Abstract
Kit is a tyrosine kinase receptor that plays an important role in human hematopoietic cell growth. The promoter elements that modulate the gene's expression have not been extensively studied. Because of c-kit's acknowledged importance in hematopoiesis, we sought to address this issue in more detail. To perform these studies we analyzed a human c-kit 5′ flanking fragment ∼1 kilobase in length. Deletion constructs showed a region ∼139 nucleotides upstream from the translation initiation site that was critical for promoter activity. A region containing a potential silencing element was also identified. Sequence analysis indicated several potential Myb- and Ets-binding sites. The functional significance of these sites was explored by showing that both wild-type Myb and Ets-2 protein, but not a DNA binding-deficient Myb mutant protein, bound to distinct 5′ flanking fragments that included these sites. Furthermore, binding of recombinant Myb and Ets-2 protein to these fragments could be competed with an excess of double stranded oligodeoxynucleotides containing canonical, but not mutated,Myb- or Ets-binding sites. We also showed that the 5′ flanking region of c-kit exhibited promoter activity in nonhematopoietic cells only when the cells were transfected with c-myb or ets-2 expression vectors. Moreover,Myb and Ets-2 coexpression in such cells augmented transactivation of c-kit promoter constructs in comparison to that observed in cells transfected with either construct alone. Promoter constructs lacking various Myb and Ets sites deleted were much less effective in this same system. Finally,Myb and Ets-2 mRNA expression was detected in CD34+, Kitlow as well as CD34+, Kitbright cells. In aggregate, these data further define the human c-kit promoter's functional anatomy and suggest that Myb and Etsproteins play an important, perhaps cooperative, role in regulating expression of this critical hematopoietic cell receptor.
THE C-KIT PROTO-ONCOGENE encodes a tyrosine kinase receptor, Kit1 whose cognate ligand2-5 is now commonly referred to as Steel Factor (SF). Stimulation of Kit by SF is required for development of normal neural crest melanocytes, gametocytes, and hematopoietic progenitor cells.6 In the latter cell type, binding of SF by the early stem/progenitor cells that express the c-kit encoded receptor is likely very important for cell survival.7,8 Binding of SF may also play a role in a number of critical events, including stromal adhesion and cell proliferation.9,10 In vitro, SF synergizes with other growth factors to stimulate in vitro colony formation by colony-forming units of mixed lineages (CFU-Mix), burst-forming units of erythrocytes (BFU-E), colony-forming units of granulocytes and macrophages (CFU-GM), and colongy-forming units of megakaryocytes (CFU-Meg).9 Erythropoiesis, in particular, appears critically dependent on the Kit/SF interaction.11-14 In vivo, mice with significant mutations at either the Kit encoding locus, w, or the SF locus,sl, are infertile, white coated, and anemic.15 In humans, defects in Kit expression result in amelanotic skin changes known as piebaldism.16 17 Major disturbances of human hematopoiesis resulting from defects of the Kit/SF axis have not yet been described.
Despite the importance of Kit in regulating hematopoietic cell development, relatively little is known about the factors that regulate its expression. Cytokines such as TNF-α and TGF-β are known to transiently decrease Kit expression, perhaps by destabilizing the mRNA.18-23 Factors that operate at the transcriptional level are even more obscure. We have hypothesized that the Mybtranscription factor may regulate c-kit expression because when c-myb expression is perturbed with antisense oligodeoxynucleotides, c-kit mRNA levels decline.12,24 We have also noted that the lethal anemia suffered by c-myb “knockout” mouse embryos25resembles a w-like hematopoietic defect and speculated that this might be the direct result of an inability of hematopoietic cells to express c-kit. Recent results from one of our laboratories,26 and from Vandenbark et al,27also suggest a role, likely complex, for Myb protein in the regulation of c-kit receptor expression. Nevertheless, although the above noted data strongly suggest a link between Myb andKit expression, it is not entirely certain if Mybdirectly regulates c-kit by binding in its promoter region, if it cooperates with other transcription factors in this regard, or if intermediate events are also required.
To pursue these fundamental questions, and to learn more about the functional organization of the c-kit promoter region, we isolated and sequenced ∼1kb of genomic DNA in the 5′ flanking region of the human c-kit gene. In support of our hypothesis that Myb might play a direct role in regulating c-kitexpression, sequence analysis of the region showed several potentialMyb-binding sites. Of interest, a number of potentialEts family protein binding sites were also uncovered. BecauseMyb and Ets have been reported to cooperate in the regulation of other hematopoietic genes, such as CD34,28,29CD4,30 and mim-1,31 the possibility that Myb and Ets might regulate Kit expression in a cooperative manner was also explored. By using a variety of complementary strategies, we found that wild-type, but not mutant,Myb and Ets protein bound specifically to expected sites within the c-kit promoter, and that cotransfection of wild-type, but not mutated, proteins could upregulate activity of chloramphenicol acetyltransferase (CAT) expression constructs driven by the c-kit promoter in a variety of cell types. These results suggest that Myb and Ets may play a physiologically important cooperative role in regulating c-kit expression. They therefore add to our understanding of the biology of these critical hematopoietic cell genes.
MATERIALS AND METHODS
Cell lines.
CHO (Chinese hamster ovarian cancer), COS (green monkey renal cancer cells), TK-ts13 (hamster fibroblasts), NIH 3T3 (mouse fibroblasts), K562, and HEL (human erythroleukemia) cells were obtained from the American Type Culture Collection (Bethesda, MD). K562 and HEL cells were maintained in RPMI (Gibco BRL, Grand Island, NY) with 10% bovine calf serum (BCS; Hyclone, Logan, UT). The other cell types were maintained in alfa MEM (Gibco BRL) supplemented with 10% BCS (Hyclone), 100 U/mL of penicillin G, 100 μg/mL of streptomycin, and 250 ng/mL of Fungizone (JRH Biosciences, Lenexa, KS).
Isolation of the 5′ flanking region of human c-kitgene.
A 4.2 kb EcoRI fragment of human genomic DNA known to contain the first exon of c-kit (a kind gift from R. Spritz, University of Wisconsin) was digested with various frequent cutting restriction enzymes. The digestion products were visualized on a 1% agarose gel and then transferred to a nitrocellulose filter. c-kit gene fragments were identified by hybridization with a 32P–end-labeled c-kit receptor antisense oligodeoxynucleotide (ODN) complementary to codons 1 to 6 with standard methods. AnRsaI digest of the original fragment generated suitably sized fragments for sequencing. A 1064 nt fragment was verified by hybridization and selected for further analysis.
Sequencing reactions.
DNA fragments to be sequenced were subcloned into pT7 Blue Vector (Novagen, Madison, WI). Reactions were then performed by using a Sequenase V 2.0 Kit essentially as directed by the manufacturer (USB, Cleveland, OH).
Plamid expression constructs.
Myb expression constructs were generated as follows: A full-length human c-myb cDNA (kindly provided by Dr E. Prochownik, University of Pittsburgh, Pittsburgh, PA) was digested withNcoI/DraI, yielding a 2116 nt fragment containing the entire open reading frame. The NcoI site was blunt ended with Klenow, allowing ligation of EcoRI adapters (Stratagene, La Jolla, CA) to both ends. The adapter ends were phosphorylated with T4 polynucleotide kinase (Promega, Madison, WI) and then subcloned into the pcDNA3 expression vector, thereby confering neomycin resistance to transfected cells (InVitrogen, San Diego, CA). This construct was named pCMV-myb. Construction of a plasmid engineered to express a mutated Myb protein lacking the carboxy terminal 46 amino acids of the R1 binding domain and the aminoterminal 23 amino acids of the R2 domain, has recently been reported.32
An Ets-2 expression construct was generated in a similar manner with pSVets-2, a plasmid kindly provided by Dr S. Reddy (Allegheny University of Health Sciences, Philadelphia, PA). The plasmid was digested with EcoRI, releasing a fragment containing the full-length human ets-2 cDNA (2269 bp). This fragment was gel-purified and then subsequently subcloned in a sense or antisense orientation into EcoRI-digested pcDNA3 (Invitrogen Inc). The former construct was designated pCMV ets-2.
Synthesis of recombinant Myb protein.
TK-ts13 fibroblasts (2 × 106 ) were transfected with pCMV-myb (20 μg) by using a Calcium Phosphate Transfection Kit (InVitrogen, San Diego, CA), as directed by the manufacturer. After 24 hours, G-418 selection (2 mg/mL) (GIBCO BRL, Grand Island, NY) was performed. Medium was exchanged every 2 to 3 days and after 7 to 10 days resistant clones were replated. After 2 weeks, surviving clones were screened for Myb protein expression by Western blot analysis (see below). A single clone expressing a high level of Myb was expanded and maintained.
Western blot detection of Myb protein.
TK-ts13 cells (2 × 106) were detached from culture dishes with ethylenediaminetetraacetic acid [EDTA] (0.53 mmol/L)-trypsin (0.05%) in phosphate-buffered saline (PBS), washed twice in PBS, resuspended in 1× sodium dodecyl sulfate (SDS) gel-loading buffer [50 mmol/L Tris-Cl (pH 6.8), 100 mmol/L dithiothretiol (DTT), 2% SDS, 0.1% bromophenol blue, 10% glycerol], boiled for 2 minutes, and then chilled on ice. Resulting cell extracts were centrifuged briefly (14,000g for 1 minute) and then analyzed for Myb protein content. Clarified cell extracts, protein size markers, and a positive Myb protein control were loaded onto gels and then electrophoresed in the Pharmacia LKB Phast gel apparatus (Pharmacia, Uppsala, Sweden). Separated proteins were transferred to a nitrocellulose membrane filter (Gibco BRL, Gaithersburg, MD), which were probed with murine antihuman Mybmonoclonal antibodies (MoAbs) (UBI, Lake Placid, NY). Binding of the anti-Myb antibody was detected by enhanced chemiluminescence (ECL Western blot Kit, Amersham, Buckinghamshire, UK) by using antimouse IgG (L+H) (Boehringer-Mannheim, Mannheim, Germany) conjugated with peroxidase. Positive reactions were visualized by exposure to high-sensitivity radiograph film (DuPont, Boston, MA).
DEAE-dextran–mediated transient transfections.
Briefly, TK-ts13 cells were plated in 10 cm plastic Petri dishes and grown to 70% to 80% confluence. The medium was exchanged 2 hours before transfection. Four mL of culture medium containing 40 μL of DEAE-Dextran (50 mg/mL stock) (Sigma, St. Louis, MO), 5 μg of reporter pSV2pap plasmid (kind gift from Dr T. Kadesh, University of Pennsylvania, Philadelphia, PA), and 50 μg of different CAT construct plasmids was added to each plate. After 4 hours of incubation at 37°C, the transfection medium was removed. Cells were then shocked with 10% DMSO (Sigma, St. Louis MO) for 2 minutes, and then washed three times with fresh medium. After 48 hours of incubation (37°C, 5% CO2) in a fully humidified incubator, the cells were detached by using TEN buffer (40 mmol/L Tris-HCl pH 7.5, 1 mmol/L EDTA, 15 mmol/L NaCl), harvested, and analyzed for PAP and CAT activity.
Lipofectamine-mediated transient transfections.
K562 cells were transfected with pCAT constructs by using Lipofectamine, as directed by the manufacturer (GIBCO BRL). Briefly, exponentially growing K562 cells were washed once in the serum-free OPTI MEM (GIBCO BRL), and thereafter plated in six-well culture plates at a density 2 × 106 cells/mL in 0.8 mL. Two hundred μL of OPTI MEM containing 10 μL of lipofectamine and 11 μg of plasmid DNA (10 μg of PSV2pap and 1 μg of pCAT) was then added to the cells that were subsequently incubated for 12 hours at 37°C in a humidifed atmosphere containing 5% CO2 . The cells were then diluted in 4 mL of RPMI containing 15% BCS. After an additional 48-hour incubation, the cells were harvested for PAP and CAT assays, as described below.
PAP assays.
PAP assays were performed to evaluate transfection efficiencies in a given set of experiments, thereby allowing better standardization of subsequently performed CAT assays. Target cells were cotransfected with a given pCAT construct and the pSVpap plasmid that expresses human placental alkaline phosphatase.33 Briefly, 1/15 of the extracts prepared from transfected cells were heated to 65°C for 15 minutes, then resuspended in 1 mL of reaction buffer (0.5 mmol/L p-nitro-phenyl-phosphate, 1mol/L diethanolamine), and incubated at 37°C for 15 minutes. Postincubation, the reaction mixture was clarified by centrifugation and accumulation of dye product quantitated spectrophotometrically at a wavelength of 405 nm. Preparations containing equal levels of PAP activity were then used for CAT assays, as described below.
CAT assay.
CAT assays were employed to map potential promoter elements in the ∼1 kb of DNA upstream of the c-kit translational start site. DNA fragments of varying sizes were generated by using the following restriction enzyme combinations: RsaI+BamHI [929 nt fragment]; XhoI+BamHI [461 nt fragment];SmaI+BamHI [158 nt fragment]; andNaeI+BamHI [118 nt fragment]. Fragments generated by these digestions were initially subcloned into pGEM4Z or pGEM7Z (Promega) in order to facilitate cloning of the fragments into the multiple cloning site of the pCAT basic (pCAT B) vector (Promega). AnRsaI fragment with a BamHI/NaeI deletion was also subcloned into the pCAT B vector in a similar manner.
Kit promoter constructs containing Myb orMyb/Ets binding site deletions were constructed as follows: A 910bp and a 731bp fragment were generated by PCR amplification on the c-kit–XhoI/BamHI plasmid by using two different upstream primers located inside the c-kit 5′ flanking region, and a common downstream (3′) primer mapping inside the pCAT basic vector at nts [+2819 to +2840]. The 5′ primers employed were complementary to nts [-405 to -385], and nts [-224 to -205]. The former allowed amplification of a c-kit promoter fragment missing theMyb binding site located between nts -420 and -410 that we designated M1, whereas the latter 5′ primer amplified a c-kit promoter fragment missing M1, an additional downstreamMyb binding-site designated M2, and two putativeEts-binding sites designated E1 and E2 (Fig 1). The amplified products were subcloned into the pCR2.1 vector by using the TA cloning kit(Invitrogen Corp) to generate constructs called pCR D M1 and pCRE3, respectively. These plasmids were digested withHindIII/NcoI restriction endonucleases and the fragments released (containing the c-kit flanking region from nucleotide -405, or from -224, to the BamHI site in the c-kit 5′UTR plus the indicated portion of the CAT gene) and subcloned into pCAT basic vector previously digested with theHindIII/NcoI restriction enzymes. The resulting plasmids were designated pc-kit D M1, and pc-kit–E3. Plasmids sequences were confirmed by automated sequencing with the Taq Dye Deoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA).
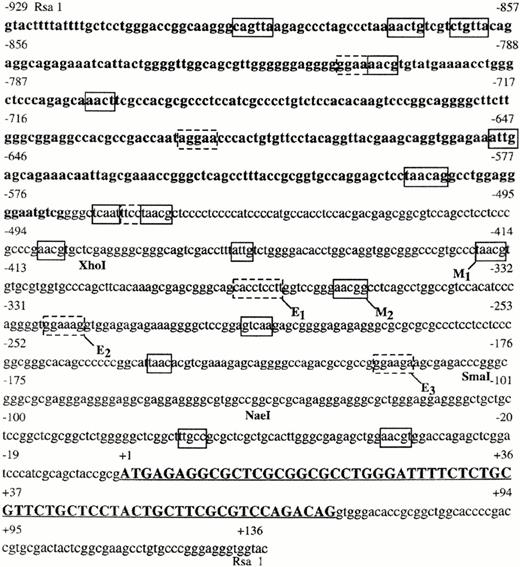
Nucleotide sequence of human c-kit gene spanning 929 bases upstream of the initiating methionine to 69 bases within the first intron. Coding sequence is indicated by capital letters and is underlined. Potential Myb-binding sites, on the + or − strand, in the region upstream of the ATG are enclosed by solid line boxes. Potential Ets-binding sites, on the + or − strand, are enclosed by dashed boxes. Location of restriction sites are indicated below the sequence.
Nucleotide sequence of human c-kit gene spanning 929 bases upstream of the initiating methionine to 69 bases within the first intron. Coding sequence is indicated by capital letters and is underlined. Potential Myb-binding sites, on the + or − strand, in the region upstream of the ATG are enclosed by solid line boxes. Potential Ets-binding sites, on the + or − strand, are enclosed by dashed boxes. Location of restriction sites are indicated below the sequence.
CAT assays were performed according to the manufacturer's protocol (Promega). Products of the CAT assay, 14 C-chloramphenicol, and its acetylated derivatives, were detected by thin layer chromatography. After drying, gels were exposed to high-sensitivity films (NEF 496, DuPont, Boston, MA). Reaction intensity, a direct function of the amount of 14 C-labeled product present in the gel, was quantitated by densitometry with a Personal Densitometer (Molecular Dynamics, Sunnyvale, CA).
Nuclear extracts and electrophoretic mobility shift (EMSA) assay.
Nuclear protein extracts and EMSA assays were performed, as previously described.34 γ32 P-labeled double-stranded DNA probes (∼5 × 104 cpm) corresponding to three different segments of the human c-kit promoter from nucleotides -724 to -480; -465 to -220; and -196 to -20 upstream from the translation start site (Fig 1) were synthesized by PCR. Oligonucleotides corresponding to the canonical MYB-binding sites (M1 [nucleotides from -426 to -404] and M2 [nucleotides from -367 to -343]; or to Ets-binding sites (E1 [nucleotides from -384 to -360], E2 [nucleotides from -257 to -233] and E3 [nucleotides from -203 to -179] were synthesized in an Applied Biosystems automated synthesizer and end labeled with γ-[32 P] ATP.
Five hundred ng of GST-MYB fusion protein35 or 15 μg of bacterial lysate from HB101 cells transformed with a pFlag-c-myb vector36 or a pFlag-ets–2 vector37 were used in EMSA, as described,36with some modifications. Briefly, proteins (purified fusion protein or bacterial lysate) in binding buffer (10 mmol/L Tris-HCl [pH 7.5], 50 mmol/L NaCl, 1 mmol/L EDTA, 10% glycerol, and 1 mmol/L dithiothretiol) were incubated with 0.12 mg/mL of poly (dI-dC) for 10 minutes on ice. γ-32 P-end labeled double-stranded oligonucleotide probes were added to the binding reactions, which were incubated for an additional 15 minutes at room temperature. When indicated, EMSA were also performed in the presence of a 100–fold molar excess of double-stranded oligonucleotides used as specific or nonspecific competitors. Binding reactions were electrophoresed in native 5% polyacrylamide gel electrophoresis (PAGE) in low ionic strength (0.25 × Tris borate EDTA). Gels were dried and exposed to radiograph films for autoradiography. Double-standed oligonucleotides of the human cdc2 promoter containing an Ets binding site37 (wtEts binding site= gagagggggaAGGAAagaacaagaa) and of the chickenmim-1 promoter containing a Myb-binding site (wtMyb tcgacacattaTAACGGttttttagc) were employed as specific competitors in EMSA. Double-stranded oligonucleotides carrying mutations (mut) in the core of either MYB (mut MYB binding site: tcgacacattaTGGGGGttttttagc) or Ets (mut Ets binding site: gagagggggcGTAGCagaacaagaa) consensus sequence, were used as nonspecific competitors in EMSA.
Fluorescence-activated cell sorting (FACS) analysis and cell sorting.
FACS analysis and cell sorting were performed as previously reported.8 In brief, MNC (∼3 to 6 × 107) were simultaneously labeled with fluorescein conjugated anti-CD34 MoAb (Becton-Dickinson, Mountain View, CA), and a phycoerythrin-labeled antihuman Kit receptor MoAb (kind gift of Dr V. Broudy, University of Washington, Seattle, WA). Cells categorized as CD34+, Kitlow (defined as the dimmest 50% of labeled cells) and CD34+, Kitbright (defined as the brightest 50% of labeled cells) were isolated by FACS. RNA was extracted from these cells, reverse transcribed, and then PCR amplified to detect Ets, Myb, and β-actin mRNA expression, as previously reported.8 12
RESULTS
Sequence analysis of the 5′ flanking region of the human c-kit gene.
RsaI digestion of a 4.2 kb genomic fragment containing the c-kit 5′ flanking region generated a number of fragments that were suitably sized for sequencing. One of these, ∼1064 nts in length, hybridized with a c-kit oligonucleotide probe complementary to codons 1 to 6. This fragment was cloned into the pT7 blue sequencing vector for complete sequencing. Comparison of the sequence derived with those previously reported by Giebel et al,38 and Yamamoto et al39 showed that the fragment contained the entire first exon of c-kit flanked downstream by a small part of the first intron, and upstream by 925 nts of sequence (Fig 1). In the areas in which the reported sequences overlap, they are virtually identical. As reported,38 we too noted the presence of numerous potential binding sites for transcription factors including two AP 2-binding sites (nts 218- and nts 420-), several SP 1 sites, and a number of cis elements for melanocyte-specific genes including melanocyte-specific upstream (MEL-US) and downstream (MEL-DS) elements. In addition, we have also identified several potential Myb (Py-AAC G/T G)40,41 and Ets family protein (AGGAA)42 consensus binding sites (Fig 1).
Functional analysis of c-kit 5′-flanking region.
CAT assays were employed to map potential promoter elements in the ∼1 kb of DNA upstream of the c-kit translational start site. As noted, the 1064 nts RsaI fragment was restriction digested with various enzyme combinations to yield a 909 ntsRsaI/BamHI fragment, a 461 ntsXhoI/BamHI fragment, a 158 ntsSmaI/BamHI fragment, and a 118 ntsNaeI/BamHI fragment. The fragments positions relative to the translational start site are shown in Figure 2. They were subcloned into pCAT B, which contains no promoter or enhancer sequence, and then transiently transfected into K562 cells. FACS analysis of these human leukemia cells shows that ∼70% of the cells employed in our laboratory express Kit at low level (data not shown). Reporter constructs were also transfected into nonhematopoietic NIH3T3, TKts-13, CHO, and COS cells. Activity in each case was compared with that obtained with a pCAT positive control vector driven by the SV40 promoter and enhancer, which was arbitrarily assigned a value of 100%.
Restriction fragments used to functionally characterize the c-kit promoter. Fragments were derived from a 1064 ntsRsaI fragment as detailed in the methods section and ultimately subcloned into pCAT vectors to assess their promoter activity. Length and position of the various fragments relative to the translational start site (+1) is indicated in the cartoon.
Restriction fragments used to functionally characterize the c-kit promoter. Fragments were derived from a 1064 ntsRsaI fragment as detailed in the methods section and ultimately subcloned into pCAT vectors to assess their promoter activity. Length and position of the various fragments relative to the translational start site (+1) is indicated in the cartoon.
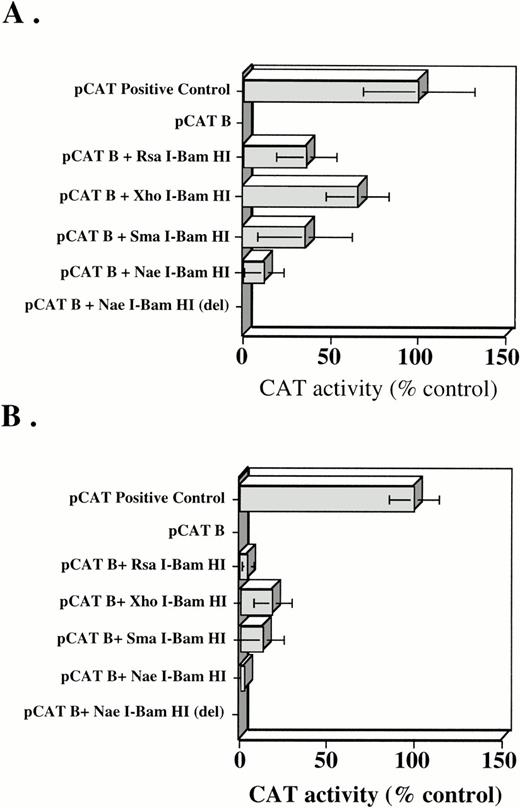
In K562 cells, no CAT activity was detected after transfection of the empty pCAT B vector (Fig 3A). However, when theRsaI-BamHI fragment was subcloned into the pCAT B vector, CAT activity approximately one third that of the positive control was observed suggesting that promoter elements were contained within this region. Assays with additional constructs were also performed in hopes of deriving a more detailed functional map of the promoter region. The XhoI-BamHI fragment had nearly twice the activity of the longer RsaI-BamHI fragment, suggesting that an unidentified transcription inhibitory protein might bind upstream of the XhoI site. The SmaI-BamHI fragment gave promoter activity similar to theXhoI-BamHI fragment, but the NaeI-BamHI fragment missing the intervening 40 nucleotides between theSmaI and NaeI sites (NaeI-BamHI [del]) lost ∼50% of the promoter activity. Because theNaeI/BamHI fragment had the weakest measureable activity, we then determined if this fragment might represent a minimal promoter unit. To test this possibility, the RsaI/BamHI fragment was restriction digested with NaeI. This yielded a 790 nt fragment missing the proximal (relative to the start site) 118 nt piece (Fig 2). When the 790 nt fragment was subcloned into pCAT B and expressed in K562 cells, no promoter activity was detected in the CAT assay (Fig 3A). Therefore, the promoter and transcription initiation site likely reside within this fragment of genomic DNA.
(A) CAT activity in K562 cells transiently transfected with a pCAT positive control vector, pCAT B negative control vector, and pCAT B containing RsaI-BamHI,XhoI-BamHI, SmaI-BamHI, NaeI-BamHI, and NaeI-BamHI (del) inserts. (B) CAT activity in CHO cells transiently transfected with a pCAT positive control vector, pCAT B-negative control vector, and pCAT B containingRsaI-BamHI, XhoI-BamHI, SmaI-BamHI, NaeI-BamHI, andNaeI-BamHI (del) inserts. Transfection reactions were normalized by use of PAP assays, as detailed in Materials and Methods. Mean ±SD of six different experiments, each performed in duplicate, are shown. CAT activity was normalized to the CAT control transfected cells, which were arbitrarily set at 100%.
(A) CAT activity in K562 cells transiently transfected with a pCAT positive control vector, pCAT B negative control vector, and pCAT B containing RsaI-BamHI,XhoI-BamHI, SmaI-BamHI, NaeI-BamHI, and NaeI-BamHI (del) inserts. (B) CAT activity in CHO cells transiently transfected with a pCAT positive control vector, pCAT B-negative control vector, and pCAT B containingRsaI-BamHI, XhoI-BamHI, SmaI-BamHI, NaeI-BamHI, andNaeI-BamHI (del) inserts. Transfection reactions were normalized by use of PAP assays, as detailed in Materials and Methods. Mean ±SD of six different experiments, each performed in duplicate, are shown. CAT activity was normalized to the CAT control transfected cells, which were arbitrarily set at 100%.
To determine tissue specificity of the putative promoter unit, the CAT constructs were also expressed in nonhematopoietic NIH3T3 cells and TK-ts13 fibroblasts. Neither of these cell types express the Kit receptor. In distinct contrast to results obtained with K562 cells, none of the pCAT B constructs showed promoter activity in 3T3 or TK-ts13 fibroblasts (not shown). Slightly different results were obtained when the same constructs were transfected into COS and CHO (Fig 3B) cells, both of which constitutively express c-kit at low levels. All of the fragments cloned into the pCAT B vector showed weak promoter activity in these cells. Again, theNaeI/BamHI fragment appeared to have the weakest activity, suggesting it might represent the minimal promoter unit.
Effect of Myb and Ets protein on c-kitpromoter activity.
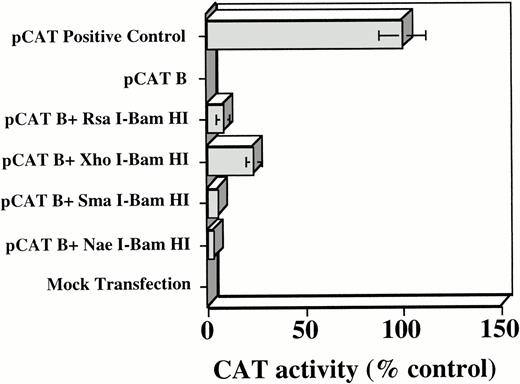
Our previous studies,12 and the presence of several potential Myb-binding sites in the c-kit 5′ flanking region (Fig 1), suggested that Myb might play a role in regulating Kit expression. To address this question, we first sought to determine if Myb protein expression induced transactivation of the Kit promoter. TK-ts13 cells were therefore transfected with Myb protein expression constructs. Stable lines were screened for protein expression and a positive clone was expanded. Cells in this clone were then transiently cotransfected with the various pCAT B-kit promoter constructs. In distinct contrast to results obtained with Myb negative TK-ts13 cells, transient transfection of the various c-kit promoter constructs into the Myb-positive TK-ts13 cells led to clearly detectable, albeit weak, CAT activity in the cell extracts. A histogram of the densitometric quantitation of five separate CAT assays, each performed in duplicate, with each of the constructs is shown in Figure 4. As previously observed, theXhoI/BamHI fragment had greater activity than the intact RsaI-BamHI fragment. This result again supports the possibility that a negative regulatory element might reside upstream of the XhoI site between nts -929 and -481. Finally, as an additional proof that Myb may regulate c-kitpromoter activity, wild-type TK-ts13 cells were cotransfected with a pCDNA3 construct expressing either the wild-typeMyb protein or a DNA binding-deficient mutant,35 as well as each of the various pCAT B c-kit promoter constructs (Fig 2). CAT activity was only observed in cells expressing the wild-type protein (data not shown), again strongly suggesting that a functional Myb protein was necessary for the observedKit promoter activity .
Densitometric quantitation of CAT activity in stable, MYB-expressing TK-ts13 cells cotransfected with a pCAT B plasmid containing various portions of the c-kit 5′ flanking region. CAT activity was normalized to the CAT positive-control transfected cells, which were arbitrarily set at 100%. Mean ±SD of five different experiments, each performed in duplicate are shown. Transfection reactions were normalized by use of PAP assays, as detailed in Materials and Methods.
Densitometric quantitation of CAT activity in stable, MYB-expressing TK-ts13 cells cotransfected with a pCAT B plasmid containing various portions of the c-kit 5′ flanking region. CAT activity was normalized to the CAT positive-control transfected cells, which were arbitrarily set at 100%. Mean ±SD of five different experiments, each performed in duplicate are shown. Transfection reactions were normalized by use of PAP assays, as detailed in Materials and Methods.
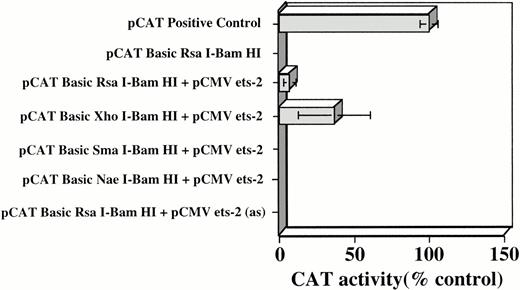
As noted in Figure 1, in addition to Myb-binding sites, a number of potential Ets-binding sites were also identified in the c-kit flanking region under study. The potential significance of these sites was evaluated with the experimental approach employed to determine Myb's role in regulatingKit expression. The same sets of pCAT B-Kit promoter constructs were cotransfected into TK-ts13 cells along with an Ets-2 expression vector, pCMV-ets-2. Ets-2 was chosen for these studies because Myb/Ets-2 cooperation has previously been shown important in regulating promoter activity of the CD34 and mim-1 genes.28 31 As shown in Figure 5, no CAT activity was detectable in the TK-ts13 cells transfected with the pCAT BRsaI-BamHI construct alone. However, when cells were cotransfected with an Ets expression construct, weak transactivation relative to the pCAT positive control, of theRsaI/BamHI promoter fragment, and even stronger activity with the XhoI/BamHI fragment was observed. Of interest, in contrast to results obtained with Myb expressing cells, Ets-2 did not transactivate the promoter units contained within the smaller SmaI/BamHI andNaeI/BamHI fragments. These latter results were expected because neither of these fragments contain Ets-binding sites. As an additional control, cells were cotransfected with the pCAT B RsaI-BamHI construct and pCMV ets-2 in which the Ets expression cassette was cloned into pCMV in an antisense orientation [pCMV ets-2 (as)]. CAT activity was undetectable in extracts prepared from these cells (Fig 5).
Densitometric quantitation of CAT activity in TK-ts13 cells cotransfected with pCMV-Ets-2 and pCAT B constructs containing various portions of the c-kit5′ flanking region. pCAT B RsaI-BamHI + pCMV ets-2 in antisense (as) orientation was employed as an additional control. Transfection reactions were normalized by use of PAP assays, as detailed in Materials and Methods. Mean ±SD of five different experiments, each performed in duplicate, are shown. CAT activity was normalized to the CAT control transfected cells that were arbitrarily set at 100%.
Densitometric quantitation of CAT activity in TK-ts13 cells cotransfected with pCMV-Ets-2 and pCAT B constructs containing various portions of the c-kit5′ flanking region. pCAT B RsaI-BamHI + pCMV ets-2 in antisense (as) orientation was employed as an additional control. Transfection reactions were normalized by use of PAP assays, as detailed in Materials and Methods. Mean ±SD of five different experiments, each performed in duplicate, are shown. CAT activity was normalized to the CAT control transfected cells that were arbitrarily set at 100%.
Myb and Ets-2 protein have additive effects on c-kit promoter activity.
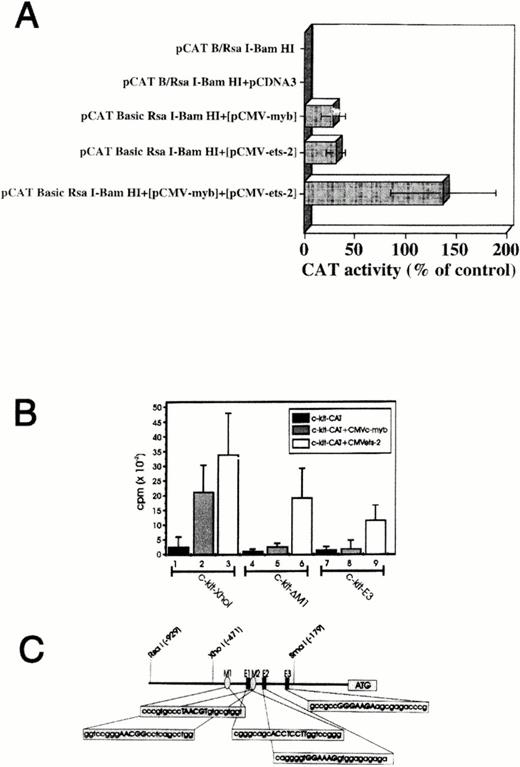
We also investigated if Myb and Ets-2 could cooperate in transactivating the Kit promoter by using several different strategies. TK-ts13 fibroblasts were transfected withMyb (pCMV-myb), Ets-2 (pCMV-ets-2), orMyb + Ets expression vectors, along with the pCAT BRsaI/BamHI fragment construct (Fig 6 A). No CAT activity was detected in extracts from cells transfected with the pCAT BRsaI-BamHI construct alone, or in cells cotransfected with this Kit promoter construct and an insertlessMyb/Ets expression vector. As noted previously, CAT activity was detected in cells cotransfected with pCAT BRsaI-BamHI and either the Myb or Ets-2 expression vector. Cotransfection of pCAT B RsaI-BamHI with both the Myb and Ets-2 expression vectors into the TK-ts13 cells led to CAT activity that was approximately threefold greater then that observed with the Myb or Etsexpression constructs alone.
Myb and Ets-2 transactivate c-kitpromoter in a cooperative manner. (A) CAT activity in TK-ts13 fibroblasts cotransfected with Myb(pCMV-myb), Ets-2 (pCMV-ets-2), orMyb+Ets expression vectors, plus a pCAT B-RsaI/BamHI fragment construct. Transfection of all three vectors into the TK-ts13 cells led to CAT activity ∼threefold greater than that observed with the Myb or Etsexpression constructs alone, and ∼140% that of the pCAT positive control (not shown). (B) Ability of Myb and Ets protein to transactivate wild-type, or mutated forms of theXhoI-BamHI Kit promoter fragment in TK-ts13 cells. In the absence of Myb or Ets, the construct was without CAT activity (lane 1). Cotransfection of Myb orEts-2 expression constructs along with the pCAT BXhoI-BamHI promoter construct significantly increased CAT activity (lanes 2 to 3). Deletion of a single Myb binding site (c-kit-D M1) resulted in loss of Myb's (lane 5), but not Ets's (lane 6) ability to augment CAT activity. Deletion of both Myb sites and two of three Ets sites diminished, but did not entirely abolish the ability of Ets to transactivate the promoter construct (lane 9). Neither c-kit-D M1, nor c-kit-E3 alone elicited CAT activity in the TK-ts13 cells (lanes 4 and 7, respectively). See text for complete details. (Panel C) Schematic representation of Myb andEts-2–binding sites within the XhoI c-kitpromoter region (-481/-24).
Myb and Ets-2 transactivate c-kitpromoter in a cooperative manner. (A) CAT activity in TK-ts13 fibroblasts cotransfected with Myb(pCMV-myb), Ets-2 (pCMV-ets-2), orMyb+Ets expression vectors, plus a pCAT B-RsaI/BamHI fragment construct. Transfection of all three vectors into the TK-ts13 cells led to CAT activity ∼threefold greater than that observed with the Myb or Etsexpression constructs alone, and ∼140% that of the pCAT positive control (not shown). (B) Ability of Myb and Ets protein to transactivate wild-type, or mutated forms of theXhoI-BamHI Kit promoter fragment in TK-ts13 cells. In the absence of Myb or Ets, the construct was without CAT activity (lane 1). Cotransfection of Myb orEts-2 expression constructs along with the pCAT BXhoI-BamHI promoter construct significantly increased CAT activity (lanes 2 to 3). Deletion of a single Myb binding site (c-kit-D M1) resulted in loss of Myb's (lane 5), but not Ets's (lane 6) ability to augment CAT activity. Deletion of both Myb sites and two of three Ets sites diminished, but did not entirely abolish the ability of Ets to transactivate the promoter construct (lane 9). Neither c-kit-D M1, nor c-kit-E3 alone elicited CAT activity in the TK-ts13 cells (lanes 4 and 7, respectively). See text for complete details. (Panel C) Schematic representation of Myb andEts-2–binding sites within the XhoI c-kitpromoter region (-481/-24).
We then examined the ability of Myb and Ets protein to transactivate wild-type, or mutated forms of theXhoI-BamHI Kit promoter fragment in TK-ts13 cells (Fig 6B). As was shown above, in the absence of Myb orEts, the construct was without significant activity (Fig 6B, lane 1). Again, cotransfection of Myb or Ets-2 expression constructs along with the pCAT B XhoI-BamHIkit promoter fragment construct led to a significant increase in CAT activity (Fig 6B, lanes 2 to 3). Deletion of a singleMyb binding site (c-kit-D M1) located within nts -427 to -402 of the promoter fragment (see M1, Fig 6C) resulted in almost a complete loss of the ability of Myb protein to augment CAT activity (Fig 6B, lane 5). This Myb binding site appears then to be crucial for transactivation of this promoter fragment. Nevertheless, in spite of this deletion, Ets-2 protein remained active as a transactivating factor (Fig 6B, lane 6). When bothMyb sites (nts -427 to -402; -367 to -390) and two (-384 to -360 and -332 to -309) of three Ets sites contained within this fragment were deleted (c-kit-E3; see Fig 6C- M1, M2, E1, and E2), only minimal CAT activity was observed in the presence ofMyb (Fig 6B, lane 8), whereas activity could be detected in cells cotransfected with the Ets-2 construct (Fig 6B, lane 9), presumably due to the remaining Ets-2 binding site (E3). c-kit-D M1, and c-kit-E3 elicited only minimal CAT activity in the TKts-13 cells (Fig 6B, lanes 4 and 7, respectively).
The specificity of these interactions was further confirmed by competition experiments performed in intact cells. In these studies, TK-ts13 cells were again cotransfected with the Myb(pCMV-myb) or Ets-2 (pCMV-ets) expression vectors previously shown capable of transactivating the pCAT BXhoI/BamHI Kit expression construct (Fig 7A, lanes 2 and 5 respectively; Fig 7B as labeled). CAT activity in cells in which a 200-fold molar excess of double-stranded mutated or wild-type Myb binding site oligonucleotides had been added was either unchanged or significantly reduced, respectively (Fig 7A, lanes 3 and 4; Fig 7B, as labeled). Similar results were obtained when this experiment was repeated with a 200-fold molar excess of mutated or wild-type double-strandedEts-2 binding site oligonucleotides Ets-2 (Fig 7A, lanes 6 and 7; Fig 7B, as labeled).
(A) CAT activity competition assay of double stranded oligomers containing wild-type (WT) or mutated (mut) Myb orEts-2 binding motifs in the transient transfections experiments in TK-ts13 fibroblasts. pCAT B XhoI-BamHI (lane 1), pCAT B XhoI-BamHI + pCMV-myb (lane 2), pCAT B XhoI-BamHI + pCMV-myb + mut oligomers (lane 3), pCAT B XhoI-BamHI + pCMV-myb + wt oligomers (lane 4), pCAT BXhoI-BamHI + pCMV-ets-2 (lane 5), pCAT BXhoI-BamHI + pCMV-ets-2 + mut oligomers (lane 6), pCAT B XhoI-BamHI + pCMV-ets-2 + wt oligomers (lane 2; lane 7), pCAT B XhoI-BamHI + pCDN3 (lane 9). (B) Densitometric quantitation of CAT activity in transiently, MYB and Ets-2 expressing TK-ts 13 cells cotransfected with a pCAT B plasmids containing theXhoI-BamHI c-kit 5′ flanking region and 200 molar excess of wt or mut oligomers. CAT activity present in cells transfected with the pCAT B XhoI-BamHI + pCMV-myb constructs was arbitrarily given a value of 100%. Mean ±SD of six different experiments, each performed in duplicate, are shown.
(A) CAT activity competition assay of double stranded oligomers containing wild-type (WT) or mutated (mut) Myb orEts-2 binding motifs in the transient transfections experiments in TK-ts13 fibroblasts. pCAT B XhoI-BamHI (lane 1), pCAT B XhoI-BamHI + pCMV-myb (lane 2), pCAT B XhoI-BamHI + pCMV-myb + mut oligomers (lane 3), pCAT B XhoI-BamHI + pCMV-myb + wt oligomers (lane 4), pCAT BXhoI-BamHI + pCMV-ets-2 (lane 5), pCAT BXhoI-BamHI + pCMV-ets-2 + mut oligomers (lane 6), pCAT B XhoI-BamHI + pCMV-ets-2 + wt oligomers (lane 2; lane 7), pCAT B XhoI-BamHI + pCDN3 (lane 9). (B) Densitometric quantitation of CAT activity in transiently, MYB and Ets-2 expressing TK-ts 13 cells cotransfected with a pCAT B plasmids containing theXhoI-BamHI c-kit 5′ flanking region and 200 molar excess of wt or mut oligomers. CAT activity present in cells transfected with the pCAT B XhoI-BamHI + pCMV-myb constructs was arbitrarily given a value of 100%. Mean ±SD of six different experiments, each performed in duplicate, are shown.
Myb and Ets-2 protein bind to their predicted sites in the Kit promoter region.
The above studies clearly suggest a role for the Myb andEts proteins in the regulation of c-kit promoter activity. They do not indicate, however, whether this effect is a direct one. This issue was explored by electrophoretic mobility shift assays (EMSA) designed to determine if these respective proteins could bind to their predicted sites within the promoter region, and whether such binding was specific. Our initial experiments employed three different 32 P-labeled fragments of the c-kitflanking region that were incubated with bacterial lysates that either contained, or did not contain, Myb protein (Fig 8A). With each fragment tested, one complex was retarded in the gel by lysates that contained Mybprotein. In contrast, there was no retardation in the lysates that did not contain Myb protein. The specificity of these reactions was confirmed in competition experiments. When a 100-fold molar excess of an unlabeled specific competitor (a 36 bp fragment of the CD34 gene 5′flanking region containing two Myb-binding sites [20]) was added to the Myb containing binding mixture, the previously noted band shifts were no longer observed. When the twoMyb-binding sites were mutated, and a 100-fold molar excess of this nonspecific competitor was added to the mixtures, the shifted bands were noted again. Finally, in the presence of bacterial lysate containing mutated Myb protein lacking portions of the R1 and R2 DNA binding domains, no band shift could be detected (Fig 8B). These results provided highly suggestive evidence that Myb protein bound specifically to sites within the promoter region.
Purified Myb protein binds in the c-kitpromoter and can be specifically competed. Three different fragments of the c-kit promoter, with nt positions as indicated, were labeled with γ-32 P and then used as probes in EMSA assays performed with bacterial lysates containing, or not containing,Myb protein. (A) Lanes 3, 8, and 13 contain Mybprotein. Lanes 1, 2, 6, 7, 11, and 12 contain no Myb protein. Lanes 4, 9, and 14 contain 100 times excess of cold Myb-binding site competitor (CD34 promoter). Lanes 5, 10, and 15 contain a competitor oligonucleotide with mutated Myb-binding sites. (B) Lanes 1 and 2, no Myb protein in lysate. Lane 3, lysate withMyb protein. Lane 4, lysate with mutated Myb protein (R1 and R2 partially deleted).
Purified Myb protein binds in the c-kitpromoter and can be specifically competed. Three different fragments of the c-kit promoter, with nt positions as indicated, were labeled with γ-32 P and then used as probes in EMSA assays performed with bacterial lysates containing, or not containing,Myb protein. (A) Lanes 3, 8, and 13 contain Mybprotein. Lanes 1, 2, 6, 7, 11, and 12 contain no Myb protein. Lanes 4, 9, and 14 contain 100 times excess of cold Myb-binding site competitor (CD34 promoter). Lanes 5, 10, and 15 contain a competitor oligonucleotide with mutated Myb-binding sites. (B) Lanes 1 and 2, no Myb protein in lysate. Lane 3, lysate withMyb protein. Lane 4, lysate with mutated Myb protein (R1 and R2 partially deleted).
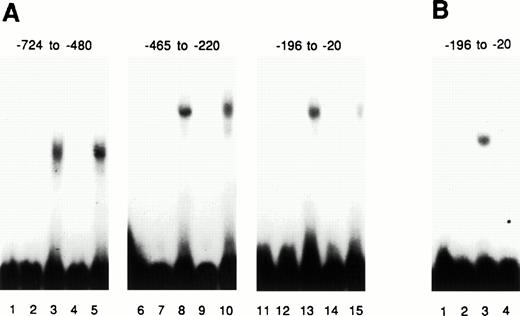
Additional experiments were also performed to further determine ifMyb and Ets-2 protein could bind to specific motifs identified in the 5′ flanking region of c-kit gene (Fig1). Because the strongest promoter activity appeared to reside in theXhoI/BamHI fragment, and because this fragment also contained three Ets-2 and two Myb canonical binding sites, we selected this fragment for analysis. EMSA assays employing oligonucleotides corresponding to the Myb- (M1 and M2) (Fig 9A) and Ets (E1, E2, E3) (Fig9B)-binding sites in the XhoI/BamHI promoter fragment are shown in Figure 9. Binding of bacterially expressed Myb(Fig 9, Panel A) or Ets proteins (Fig 9, Panel B) was specifically competed by wild-type, but not mutated, double-stranded oligonucleotides. Moreover, EMSA assays performed with nuclear proteins derived from K562 cells also showed the formation of specific complexes with Myb M1- and M2-binding sites (Fig 9C), as well as the E1 and E2 Ets-2–binding sites (Fig 9D).
Myb and Ets-2 proteins bind specifically to their predicted sites in the kit promoter. γ-32 P-end labeled double-stranded oligonucleotide probes corresponding either to the canonical Myb (M1 and M2) or Ets-2 (E1, E2 and E3) binding sites within the XhoI-SmaI promoter region fragment (nts-461 to -24) (see Fig 6C) were employed in EMSA. Binding of bacterially expressed Myb (A) or Ets proteins (B) was competed with either wild-type (wt) or mutated (mut) double-stranded oligonucleotides as indicated above each lane. Similar experiments were also performed with nuclear extract derived from K562 cells. Fifteen μg of K562 nuclear extract was incubated in binding buffer with γ-32 P-end labeled double-stranded oligonucleotide probes (lane 1; [C and D]) corresponding either to the canonical Myb (M1 and M2; [C]) or Ets-2– (E1, E2; [D]) binding sites as described above. Specificity of binding was assessed using wild type (lane 2; [C and D]) or mutated (lane 3; [C and D]) double-stranded oligonucleotides as indicated above each lane. Sequences of the double-stranded oligodeoxynucleotides for both mutated and wild-type Myb and Ets-2 binding sites are given in the Materials and Methods section.
Myb and Ets-2 proteins bind specifically to their predicted sites in the kit promoter. γ-32 P-end labeled double-stranded oligonucleotide probes corresponding either to the canonical Myb (M1 and M2) or Ets-2 (E1, E2 and E3) binding sites within the XhoI-SmaI promoter region fragment (nts-461 to -24) (see Fig 6C) were employed in EMSA. Binding of bacterially expressed Myb (A) or Ets proteins (B) was competed with either wild-type (wt) or mutated (mut) double-stranded oligonucleotides as indicated above each lane. Similar experiments were also performed with nuclear extract derived from K562 cells. Fifteen μg of K562 nuclear extract was incubated in binding buffer with γ-32 P-end labeled double-stranded oligonucleotide probes (lane 1; [C and D]) corresponding either to the canonical Myb (M1 and M2; [C]) or Ets-2– (E1, E2; [D]) binding sites as described above. Specificity of binding was assessed using wild type (lane 2; [C and D]) or mutated (lane 3; [C and D]) double-stranded oligonucleotides as indicated above each lane. Sequences of the double-stranded oligodeoxynucleotides for both mutated and wild-type Myb and Ets-2 binding sites are given in the Materials and Methods section.
Myb and Ets-2 mRNA expression in primitive hematopoietic cells.
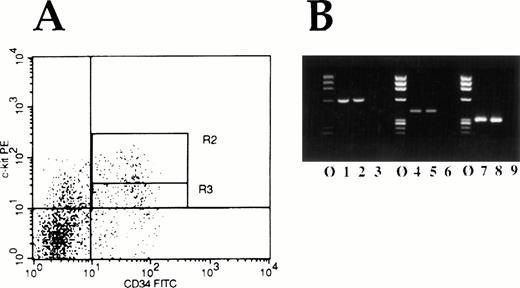
Because c-kit is expressed on the most primitive of human hematopoietic cells, one might expect to find that Myb andEts are also expressed in these cells if they play an important physiologic role in regulating Kit expression. Accordingly, we FACS isolated normal human CD34+Kitbright cells and CD34+Kitdull cells (Fig 10A) and determined whether c-myb and ets-2 mRNA could be detected in these primitive cells by using reverse-transcriptase polymerase chain reaction (RT-PCR). As shown in Figure 10B, both mRNA species, as anticipated, were detected in these very primitive human hematopoietic cells.
(A) FACS histogram of bone marrow MNC double-labeled for expression of CD34 antigen (FITC) and c-kit receptor (PE). Cells in region R2 are defined as being Kit “bright,” whereas cells in the region labeled R3 are defined as Kit“low.” (B) RT-PCR analysis of FACS isolated cells for Etsand Myb mRNA expression. Molecular weight marker lanes are indicated by ø. Lanes 1 and 2 show Ets-2 mRNA expression, lanes 4 and 5 c-myb expression, and lanes 7 and 8, β-actin expression in Kit “bright” and Kit “dull” cells respectively. Lanes 3, 6, and 9 represent water control lanes for the PCR reactions.
(A) FACS histogram of bone marrow MNC double-labeled for expression of CD34 antigen (FITC) and c-kit receptor (PE). Cells in region R2 are defined as being Kit “bright,” whereas cells in the region labeled R3 are defined as Kit“low.” (B) RT-PCR analysis of FACS isolated cells for Etsand Myb mRNA expression. Molecular weight marker lanes are indicated by ø. Lanes 1 and 2 show Ets-2 mRNA expression, lanes 4 and 5 c-myb expression, and lanes 7 and 8, β-actin expression in Kit “bright” and Kit “dull” cells respectively. Lanes 3, 6, and 9 represent water control lanes for the PCR reactions.
DISCUSSION
The human c-kit gene is located on the long arm of chromosome 4 [4q11 - 12], close to the locus of the platelet-derived growth factor receptor-alpha (PDGF-Rα) gene.43 Several groups have reported genomic analyses of the c-kit gene43-45and a few have begun to study the promoter region as well.27,39,44 By providing additional functional analyses of the human c-kit promoter we usefully extend these earlier works and provide new insights into the regulation of this critical hematopoietic gene. For example, in agreement with the previously published studies, we have shown that the ∼1kb genomic fragment we analyzed contained weak promoter activity akin to the 5′ flanking region of the c-fms gene to which c-kit is highly related. Also in agreement with previously published reports,27,39 we found that CAT activity in Kitexpressing cells was highly dependent on the presence of a relatively small region of the promoter immediately upstream of the translational start site. Our experiments showed that deletion of the distalNaeI/BamHI fragment (nts -138 to -20) resulted in almost complete loss of promoter activity in K562 cells. This fragment was required for Kit promoter activity in nonhematopoietic cells as well, even if, like the K562 cells employed in our laboratory, they expressed Kit at low level. Whether distal genomic elements modulate Kit expression in a cell type specific manner, as has been suggested by Vandenbark et al,27 was not evaluated.
Perhaps of greater importance we have also begun to identify important regulatory factors that govern the gene's expression (Fig 1). Previously, a TATA-like element (-ATTAA-; nts -232 to -228) was identified by Giebel et al38 but its functional significance was not explored. Our deletion experiments suggest that it is not critical for Kit expression. For example, the data presented in Figure 4 show that expression constructs devoid of this element do not lose significant promoter activity relative to those that contain it. Whereas the more quantitative data presented in Figures 4 and 7 do suggest some weakening of activity in the absence of this element, a deletion construct that contains the TATA-like element, but not the proximal ∼118 nts of the putative promoter, has no activity at all in our CAT reporter assays. Further analysis of the available 5′ flanking sequence suggests that a number of other potentially important regulatory sites, both positive and negative, have been identified. For example, deletions from the 5′ end of the RsaI fragment appeared to yield fragments with stronger promoter activity. Potential negative regulatory elements present in the deleted region include an MZF1 binding site at nucleotides -820 to -808.30 Studies to directly confirm the importance of these sites in the expression of the Kit receptor in human hematopoietic cells are presently ongoing in our laboratory.
We have also explored the potential functionality of several putativeMyb-and Ets-2–binding sites in the 5′ flanking region of the c-kit gene. This was of great interest to us because we have previously shown that when Myb expression is perturbed in human CD34+ hematopoietic cells exposed to c-myb antisense oligodeoxynucleotides12 or murine embryonic stem cells,24 c-kit mRNA levels also decline. In addition, Vandenbark et al27 identified and evaluated the effect of two potential Myb-binding sites, beginning at nts -900 (MYB2) and -1329 (MYB1), on Kit promoter activity in K562 cells. They found that only one of these sites (MYB1) actually bound c-Myb protein and further analysis suggested that this site functioned as a transcriptional repressor. Although MYB2 appeared to be transcriptionally active, it could not be shown that c-Myb protein actually bound to the site. Accordingly, this work did not establish a direct effect of c-Myb protein on upreguation of Kit expression. We now provide experimental data that supports the hypothesis that Myb is a direct, positive regulator of c-kit expression and that it may function in this capacity through an Ets-2 cooperation. In this regard, we have shown by gel retardation assays that Myb and Ets-2 bind to segments of the c-kit 5′ flanking region containing putative Myb and Ets-2–binding sites. We have also shown that CAT constructs driven by the 5′ flanking region of c-kit have increased activity in the presence of functionalMyb and Ets-2 proteins. Moreover, we showed that both proteins have a cooperative effect on transactivation of the human c-kit promoter. Additional supporting studies showed that mutated Myb protein does not bind to the c-kitpromoter, nor does it have activity in the reporter assays. Finally, although c-myb and ets have previously been shown to be expressed in CD34+ cells, we now show that the mRNAs for these respective genes are also expressed in very primitive CD34+, Kit+ and CD34+,Kit- cells. Accordingly, whereas it remains possible that Myb and/or Ets-2 indirectly regulate Kit expression, we consider this possibility less likely in view of the overwhelming evidence that these transcription factors interact directly with c-kit's promoter.
The finding that Myb and Ets-2 may play a role in regulating Kit expression is of interest from several points of view. First, because c-kit expression is clearly important for hematopoietic cell development6,12-15 another critical function for Myb, in addition to its role in regulating hematopoietic cell development,25,46 G1/S transition,47 transactivating CD4,30,48CD13/APN,49 and CD34,28,29,50 may now have been discerned. In this regard, it is interesting to postulate that the anemia noted in Myb “knock-out” mice, which phenotypically resembles that observed in w or slmutations of c-kit and SF, respectively, may have been mechanically caused by failure of these developing embryos to expressKit. Second, and perhaps more speculative, is Myb's potential role in the regulation of apoptosis via its effects onKit receptor expression. A number of laboratories, including our own, have suggested that Kit's ligand, SF, prevents hematopoietic cells from undergoing apoptosis.7,8 There are also suggestive data that the mechanism for this effect involves upregulation of bcl-2 because in at least one system, expression of bcl-2 decreases when Kit expressing cells are deprived of SF.51 If Myb proves to be physiologically required for Kit expression, it may then assist in early hematopoietic cell survival by permiting the Kitreceptor/ligand interaction to occur. Of equal interest, we have reported that malignant hematopoietic cells are less tolerant of transient interruption of Myb expression than normal cells.52,53 Downregulation of Kit expression in cells that are dependent on Kit ligand for viability may be at least a partial explanation of this observation. Finally, although recent studies suggest that c-ets and c-myb may have opposing function in the regulation of c-fmsexpression,54 their activities appear to be entirely cooperative in regulating Kit expression.
Besides Myb and Ets, additional transcription factors, such as one encoded by the mi locus in mice, may also be important regulators of c-kit expression.55 It is therefore clear that knowledge of the factors that regulate Kitexpression in hematopoietic cells remains incomplete. Because the basic scientific and translational implications of these studies continues to grow, we continue to pursue these investigations as important corollaries of understanding the developmental biology of normal and malignant human hematopoietic stem cells.
ACKNOWLEDGMENT
We thank R. Spritz for c-kit genomic DNA fragment employed for sequencing, Dr H. Hung (Department of Pathology, University of Pennsylvania, Philadelphia, PA) for constructive comments, and E.R. Bien for editorial assistance.
Supported in part by grants from the National Institutes of Health to A.M.G. and B.C.
Address reprint requests to Alan M. Gewirtz, MD, Room 513B, Stellar-Chance Laboratories, University of Pennsylvania School of Medicine, 422 Curie Blvd, Philadelphia, PA, 19104.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.



![Fig. 9. Myb and Ets-2 proteins bind specifically to their predicted sites in the kit promoter. γ-32 P-end labeled double-stranded oligonucleotide probes corresponding either to the canonical Myb (M1 and M2) or Ets-2 (E1, E2 and E3) binding sites within the XhoI-SmaI promoter region fragment (nts-461 to -24) (see Fig 6C) were employed in EMSA. Binding of bacterially expressed Myb (A) or Ets proteins (B) was competed with either wild-type (wt) or mutated (mut) double-stranded oligonucleotides as indicated above each lane. Similar experiments were also performed with nuclear extract derived from K562 cells. Fifteen μg of K562 nuclear extract was incubated in binding buffer with γ-32 P-end labeled double-stranded oligonucleotide probes (lane 1; [C and D]) corresponding either to the canonical Myb (M1 and M2; [C]) or Ets-2– (E1, E2; [D]) binding sites as described above. Specificity of binding was assessed using wild type (lane 2; [C and D]) or mutated (lane 3; [C and D]) double-stranded oligonucleotides as indicated above each lane. Sequences of the double-stranded oligodeoxynucleotides for both mutated and wild-type Myb and Ets-2 binding sites are given in the Materials and Methods section.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/6/10.1182_blood.v91.6.1934/2/m_blod4063909a.jpeg?Expires=1769086005&Signature=MgjBILoQVw8niBIszNktVIMFnu6JpW7M3urFls25WrYI0wq9mrc4p8SyZZOJMBOUtBT~XbWAgpHe78KdjF5PRjQ~YMlTdIIF3xygIoBZKMEDn1vkgO933HqUcgptlBmBcAg-emFfWjBB3bqEm~kXrMfu8CrHdoDQ6GTOqKzenoZ1hqb7GhIn1~aPvKx6Xdi6CeK0mB2Lk5WrK5D9r6cuizjIBhI3~ne3ZWppk1zoOYrRSa4W1v2yJCpnKzmaGCUe49FCnOjIJi87~y4EunsRWTWSCVyLc4giWpSvRCGGpEhR7yDMzXHowfjnx4KYAvr6XMnpuI6bEYCFHQcBnYibvg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 9. Myb and Ets-2 proteins bind specifically to their predicted sites in the kit promoter. γ-32 P-end labeled double-stranded oligonucleotide probes corresponding either to the canonical Myb (M1 and M2) or Ets-2 (E1, E2 and E3) binding sites within the XhoI-SmaI promoter region fragment (nts-461 to -24) (see Fig 6C) were employed in EMSA. Binding of bacterially expressed Myb (A) or Ets proteins (B) was competed with either wild-type (wt) or mutated (mut) double-stranded oligonucleotides as indicated above each lane. Similar experiments were also performed with nuclear extract derived from K562 cells. Fifteen μg of K562 nuclear extract was incubated in binding buffer with γ-32 P-end labeled double-stranded oligonucleotide probes (lane 1; [C and D]) corresponding either to the canonical Myb (M1 and M2; [C]) or Ets-2– (E1, E2; [D]) binding sites as described above. Specificity of binding was assessed using wild type (lane 2; [C and D]) or mutated (lane 3; [C and D]) double-stranded oligonucleotides as indicated above each lane. Sequences of the double-stranded oligodeoxynucleotides for both mutated and wild-type Myb and Ets-2 binding sites are given in the Materials and Methods section.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/6/10.1182_blood.v91.6.1934/2/m_blod4063909c.jpeg?Expires=1769086005&Signature=Yq~5QpdIvn0Sk~-YLmEIasGdWSIaej-cU1zc3gw29d5Cxy2SOKoG12-WvD3YNygnjylr9dZwVUjLL2JVe1ik23OrAD~xTmqwwDvoxS~RaMkq48fFChsxdfr5zacfv8VsUFPFSay3TY7SEZj-kEMZH30gKvnJLCypjeUkqjj1lVCRhFKQe74anUHE3kWoAdCe-Dgb1kv1WPA-xGVThQ5RXHwae3zp1xZhg9FFJlOkrVPY4jp9afUZvKlAMeli7ZupWfftI2oJAO22p0icwjO6cUUo~VbK3jIPo9E57c1Fqk8o2ucYOYGOb9BPx3X~QC15ye6fPjIjs9xPxspZ~iTgkg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 9. Myb and Ets-2 proteins bind specifically to their predicted sites in the kit promoter. γ-32 P-end labeled double-stranded oligonucleotide probes corresponding either to the canonical Myb (M1 and M2) or Ets-2 (E1, E2 and E3) binding sites within the XhoI-SmaI promoter region fragment (nts-461 to -24) (see Fig 6C) were employed in EMSA. Binding of bacterially expressed Myb (A) or Ets proteins (B) was competed with either wild-type (wt) or mutated (mut) double-stranded oligonucleotides as indicated above each lane. Similar experiments were also performed with nuclear extract derived from K562 cells. Fifteen μg of K562 nuclear extract was incubated in binding buffer with γ-32 P-end labeled double-stranded oligonucleotide probes (lane 1; [C and D]) corresponding either to the canonical Myb (M1 and M2; [C]) or Ets-2– (E1, E2; [D]) binding sites as described above. Specificity of binding was assessed using wild type (lane 2; [C and D]) or mutated (lane 3; [C and D]) double-stranded oligonucleotides as indicated above each lane. Sequences of the double-stranded oligodeoxynucleotides for both mutated and wild-type Myb and Ets-2 binding sites are given in the Materials and Methods section.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/6/10.1182_blood.v91.6.1934/2/m_blod4063909b.jpeg?Expires=1769086005&Signature=i4TdVA2GIkXjMvg5QKNY2WVhzjRFN1xcXJNzSAPOLoWSV-j42wRzhZIZYUqC6IhwTcY8P13btfmab99UEXXkSpZ6G3o8dJ-4bwb-YuVRnH3tSmcoZzL1mMNaqwYFFiMv7z3euEMvPhJHWwONDdKtX-zBZZp6xP4V3V8b1fvl9Q3dZcPRsMX0LoVRJ4F-F6REqg2MVjLSZoIPzqfQNs5mIRxR6UAC8Z-zT4YnKJ47diSOE1oG0IbAbFsx5O5X-ebooha2TLyJjj9pbAM~0ndNHh4ZAgOWf1B1zzvVVDw148ccL~hIzdox4c4ONEIRiW-REILv979NFdXiF1WMxWFJFA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 9. Myb and Ets-2 proteins bind specifically to their predicted sites in the kit promoter. γ-32 P-end labeled double-stranded oligonucleotide probes corresponding either to the canonical Myb (M1 and M2) or Ets-2 (E1, E2 and E3) binding sites within the XhoI-SmaI promoter region fragment (nts-461 to -24) (see Fig 6C) were employed in EMSA. Binding of bacterially expressed Myb (A) or Ets proteins (B) was competed with either wild-type (wt) or mutated (mut) double-stranded oligonucleotides as indicated above each lane. Similar experiments were also performed with nuclear extract derived from K562 cells. Fifteen μg of K562 nuclear extract was incubated in binding buffer with γ-32 P-end labeled double-stranded oligonucleotide probes (lane 1; [C and D]) corresponding either to the canonical Myb (M1 and M2; [C]) or Ets-2– (E1, E2; [D]) binding sites as described above. Specificity of binding was assessed using wild type (lane 2; [C and D]) or mutated (lane 3; [C and D]) double-stranded oligonucleotides as indicated above each lane. Sequences of the double-stranded oligodeoxynucleotides for both mutated and wild-type Myb and Ets-2 binding sites are given in the Materials and Methods section.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/91/6/10.1182_blood.v91.6.1934/2/m_blod4063909d.jpeg?Expires=1769086005&Signature=sTTgU7eJCvkAD3YmNtB~iVUnx2qOMY3Lp186eY6~~Bm1BNYzE8~S4lUWgLXKwcfoMl9jcNLCXHT7OE2U2EL730EqJ0~NrrzJWVQo2ASHxgh0eAitQ6hRdMvmEpLJkwXNpAyqXFBmuLXVec1tk1rvYHiwZtZ7M3R61ivYBadS685oJQwLathnsocvRKNggDYXcZ-PxqG0PzmOAUTdO2aTnUhBw0YUHDpMwzNTdDZjh7BoEXBPA3uFiVbh~oMgUNiTbfi03EmQWXkoNEQTAAbSAJC8FcX1yi1z7ctF1P1Wkf-RXcq35u~ER3Kwye848JYjdkSSAolHU8o~G2zvpNFZRg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal