, previously unreported CBFB-MYH11 fusion mRNA was also identified in a case of t(16;16) AML.
MATERIALS AND METHODS
Patient data and samples.
Patients enrolled in various Southwest Oncology Group (SWOG) leukemia trials (S8600,27 S9031,28 S9034, S9126, S9129,29 and S9300), with cytogenetic inv(16) or t(16;16) abnormalities, were selected for study. Control cases were randomly selected from these SWOG trials, as well as from patient records at the University of New Mexico (UNM). A total of 38 SWOG patients, 3 non-SWOG patients [including cases with inv(16) or other abnormalities], and normal blood and bone marrow samples established the study group. Pretreatment clinical and pathologic data were obtained from the SWOG Statistical Center in Seattle, WA, or the University of New Mexico Hospital. Cytogenetic data on all SWOG patients except 1 (patient no. [PN] 36, Table 1; probable chronic myelogenous leukemia [CML]) were centrally reviewed; non-SWOG cytogenetic data from two UNM patients were analyzed by Dr T. McConnell (SWOG Cytogenetics Committee and UNM Cytogenetics Laboratory). For SWOG leukemia patients, pretreatment routinely stained bone marrow and peripheral blood slides along with centrally performed cytochemical stains (Sudan black B, ANB, ANA) were reviewed by members of the SWOG Leukemia Review Panel. Cryopreserved patient bone marrow or blood samples were accessed through the SWOG Leukemia Repository and the UNM Center for Molecular and Cellular Diagnostics tissue banking facility, UNM Cancer Center. Statistical analysis was performed at the SWOG Statistical Center (Seattle, WA). Cytogenetic and morphology reviews, flow cytometry, and RT-PCR analysis were all performed in blinded fashion, without concurrent knowledge of the individual results.
Summary of Patient Data, Karyotype, RT-PCR, and Anti-inv16 Ab Flow Analysis
| PN . | Age/Sex . | Cytogenetics . | Morphology . | RT-PCR-150 . | FL-DV . | FL-MCS . |
|---|---|---|---|---|---|---|
| 1 | 24/F | 47,XX,inv(16)(p13q22),+21[14]/46,XX[6] | M2Eo | Type D | 0.46 | 42 |
| 2 | 38/F | 46,XX,inv(16)(p13q22) [6] | M2Eo | Type A | 0.66 | 65 |
| 3 | 34/F | 46,XX,inv(16)(p13q22)[8]/46,XX[32] | AML-NOS | Type A | 0.77 | 87 |
| 4 | 51/F | 46,XY,inv(16)(p13q22)[19]/47,idem,+22[1] | M4Eo | Type A | 0.7 | 85 |
| 5 | 37/M | 46,XY,add8(q24.3),inv(16)(p13q22)[42] | M2 | Type A | 0.71 | 57 |
| 6 | 45/M | 46,X,−Y,+8,inv(16)(p13q22)[17]/46,idem,del(7)(q32)[3] | M4Eo | Type A | 0.6 | 49 |
| 7 | 48/F | 46,XX,inv(16)(p13q22)[20] | M4Eo | Type A | 0.56 | 63 |
| 8 | 50/F | 46,XX,inv(16)(p13q22)[13]/46,XX[10] | M4 | Type A | 0.37 | 44 |
| 9 | 44/F | 46,XX,inv(16)(p13q22)[30]/47,idem,+8 [2] | M2 | Type A | 0.69 | 49 |
| 10 | 68/F | 46,XX,t(16;16)(p13;q22)[22] | M4 | Type A | 0.5 | 41 |
| 11 | NA/M | 46,XY,inv(16)(p13q22)[20] | Data unavailable | Type A | 0.64 | 63 |
| 12 | 43/M | 46,XY,inv(16)(p13q22)[21] | Inadequate sample | Type A | 0.42 | 43 |
| 13 | 27/M | 46,XY,inv(16)(p13q22)[20] | M4 | Type A | 0.68 | 60 |
| 14 | 31/M | 46,XY,inv(16)(p13q22)[20] | M4 | Type A | 0.68 | 47 |
| 15 | 20/M | 46,XY,inv(16)(p13q22)[15] | M4Eo | Type A | 0.73 | 65 |
| 16 | 19/M | 46,XY,inv(16)(p13q22)[30] | M2 | Type A | 0.74 | 79 |
| 17 | 23/M | 46,XY,t(16;16)(p13;q22)[9]/46,XY[11] | M4 | Type A | 0.66 | 65 |
| 18 | 45/M | 47,XY,inv(16) (p13q22),+22[20] | M2 | Type A | 0.42 | 38 |
| 19 | 51/F | 47,XX,+8,t(16;16)(p13;q22)[cp3]/46,XX[27] | M1 | Type A | 0.67 | 73 |
| 20 | 51/M | 46,XY,inv(16)(p13q22)[12]/47,XY,+8,inv(16)(p13q22)[12] | M2 | Neg. | 0.35 | 32 |
| 21 | 24/M | 46,XY,t(16;16)(p13;q22)[8]/47,idem,+8[1]/46,XY[2] | M2Eo | Type A | 0.7 | 64 |
| 22 | 43/M | 46,XY,t(16;16)(p13;q22)[49]/47,idem,+22[1] | M2Eo | 400 bp-151 | 0.36 | 33 |
| 23 | 67/M | 47,XY,der(13)t(11;13)(q13;q34),+der(13)t(11;13)(q13;q34), inv(16)(p13q22)[2 0] | M4 | Type C | 0.4 | 24 |
| 24 | 56/F | 46,XX,?del(16)(q22)[?7]/46,XX[17] | M3-152 | Neg. | 0.55 | 48 |
| 25 | 19/M | 46,XY,t(8;21)(q22;q22)[20] | Inadequate sample | Neg. | 0.08 | 13 |
| 26 | 37/F | 46,XX[20] | M1 | Neg. | 0.03 | −2 |
| 27 | 49/F | 46,XX[20] | M4 | Neg. | 0.06 | −3 |
| 28 | 23/F | 46,XX,?der(11)t(9;11)(p22;q23)[2]/46,XX[18] | M4 | Neg. | 0.11 | 9 |
| 29 | 26/F | 46,XX,der(2)t(2;?)(q3?7;?),t(8;21)(q22;q22),der(17)t(17;?)(p13;?) [14]/46,XX[7] | M2Eo | Neg. | −0.21 | −14 |
| 30 | 38/M | 45,XY,t(13q14q)?c[4]/45,XY,t(13q14q)?c,del(11)(q23)[16] | M4Eo | Neg. | −0.12 | −7 |
| 31 | 43/F | 46,XX,t(15;17)(q24;q21)[19]/46,XX[1] | M3 | Neg. | 0.14 | 10 |
| 32 | 3/M | complex hyperdiploid | ALL-B precursor | Neg. | −0.2 | −16 |
| 33 | 26/F | 46,XX,t(15;17)(q24;q21)[17]/46,XX[5] | M3 | Neg. | 0.04 | 1 |
| 34 | 23/M | 46,XX,t(15;17)(q24;q21)[16]/46,XX[4] | M3 | Neg. | 0.15 | 12 |
| 35 | 2/F | 46,XX,t(9;22)(q34.1;q11.2) | ALL B precursor | Neg. | 0.11 | 28 |
| 36 | 80/F | 46,XX,t(9;22)(q34;q11.2) | Probable CML | ND | 0.18 | 22 |
| 37 | 43/M | 46,XY,del(16)(q22q24)[cp4]/46,XY[16] | M2 | Neg. | −0.15 | −10 |
| 38 | 39/F | 46,XX | M2 | Neg. | 0.07 | 6 |
| 39 | 50/F | 46,XX | M1 | Neg. | 0.08 | 7 |
| 40 | 41/M | 46,XY | Data unavailable | Neg. | 0.12 | 11 |
| 41 | 43/M | 46,XY | M4 | Neg. | 0.05 | 4 |
| 42 | NA/NA | Peripheral blood donor-normal control | Not done | Neg. | −0.08 | −13 |
| PN . | Age/Sex . | Cytogenetics . | Morphology . | RT-PCR-150 . | FL-DV . | FL-MCS . |
|---|---|---|---|---|---|---|
| 1 | 24/F | 47,XX,inv(16)(p13q22),+21[14]/46,XX[6] | M2Eo | Type D | 0.46 | 42 |
| 2 | 38/F | 46,XX,inv(16)(p13q22) [6] | M2Eo | Type A | 0.66 | 65 |
| 3 | 34/F | 46,XX,inv(16)(p13q22)[8]/46,XX[32] | AML-NOS | Type A | 0.77 | 87 |
| 4 | 51/F | 46,XY,inv(16)(p13q22)[19]/47,idem,+22[1] | M4Eo | Type A | 0.7 | 85 |
| 5 | 37/M | 46,XY,add8(q24.3),inv(16)(p13q22)[42] | M2 | Type A | 0.71 | 57 |
| 6 | 45/M | 46,X,−Y,+8,inv(16)(p13q22)[17]/46,idem,del(7)(q32)[3] | M4Eo | Type A | 0.6 | 49 |
| 7 | 48/F | 46,XX,inv(16)(p13q22)[20] | M4Eo | Type A | 0.56 | 63 |
| 8 | 50/F | 46,XX,inv(16)(p13q22)[13]/46,XX[10] | M4 | Type A | 0.37 | 44 |
| 9 | 44/F | 46,XX,inv(16)(p13q22)[30]/47,idem,+8 [2] | M2 | Type A | 0.69 | 49 |
| 10 | 68/F | 46,XX,t(16;16)(p13;q22)[22] | M4 | Type A | 0.5 | 41 |
| 11 | NA/M | 46,XY,inv(16)(p13q22)[20] | Data unavailable | Type A | 0.64 | 63 |
| 12 | 43/M | 46,XY,inv(16)(p13q22)[21] | Inadequate sample | Type A | 0.42 | 43 |
| 13 | 27/M | 46,XY,inv(16)(p13q22)[20] | M4 | Type A | 0.68 | 60 |
| 14 | 31/M | 46,XY,inv(16)(p13q22)[20] | M4 | Type A | 0.68 | 47 |
| 15 | 20/M | 46,XY,inv(16)(p13q22)[15] | M4Eo | Type A | 0.73 | 65 |
| 16 | 19/M | 46,XY,inv(16)(p13q22)[30] | M2 | Type A | 0.74 | 79 |
| 17 | 23/M | 46,XY,t(16;16)(p13;q22)[9]/46,XY[11] | M4 | Type A | 0.66 | 65 |
| 18 | 45/M | 47,XY,inv(16) (p13q22),+22[20] | M2 | Type A | 0.42 | 38 |
| 19 | 51/F | 47,XX,+8,t(16;16)(p13;q22)[cp3]/46,XX[27] | M1 | Type A | 0.67 | 73 |
| 20 | 51/M | 46,XY,inv(16)(p13q22)[12]/47,XY,+8,inv(16)(p13q22)[12] | M2 | Neg. | 0.35 | 32 |
| 21 | 24/M | 46,XY,t(16;16)(p13;q22)[8]/47,idem,+8[1]/46,XY[2] | M2Eo | Type A | 0.7 | 64 |
| 22 | 43/M | 46,XY,t(16;16)(p13;q22)[49]/47,idem,+22[1] | M2Eo | 400 bp-151 | 0.36 | 33 |
| 23 | 67/M | 47,XY,der(13)t(11;13)(q13;q34),+der(13)t(11;13)(q13;q34), inv(16)(p13q22)[2 0] | M4 | Type C | 0.4 | 24 |
| 24 | 56/F | 46,XX,?del(16)(q22)[?7]/46,XX[17] | M3-152 | Neg. | 0.55 | 48 |
| 25 | 19/M | 46,XY,t(8;21)(q22;q22)[20] | Inadequate sample | Neg. | 0.08 | 13 |
| 26 | 37/F | 46,XX[20] | M1 | Neg. | 0.03 | −2 |
| 27 | 49/F | 46,XX[20] | M4 | Neg. | 0.06 | −3 |
| 28 | 23/F | 46,XX,?der(11)t(9;11)(p22;q23)[2]/46,XX[18] | M4 | Neg. | 0.11 | 9 |
| 29 | 26/F | 46,XX,der(2)t(2;?)(q3?7;?),t(8;21)(q22;q22),der(17)t(17;?)(p13;?) [14]/46,XX[7] | M2Eo | Neg. | −0.21 | −14 |
| 30 | 38/M | 45,XY,t(13q14q)?c[4]/45,XY,t(13q14q)?c,del(11)(q23)[16] | M4Eo | Neg. | −0.12 | −7 |
| 31 | 43/F | 46,XX,t(15;17)(q24;q21)[19]/46,XX[1] | M3 | Neg. | 0.14 | 10 |
| 32 | 3/M | complex hyperdiploid | ALL-B precursor | Neg. | −0.2 | −16 |
| 33 | 26/F | 46,XX,t(15;17)(q24;q21)[17]/46,XX[5] | M3 | Neg. | 0.04 | 1 |
| 34 | 23/M | 46,XX,t(15;17)(q24;q21)[16]/46,XX[4] | M3 | Neg. | 0.15 | 12 |
| 35 | 2/F | 46,XX,t(9;22)(q34.1;q11.2) | ALL B precursor | Neg. | 0.11 | 28 |
| 36 | 80/F | 46,XX,t(9;22)(q34;q11.2) | Probable CML | ND | 0.18 | 22 |
| 37 | 43/M | 46,XY,del(16)(q22q24)[cp4]/46,XY[16] | M2 | Neg. | −0.15 | −10 |
| 38 | 39/F | 46,XX | M2 | Neg. | 0.07 | 6 |
| 39 | 50/F | 46,XX | M1 | Neg. | 0.08 | 7 |
| 40 | 41/M | 46,XY | Data unavailable | Neg. | 0.12 | 11 |
| 41 | 43/M | 46,XY | M4 | Neg. | 0.05 | 4 |
| 42 | NA/NA | Peripheral blood donor-normal control | Not done | Neg. | −0.08 | −13 |
Abbreviations: FL-DV, flow cytometry KS D value (see the Materials and Methods); FL-MCS, flow cytometry median channel shift.
Type A, 415-bp product; type C, 1,200-bp product; and type D, 1,400-bp product.
Novel CBFB-MYH11 fusion PCR product.
Atypical promyelocytic leukemia; not classic M3 (refer to text).
RT-PCR for CBFB-MYH11.
Total RNA was isolated from cell pellets or suspensions using RNazol B (Tel-Test, Inc, Friendswood, TX), according to the manufacturer's directions. One to two micrograms of RNA was reverse transcribed from random hexamers in a total volume of 20 μL containing 50 mmol/L KCl, 10 mmol/L Tris (pH 8.4), 5 mmol/L MgCl2, 1 mmol/L dNTP, 50 pmol random hexamer, 1 U/μL RNAsin (Promega, Madison, WI), 5 mmol/L dithiothreitol (DTT), and 100 U Moloney's murine leukemia virus-RT (MMLV-RT; GIBCO-BRL Life Technologies, Grand Island, NY), under the following conditions: 23°C for 10 minutes, 42°C for 60 minutes, 95°C for 5 minutes, and 5°C hold. PCR amplifications were subsequently performed in a 50 μL volume (10 mmol/L Tris, 50 mmol/L KCl, 1.5 mmol/L MgCl2, 200 μmol/L dNTP, 20 pmol of each primer, and 2.5 U Taq polymerase [Perkin-Elmer/Roche, Branchburg, NJ]), using 7.5 μL of cDNA for each of inv(16) primer sets C1-M1 and C1-M213 and 5 μL for the β2-microglobulin control primer set. Cycling parameters were 95°C for 30 seconds, 59°C for 45 seconds, and 72°C for 90 seconds for 3 cycles, and then 95°C for 30 seconds, 57°C for 30 seconds, and 72°C for 60 seconds for 30 cycles. The PCR was preceded by an initial 5 minutes of denaturation at 95°C and followed by a terminal 8 minutes of extension at 72°C. Primer sequences are as follows (all 5′ to 3′): C1, GCAGGCAAGGTATATTTGAAGG; M1, CTCTTCTCCTCATTCTGCTC; M2, ACTGCAGCTCCTGCACCTGC; doM3′, CGTTCTTGCCCACGTCAT; β2M, 5′-GAAAAAGATGAGTATGCCTG; β2M, 3′-ATCTTCAAACCTCCATGATG.
PCR products were analyzed by electrophoresis in 1.5% agarose gels (Seakem ME; FMC Bioproducts, Rockland, ME), vacuum transferred to nylon membranes (Pall Biodyne, East Hills, NY), and UV cross-linked. Membranes were subsequently hybridized with a biotinylated CBFB oligonucleotide sequence (inv16 probe-ATAGAGACAGGTCTCATCGG)13,18 or a novel biotinylated junctional sequence (GACACGCGACAGCTCCAAGG) and detected by chemiluminescence (ECL System; Amersham Life Sciences, Arlington Heights, IL) on autoradiograph film (Kodak XAR; Eastman Kodak, Rochester, NY). In certain instances, a 0.8-kbCBFB cDNA13 or a PCR-generated 550-bp 3′CBFB cDNA fragment were 32P-labeled by nick translation and used as probes. All PCR primers and oligoprobes were synthesized by the UNM Protein Chemistry facility.
DNA sequence analysis.
PCR products were ligated into pCR 2.1 vector using the Invitrogen T-A cloning kit (Invitrogen, San Diego, CA) and used to transform DH5αEscherichia coli, according to the manufacturer's instructions. Transformants were spread onto kanamycin X-gal–coated plates. Several white colonies were selected for minipreparations. After plasmid isolation and EcoRI (Promega) digestion, agarose gel analysis confirmed inserts of the correct size. Selected clones were sequenced in both directions using 35S dATP and the Sequenase v 2.0 method (US Biochemicals, Cleveland, OH) with forward and reverse primers C1 and M1, respectively. Sequencing reactions were analyzed on a 6% polyacrylamide gel (Sequagel-6; National Diagnostics, Atlanta, GA) under standard conditions. Manual sequencing was repeated twice from different clones and the sequence data compared with published CBFB13 and MYH11(K. Okajima, unpublished, GenBank Accession No. X69292, 1992)cDNA sequences.
Flow cytometric analysis with anti-inv16 Ab.
On initial receipt, samples were enriched for leukemic blasts by Ficoll-Hypaque (Pharmacia LKB, Piscataway, NJ) density gradient centrifugation under sterile conditions. Cells were cryopreserved in 90% fetal calf serum/10% dimethyl sulfoxide and stored at −135°C. For this study, cryopreserved cells were defrosted rapidly, pelleted, and resuspended in PAB (phosphate-buffered saline [PBS], albumin, and Na azide) before staining. Viability was determined by trypan blue exclusion and was greater than 90% in all cases. CD34-fluorescein isothiocyanate (Becton Dickinson, Mountain View, CA) was used to allow simultaneous identification of the leukemic blast subpopulation, where applicable. After staining, cells were washed twice with Hank's balanced salt solution (HBSS), fixed with 3.7% formaldehyde/PBS for 10 minutes at room temperature, and then permeabilized with 50% acetone/HBSS for 5 minutes at 4°C or, alternatively, with 0.05% Tween 20/PBS for 15 minutes at 37°C. Both permeabilization methods were found to yield comparable flow cytometric results. After two washes with HBSS, cells were resuspended in PAB and 4 μL of anti-inv16 Ab was added for incubation at 4°C for 40 minutes. Polyclonal anti-inv16 Ab is an affinity-purified rabbit antibody recognizing an 18 amino acid epitope region spanning the fusion site in the type A CBFβ-SMMHC chimeric protein of the inv(16)/t(16;16) abnormality.26 Polyclonal rabbit IgG was used as isotype control. After incubation, cells were washed and stained with phycoerythrin-conjugated goat antirabbit Ig for 30 minutes at 4°C. Cells were then washed twice, suspended in 200 μL of PAB, and analyzed on a FACScan cytometer (Becton Dickinson) using LYSIS II software with gating to exclude the lymphocyte region. Both the median fluorescence channel intensity shift and D-value (calculated from Kolmogorov-Smirnov statistics) were used in the analysis of flow cytometry data. Validation of the D-value statistic for comparative flow cytometric assessment of samples has been previously described.30 31
Cell dilution/sensitivity studies.
Viable cells from a t(16;16)-positive AML (PN 19, Table 1) with a type A CBFB-MYH11 fusion were diluted into either normal bone marrow or negative control leukemic sample cells (PN 30, Table 1) as follows: 100% t(16;16), 1:1, 1:5, 1:10, 1:20, 1:50, 1:100, 1:103, 1:104, and 100% negative control. Samples were split proportionately for flow cytometry and RT-PCR. Total RNA was isolated immediately and RT-PCR performed as described above. After PCR and gel transfer, blots were probed with a type A junctional sequence oligoprobe (5′-GCTCATGGACCTCCATTTCC). Flow cytometric analysis with anti-inv16 Ab was performed as outlined above, with the exception that a greater number of events were collected per dilution.
Statistical methods.
Distributions of median channel shifts and D-values were compared between subgroups of patients using the Wilcoxon rank sum test, with results reported as two-tailed P values.
RESULTS
Patient pretreatment characteristics.
In total, 42 samples from leukemia patients and normal controls were selected for this study. The median patient age was 40 years (range, 2 to 80 years), and the male:female ratio was 1:1. The median presentation white blood cell count among these patients was 41.4 (range, 1.6 to 229.8), and the median bone marrow blast count was 70% (range, 3% to 95%). Cytogenetic data were reviewed in 41 patients, 23 of whom were reported to have an inv(16) or t(16;16); 11 patients had other leukemia-associated karyotypic abnormalities, including t(8;21), t(15;17), t(9;22), and del(16q). Six AML cases had normal cytogenetics. One patient (PN 24, Table 1) had a suspected chromosome 16 abnormality [?del(16q) v inv(16)] in a diagnostic specimen. Morphologic classifications were available for 36 patients. The inv(16)/t(16;16) cases included AML-M1, M2, and M4 FAB subtypes. Only 8 of 21 (38%) morphologically reviewed inv(16)/t(16;16)-positive leukemias were characterized by the presence of classical abnormal eosinophils (M4Eo or M2Eo); however, abnormal eosinophils were also noted in 2 cases not associated with these karyotypes. Controls included a spectrum of AML cases, as well as 1 case of chronic myeloid leukemia, 2 acute lymphoblastic leukemias, and normal peripheral blood and bone marrow. Pretreatment patient data, morphologic classification, and cytogenetic features on these patients are summarized in Table 1.
RT-PCR analysis for CBFB-MYH11 fusion.
Forty leukemia samples, including inv(16)/t(16;16) AML and controls and 1 peripheral blood specimen, were analyzed for the presence of the chimeric CBFB-MYH11 mRNA by RT-PCR, the results of which are presented in Table 1. Twenty-two of 23 cytogenetically confirmed inv(16)/t(16;16) cases were PCR-positive and of these, 20 (91%) demonstrated the common CBFB nt 495-MYH11 nt 1921 type A fusion transcript (415-bp PCR product, PN 2 through 19 and PN 21, Table 1). One each of type C and type D fusions were identified (1.2-kb and 1.4-kb PCR fragment lengths, PN 1 and PN 23, respectively, Table1). All positive PCR results were confirmed by hybridization with the internal CBFB oligonucleotide probe (Fig 1A and B).
Detection of CBFB-MYH11 fusion transcripts by RT-PCR. (A) Agarose gel electrophoresis of C1-M1 primer set amplification products. (B) Hybridization of PCR products with the inv(16) CBFB internal oligoprobe. In both panels, lane numbers 1 through 6 correspond to samples from PN 14, 13, 22, 1, 18, and 23, respectively (Table 1). Lane 7 represents a relapse sample from PN 23. Lane 8 is a negative control (no RNA). Sample from PN 22 (lane 3) displays a prominent, but slightly smaller PCR product than expected for a type A fusion (∼400 bp in [A]) and does not hybridize with the oligoprobe (B). This PCR product did hybridize positively with a 0.8-kb CBFB cDNA probe (not shown). Lane 4 demonstrates a type D fusion; lanes 6 and 7 show a type C fusion.
Detection of CBFB-MYH11 fusion transcripts by RT-PCR. (A) Agarose gel electrophoresis of C1-M1 primer set amplification products. (B) Hybridization of PCR products with the inv(16) CBFB internal oligoprobe. In both panels, lane numbers 1 through 6 correspond to samples from PN 14, 13, 22, 1, 18, and 23, respectively (Table 1). Lane 7 represents a relapse sample from PN 23. Lane 8 is a negative control (no RNA). Sample from PN 22 (lane 3) displays a prominent, but slightly smaller PCR product than expected for a type A fusion (∼400 bp in [A]) and does not hybridize with the oligoprobe (B). This PCR product did hybridize positively with a 0.8-kb CBFB cDNA probe (not shown). Lane 4 demonstrates a type D fusion; lanes 6 and 7 show a type C fusion.
A strong correlation between cytogenetic and PCR findings was observed, with a 97% overall concordance (38 of 39 cases analyzed by both methods). PCR positivity in the absence of a karyotypic inv(16) or t(16;16) was not encountered. Conversely, one cytogenetically confirmed inv(16) AML (PN 20, Table 1) was PCR negative with the primer sets used, despite the presence of amplifiable cDNA. One other unusual case (PN 24, Table 1), with a suspected chromosome 16 abnormality, demonstrated PCR negativity for CBFB-MYH11 transcript in both diagnostic and relapse samples. Because of the lack of an unequivocal karyotype in the diagnostic sample from PN 24, this case was not considered in the concordance analysis. These latter cases are discussed in further detail below (see “Atypical Cases”). All negative controls in this study lacked the CBFB-MYH11 fusion by RT-PCR analysis.
Flow cytometry with anti-inv16 Ab.
All 42 subjects were evaluable by flow cytometry with anti-inv16 Ab. Twenty-four anti-inv16 Ab positive cases were identified; interestingly, these included the two non-type A fusions noted by RT-PCR analysis. Representative flow cytometric results with anti-inv16 Ab are shown in Fig 2 and summarized in Table 1. Simultaneous dual-color measurement of CD34 allowed enhanced delineation of the blast population in many cases. In positive samples, both low granularity CD34(+) blasts and higher granularity CD34(−) cells had detectable CBFβ-SMMHC fusion protein, although the fluorescence intensity was slightly greater in the CD34(+) population (data not shown).
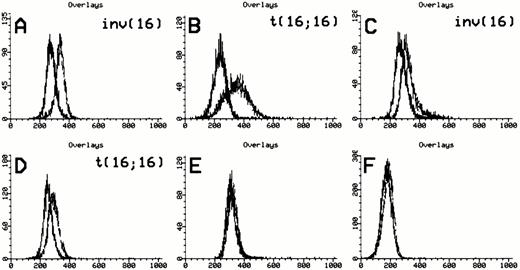
Anti-inv16 antibody detection of CBFβ-SMMHC fusions. Flow cytometric detection of CBFβ-SMMHC using anti-inv16 Ab. The black trace represents isotype control. The gray trace represents anti-inv(16) Ab. (A and B) PN 13 and PN 21, respectively, demonstrating detection of type A fusion product; (C) PN 1 with type D fusion detected by anti-inv16 Ab; (D) PN 22 with novel CBFB-MYH11fusion; (E and F) PN 28 and PN 30 negative controls. Refer to Table 1and text for details.
Anti-inv16 antibody detection of CBFβ-SMMHC fusions. Flow cytometric detection of CBFβ-SMMHC using anti-inv16 Ab. The black trace represents isotype control. The gray trace represents anti-inv(16) Ab. (A and B) PN 13 and PN 21, respectively, demonstrating detection of type A fusion product; (C) PN 1 with type D fusion detected by anti-inv16 Ab; (D) PN 22 with novel CBFB-MYH11fusion; (E and F) PN 28 and PN 30 negative controls. Refer to Table 1and text for details.
The median value of the fluorescence median channel shift (MCS) among positive cases was 53 (range, 24 to 87) versus 5 (range, −16 to 28) for negative cases (P < .0001). Similarly, the median of the calculated D-value for positive samples was 0.65 (range, 0.35 to 0.77), compared with 0.07 (range, −0.21 to 0.18) in the negative group (P < .0001). A D-value greater than 0.35 or MCS of greater than 30 with anti-inv16 Ab established a positive result in this study, although the MCS was found to exhibit greater variability in a given case, in repeated experiments. Using these criteria, positive and negative flow cytometric results were perfectly concordant with the presence or absence of the inv(16)/t(16;16) karyotypic abnormality, respectively (100% of 40 cases analyzed with combined data available). It is of note that the diagnostic AML sample from 1 case with an indeterminate chromosome 16 abnormality (PN 24, Table 1) was positive with anti-inv16 Ab, despite the lack of aCBFB-MYH11 fusion by PCR analysis (see below, “Atypical Cases”). In all positive cases, the visual degree of anti-inv16 Ab fluorescence shift compared with isotype control was adequately pronounced in the histogram curves.
Atypical cases.
Overall, 3 patients with either discordant results or atypical findings were encountered. One RT-PCR–negative AML case (PN 20, Table 1) with a reported inv(16) cytogenetic abnormality was positive with anti-inv16 Ab. This case is considered likely to have alternative CBFBand/or MYH11 breakpoint/fusion sites that are not detected with either C1-M1 or C1-M2 primer combinations. This possibility is supported by the relatively weak, but positive fluorescence intensity shift (flow DV = 0.35, MCS = 32) observed with anti-inv16 Ab compared with isotype control. Unfortunately, no further patient material was available for additional molecular analysis in this case. A second RT-PCR–negative case (PN 24, Table 1) also had a positive flow result observed in both diagnostic (Table 1) and relapse (data not shown) specimens from this patient; however,CBFB-MYH11 RT-PCR analysis was negative. This patient had a suspected morphologic diagnosis of acute promyelocytic leukemia (AML-M3) with atypical features. This case was characterized by a uniform population of CD34−/HLA-DR−/CD33+/CD13+/CD56+granular blasts. Cytogenetic analysis suggested a chromosome 16 abnormality [?del(16q) v inv(16)] in a subset of cells from the diagnostic sample; however, a subsequent study of both diagnostic and relapse samples by metaphase FISH technique (Oncor probe kit; Oncor, Gaithersberg, MD) failed to show an inv(16) or t(16;16) (data not shown). Using a partial CBFB cDNA probe, Southern hybridization analysis of the distal CBFB locus in relapse sample DNA from this second patient did not demonstrate rearrangements (data not shown). However, resolution of CBFB rearrangements in inv(16)/t(16;16) AML samples by Southern analysis, as attempted here, may be significantly compromised by the generation of very large fusion DNA fragments using typical restriction digests.32 We were unable to assess for rearrangements in the MYH11 breakpoint region, as has been described.32 In a previous series of CD56+ natural killer (NK) phenotype AML,33 inv(16) or t(16;16) karyotypes were not identified; however, the total number of reported cases was small. Thus, although case PN 24 may represent a flow cytometric false-positive with anti-inv16 Ab, the presence of a cryptic CBFB-MYH11 fusion cannot be excluded, particularly in light of the difficulty in adequately karyotyping this AML case. It is of note that no equivocal results were encountered in any of the other control cases, which included several AML morphologic subtypes.
Finally, a t(16;16) AML case (PN 22, Table 1) that which had detectable CBFβ-SMMHC protein by flow cytometric technique (Fig 2D) demonstrated a slightly smaller size PCR amplicon than the usual type A product (Fig1A, lane 3). Furthermore, this PCR product did not hybridize with the CBFB internal oligoprobe (Fig 1B, lane 3), but did hybridize with the 0.8-kb CBFB cDNA (data not shown). Sequencing of the PCR product showed a novel CBFB-MYH11 mRNA fusion at CBFB nt 455 and MYH11 nt 1893 (Fig 3A). This fusion is predicted to have a 404-bp PCR product size with primers C1 and M1, as observed. RT-PCR analysis of this leukemic sample with primer C1 and a novel tumor-specific MYH11 primer (doM3′), followed by hybridization with a junction-specific oligoprobe (doPR), confirmed the presence of this new fusion type (Fig 3B, lanes 1 and 2).
Novel CBFB-MYH11 fusion in t(16;16) AML: PN 22. (A) Partial chimeric cDNA junctional sequence of 400-bp PCR product from PN 22. Sequencing was performed in both directions using primers C1 and M1. The new CBFB-MYH11 fusion site is shown by an asterisk at CBFB nt 455 and MYH11 nt 1893. The ORF is not disrupted by this fusion, but is 12 nt shorter than the type A fusion. (B) RT-PCR analysis confirming the presence of the new fusion type in PN 22 AML. PCR was performed using primers C1 and doM3′, followed by specific oligonucleotide hybridization (do probe), as described in the Materials and Methods. Lane 1, PCR amplification of reverse transcribed cDNA from PN 22 AML sample; lane 2, seminested PCR amplification from aliquot of first round C1-M1 PCR product of PN 22 sample; lane 3, AML-M4 sample lacking the inv(16)/t(16;16) andCBFB-MYH11 fusion by RT-PCR (this case not included in study group); lane 4, PN 11 sample with type A CBFB-MYH11 fusion by C1-M1 PCR; lane 5, negative control (noRNA).
Novel CBFB-MYH11 fusion in t(16;16) AML: PN 22. (A) Partial chimeric cDNA junctional sequence of 400-bp PCR product from PN 22. Sequencing was performed in both directions using primers C1 and M1. The new CBFB-MYH11 fusion site is shown by an asterisk at CBFB nt 455 and MYH11 nt 1893. The ORF is not disrupted by this fusion, but is 12 nt shorter than the type A fusion. (B) RT-PCR analysis confirming the presence of the new fusion type in PN 22 AML. PCR was performed using primers C1 and doM3′, followed by specific oligonucleotide hybridization (do probe), as described in the Materials and Methods. Lane 1, PCR amplification of reverse transcribed cDNA from PN 22 AML sample; lane 2, seminested PCR amplification from aliquot of first round C1-M1 PCR product of PN 22 sample; lane 3, AML-M4 sample lacking the inv(16)/t(16;16) andCBFB-MYH11 fusion by RT-PCR (this case not included in study group); lane 4, PN 11 sample with type A CBFB-MYH11 fusion by C1-M1 PCR; lane 5, negative control (noRNA).
Sensitivity of anti-inv16 Ab for detection of minimal disease.
Anti-inv16 Ab flow cytometric analysis of cells from a t(16;16)-positive AML (PN 19, type A CBFβ-SMMHC fusion, Table 1) diluted serially into either normal bone marrow or AML negative control (PN 30, Table 1) cells showed a detection sensitivity in the range of 1:5 to 1:10 (Fig 4A). Selective gating of CD34+ cells did not appreciably increase the detection limit. Comparative RT-PCR dilutional analysis demonstrated detectableCBFB-MYH11 fusion transcript to a level of 1 in 104cells (Fig 4B). Thus, RT-PCR assays appear to be more sensitive than flow cytometric analysis with anti-inv16 Ab for minimal disease assessment. Despite its excellent specificity, anti-inv(16) Ab has relatively low sensitivity, possibly due to low affinity and the need for cell permeabilization to detect a cytoplasmic epitope.
Dilutional sensitivity of anti-inv16 antibody versus RT-PCR for detection of type A fusion. (A) Dilutional sensitivity of anti-inv16 Ab. PN 19 AML cells were serially diluted into normal bone marrow cells and analyzed by flow cytometry. The black trace represents isotype control. The gray trace represents anti-inv(16) Ab. (A) Undiluted PN 19 AML cells; (b and C) Cell dilutions shown to level of undetectable fluorescence shift (1:20). The D-values for results in (A), (B), and (C) are 0.71, 0.58, and 0.06, respectively. (B) Single-round RT-PCR of PN 19 AML sample [inv(16)+ with type A fusion] and serial normal bone marrow dilutions, using primers C1 and M1. PCR products were hybridized with a type A junction specific oligonucleotide probe. Lanes 1 through 6, undiluted PN 19 sample, 1:10 dilution, 1:20 dilution, 1:100 dilution, 1:103dilution., and 1:104 dilution, respectively; lane 7, pure normal marrow; lane 8, negative control (no RNA).
Dilutional sensitivity of anti-inv16 antibody versus RT-PCR for detection of type A fusion. (A) Dilutional sensitivity of anti-inv16 Ab. PN 19 AML cells were serially diluted into normal bone marrow cells and analyzed by flow cytometry. The black trace represents isotype control. The gray trace represents anti-inv(16) Ab. (A) Undiluted PN 19 AML cells; (b and C) Cell dilutions shown to level of undetectable fluorescence shift (1:20). The D-values for results in (A), (B), and (C) are 0.71, 0.58, and 0.06, respectively. (B) Single-round RT-PCR of PN 19 AML sample [inv(16)+ with type A fusion] and serial normal bone marrow dilutions, using primers C1 and M1. PCR products were hybridized with a type A junction specific oligonucleotide probe. Lanes 1 through 6, undiluted PN 19 sample, 1:10 dilution, 1:20 dilution, 1:100 dilution, 1:103dilution., and 1:104 dilution, respectively; lane 7, pure normal marrow; lane 8, negative control (no RNA).
DISCUSSION
The importance of defining the molecular pathology of leukemic subtypes for rapid diagnosis, stratification, prognostication, and, more fundamentally, delineating unique biologic features is being increasingly recognized. From the diagnostic standpoint, identification of particular leukemic entities should be rapid, sensitive, and reproducible. The inv(16) and related t(16;16) cytogenetic abnormalities occur in approximately 10% of de novo AML and are highly associated with FAB AML-M4Eo subtype. However, as demonstrated by this and previous studies,9,16,17,21,22,34,35 these cytogenetic findings may be associated with several other FAB AML types, with or without abnormal eosinophils, and the pathologic-cytogenetic correlation would appear to be somewhat dependent on the case selection bias in various studies. In addition, these karyotypic events may be subtle and difficult to discern at the cytogenetic level of resolution.36
Molecular analysis of the CBFB-MYH11 fusion is a specific method for diagnosis and potential monitoring of inv(16)/t(16;16) AML. Several techniques have been described for identification of this genetic anomaly, including FISH,13,21,37 Southern hybridization analysis for CBFB or MYH11rearrangements,13,32,38,39 and, most commonly, RT-PCR.13 16-22 The latter methodology is relatively rapid and can be used for residual disease assessment due to the enhanced sensitivity of PCR. However, the requirements for high-quality RNA, multiple procedural steps, and the possibility of sample contamination currently remain the most important shortcomings of this technique. Additionally, RT-PCR approaches must be capable of detecting the numerous CBFB-MYH11 fusion variants. Immunologic methods of detection have the potential advantages of rapidity, relative technical ease, and the ability to use smaller numbers of viable cells.
The type A CBFβ-SMMHC fusion protein has previously been studied in a small number of inv(16) AML cases by Western blot analysis and indirect immunofluorescence using anti-inv16 Ab.26 More recent evidence has convincingly demonstrated cytoplasmic distribution of the CBFβ-SMMHC protein in transfected cells with this antibody.40 We have also performed indirect immunofluorescence experiments on a limited number of primary inv(16) AML samples using anti-inv16 Ab, confirming cytoplasmic localization of the CBFβ-SMMHC protein (data not shown). These studies thus suggest that one mechanism by which the CBFβ-SMMHC protein may act in a dominant negative fashion is by sequestration of AML1 (CBFα) protein within the cytoplasm.
This is the first report to examine the use of this polyclonal antibody as an investigational and potentially a diagnostic tool in a larger group of AML samples. The method presented here allows for rapid fixation and permeabilization of cells to detect the CBFβ-SMMHC protein and simultaneously assess surface markers, such as CD34. Although cell permeabilization techniques are often used in experimental situations requiring characterization of intracellular molecules, this study further demonstrates the potential for using novel antibodies to detect specific intracytoplasmic or intranuclear leukemic fusion proteins in a relatively routine flow phenotyping setting. An excellent detection rate and specificity were observed using anti-inv16 Ab in our study group of leukemic patients. Of 23 cytogenetically confirmed inv(16)/t(16;16) AML cases, flow cytometric detection of the CBFβ-SMMHC protein was positive in all. Accordingly, no positive results were observed within the group of 18 control leukemias and normal peripheral blood and bone marrow specimens. Furthermore, although anti-inv16 Ab was raised against the type A common CBFβ-SMMHC fusion protein (junctional sequence epitope), the antibody proved capable of detecting one each of type C and type D fusions present within the group of positive cases. This antibody has been previously reported to be nonreactive with either endogenous CBFβ or SMMHC proteins.26 These foregoing observations suggest that this polyclonal rabbit antibody is capable of identifying multiple CBFβ-SMMHC fusion types, including apparently rare novel inv(16)/t(16;16) events. In part, this broader spectrum of specificity may be related to a combination of shared epitope recognition [ie, the relative invariability of the CBFβ fusion site in most inv(16)/t(16;16) AML] and some degree of cross-affinity for the repetitive coiled-coil regions of the variable SMMHC tail segments in the different fusions. In support of this latter concept is the finding that anti-inv16 Ab was also capable of recognizing a novel CBFβ-SMMHC fusion in a case of t(16;16) AML (PN 22, Table 1). RT-PCR and sequence analysis of the PCR product in this case showed a previously undescribed CBFB-MYH11 transcript, with fusion sites atCBFB nt 455 and MYH11 nt 1893. This fusion is predicted to result in an in-frame CBFB-MYH11 mRNA, differing in size from the type A fusion transcript by a deletion of 12 nt. Examination of the reported CBFB13 and MYH11 (K. Okajima, unpublished findings, GenBank Accession No.X69292, 1992) cDNA sequences did not show the presence of classical flanking donor and acceptor splice sites, respectively; thus, the mechanism underlying the generation of this fusion type is presently unclear.
RT-PCR analysis for minimal residual disease (MRD) assessment in inv(16)/t(16;16) AML has been previously described,18,19,21,22 with relatively short median follow-up intervals and somewhat inconclusive results. Two recent reports have suggested that the semiquantitative assessment ofCBFB-MYH11 transcript after standard induction chemotherapy may be correlated with outcome and relapse risk post-therapy.41,42 In the present study, the dilutional sensitivity of anti-inv16 Ab in detecting the CBFβ-SMMHC protein was determined to be in the range of only 1:5 to 1:10 leukemic cells to normal cells, in comparison to RT-PCR technique, which consistently detects chimeric CBFB-MYH11 type A fusion events at the level of at least 1 in 104cells.19,21,22,41 The flow cytometric findings obtained here may reflect differences in the relative capability of identifying intracellular targets, as opposed to more accessible surface molecules, the need for cell permeabilization, and a potentially low antibody affinity. Thus, the use of this antibody for monitoring posttherapeutic at risk patients at an appropriately sensitive predictive level does not appear to be practical, as evaluated with the current protocol. However, because the specificity and localization of anti-inv16 Ab has been previously established26 40 and corroborated in this study, these results should provide the impetus for development of a high-quality monoclonal antibody.
In summary, we have demonstrated the flow cytometric use of an antibody, anti-inv16 Ab, that recognizes the common type A CBFβ-SMMHC leukemic fusion protein and apparently rarer fusion types occurring in inv(16)/t(16;16) myeloid leukemia. In addition, we describe a previously unreported CBFB-MYH11 chimeric transcript with unique fusion sites at the mRNA level in both genes. The method described here is rapid, and the use of this antibody should enhance investigational efforts and, potentially, clinical diagnosis in inv(16)/t(16;16) AML.
M.L.S., C.R., D.R.H., and C.L.W. are members of the Southwest Oncology Group Leukemia Biology and Cytogenetics Programs, San Antonio, TX. D.S.V. was a Fellow of the R. Samuel McLaughlin Foundation of Canada during the course of this work. P.P.L. is a Special Fellow of the Leukemia Society of America. Supported by Department of Health and Human Services National Institutes of Health Grant No. U01 CA32102 supporting the SWOG Leukemia, Leukemia Biology and Cytogenetics Programs.
Address correspondence to Cheryl L. Willman, MD, University of New Mexico Cancer Center, 900 Camino de Salud NE, Albuquerque, NM 87131. Address reprint requests to SWOG Operations Office, 14908 Omicron Dr, San Antonio, TX 78245-3217.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 1. Detection of CBFB-MYH11 fusion transcripts by RT-PCR. (A) Agarose gel electrophoresis of C1-M1 primer set amplification products. (B) Hybridization of PCR products with the inv(16) CBFB internal oligoprobe. In both panels, lane numbers 1 through 6 correspond to samples from PN 14, 13, 22, 1, 18, and 23, respectively (Table 1). Lane 7 represents a relapse sample from PN 23. Lane 8 is a negative control (no RNA). Sample from PN 22 (lane 3) displays a prominent, but slightly smaller PCR product than expected for a type A fusion (∼400 bp in [A]) and does not hybridize with the oligoprobe (B). This PCR product did hybridize positively with a 0.8-kb CBFB cDNA probe (not shown). Lane 4 demonstrates a type D fusion; lanes 6 and 7 show a type C fusion.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/6/10.1182_blood.v91.6.1882/2/m_blod4064601.jpeg?Expires=1769093143&Signature=ic2HizkObgZJqZwClHnKQAVcyoWHMgXXNfFDERF-C6i8C9YRpuB7I1wd0qmlliljDSKL0hLtzYbNqCPK148F5euD8qA8i66jLzkq2jf~zBObgdFJaeHmq3JxDSnnX-POL8Ur7ysrwn6txzszu8lri4DJl9-n~G-UR1XtiQjTB0-a5G92~H7iOzQgKvp0sem3CBtbGUrJNCImnf92poYZW7YdvBe0Dg6Undo~HyJU~OH3GJX-nto~N5d9sFlp1x45zX4EJRhP3kqLLGVq3ygGw5iLu-d5xWQWroiqwI1bpId-m6CWWo6ksgAGYM~MGaYll5qMvLaM~1t3oM1BnYsecw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Fig. 4. Dilutional sensitivity of anti-inv16 antibody versus RT-PCR for detection of type A fusion. (A) Dilutional sensitivity of anti-inv16 Ab. PN 19 AML cells were serially diluted into normal bone marrow cells and analyzed by flow cytometry. The black trace represents isotype control. The gray trace represents anti-inv(16) Ab. (A) Undiluted PN 19 AML cells; (b and C) Cell dilutions shown to level of undetectable fluorescence shift (1:20). The D-values for results in (A), (B), and (C) are 0.71, 0.58, and 0.06, respectively. (B) Single-round RT-PCR of PN 19 AML sample [inv(16)+ with type A fusion] and serial normal bone marrow dilutions, using primers C1 and M1. PCR products were hybridized with a type A junction specific oligonucleotide probe. Lanes 1 through 6, undiluted PN 19 sample, 1:10 dilution, 1:20 dilution, 1:100 dilution, 1:103dilution., and 1:104 dilution, respectively; lane 7, pure normal marrow; lane 8, negative control (no RNA).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/6/10.1182_blood.v91.6.1882/2/m_blod4064604a.jpeg?Expires=1769093143&Signature=wrxqAY~AYZ0zy62R85dFB2TTlVpxlsw4~B0Cnx8rzUuMhgw89T4xQO21KURkH7gJ5qlZZYKWAd1dAbglczKzp5RSA55QPelzZEEpq0AWCjYq5nn0nMBuRk9rV627mx0Bqn6FulfkgjbXAFfE-O0WHD3C9dkx7Ik55r6-fn57kaUiHxMt8ReK1yH14p3qHIRcEe9anBWuGGtTnMPonYMUVZMJzk9D-NWfpxgNC53XmIVjKWu62zDhz9pwDpoV9fLcTGb7AR6cnH5Oa-gYNqshXm59mXrzFmtfOrggGkZ7L4KtKarr2l5wEIyO7-3LbbBxKXax-GdU3qfYVRkV9wHwZw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Dilutional sensitivity of anti-inv16 antibody versus RT-PCR for detection of type A fusion. (A) Dilutional sensitivity of anti-inv16 Ab. PN 19 AML cells were serially diluted into normal bone marrow cells and analyzed by flow cytometry. The black trace represents isotype control. The gray trace represents anti-inv(16) Ab. (A) Undiluted PN 19 AML cells; (b and C) Cell dilutions shown to level of undetectable fluorescence shift (1:20). The D-values for results in (A), (B), and (C) are 0.71, 0.58, and 0.06, respectively. (B) Single-round RT-PCR of PN 19 AML sample [inv(16)+ with type A fusion] and serial normal bone marrow dilutions, using primers C1 and M1. PCR products were hybridized with a type A junction specific oligonucleotide probe. Lanes 1 through 6, undiluted PN 19 sample, 1:10 dilution, 1:20 dilution, 1:100 dilution, 1:103dilution., and 1:104 dilution, respectively; lane 7, pure normal marrow; lane 8, negative control (no RNA).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/6/10.1182_blood.v91.6.1882/2/m_blod4064604b.jpeg?Expires=1769093143&Signature=Ij3G~xZZwkG1INQG8EFEPyTUvXnpfxQ384FE37HrZRUVWXAUJSmboKUm0FajJbYxxr0CYu3IH8xqybbtW7GZTA3E7DytYV67RDPlCoouQXEE5g18UbmWnzu-pVe7wdNxIL0LTOkiW8ERG5wsHooZp0sRCSUNMqzWc7oH7KxgO87orPqvGC3uWAjasnBZhXStw~pWrzzGGkMAM-R1etMV1xAWRNlQnRFONE2BW3K7BFO67Z1bNmSP4VlnqhNN1UL1k1k08DffoVkrf378r8NzbdrICwQkaK64jlNqItXSBYaBdL3xInbB5HdkJ8sbyCmWLzrVTnmOQuzlzIQZMdB5ow__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal