Abstract
von Willebrand factor (vWF) is a large multimeric, multidomain glycoprotein found in platelets, endothelial cells and plasma. The A1, A2, and A3 domains in vWF mediate binding to glycoprotein Ib, ristocetin, botrocetin, collagen, sulphatides, and heparin and provide a protease cleavage site. Mutations causing types 2B, 2M, and 2A von Willebrand's disease (vWD) are located in the A1 and A2 domains. Homology modeling was performed to provide a molecular interpretation of vWF function and mutation sites. This was based on our previous alignment of 75 vWF-A sequences, the doubly wound α/β fold seen in recent vWF-A crystal structures from complement receptor type 3 and lymphocyte function-associated antigen-1, and our new alignment of 28 vWF A1 and A2 sequences from different species. The active site in doubly-wound α/β folds forms a crevice that is located at the switch point between the two halves of the central β-sheet, and usually contains two metal-binding Asp residues in the vWF-A superfamily. Although one of these Asp residues is absent from the A1, A2, and A3 domains, this crevice is shown to correspond to the ristocetin binding site in the A1 domain and the protease cleavage site in the A2 domain. The residues R571-K572-R578-R579-K585 are found to be conserved in 28 A1 sequences and are predicted to constitute the heparin binding site in the A1 domain. Inspection of the type 2M vWD mutation sites that are involved in downregulation of glycoprotein Ib (GpIb) binding to vWF shows that these are spatially clustered at the carboxyl-edge of the β-sheet and above it in the A1 domain and may directly perturb GpIb binding. In contrast, the type 2B vWD mutation sites that are involved in upregulation of GpIb binding to vWF are spatially clustered at the amino edge of this β-sheet and below it and are located on the opposite side of the A1 domain from the type 2M mutation sites. The type 2B mutations are located between the heparin and GpIb binding sites. Because heparin binding inhibits the interaction with GpIb, this provides an explanation of vWF upregulation. The type 2A vWD mutation sites in the A2 domain correspond to buried residues that are otherwise 100% conserved across all 28 species, and are likely to be important for the correct folding of the A2 domain and its physiologically important protease site.
VON WILLEBRAND FACTOR (vWF) is a large glycoprotein that is found in plasma and platelets and is synthesized by megakaryocytes and endothelial cells.1-3 vWF plays at least two essential roles in hemostasis. It is involved in platelet adhesion to the damaged vascular endothelium, and it stabilizes factor VIII in plasma by acting as its carrier molecule. Precursor vWF contains 13 domains in its monomeric structure, which are multiples of four domain types A to D in the arrangement D1-D2-D′-D3-A1-A2-A3-D4-B1-B2-B3-C1-C2.4 After synthesis, the precursor is modified by post-translational processing, in which it dimerizes, and cleavage of the D1-D2 domain pair occurs. The monomer contains 2,050 residues of molecular weight 220,000. Multimeric vWF contains between 2 to 100 subunits with molecular weights ranging up to 20 ×106.
The A1, A2, and A3 domains mediate key macromolecular interactions by vWF. The A1 domain corresponds to residues 497 to 716 in mature vWF, whose N- and C-termini are joined by a disulfide bridge at Cys509 and Cys695. It is involved in vWF binding to the platelet receptor glycoprotein Ib (GpIb). This is the primary mechanism by which vWF binds to platelets under conditions of high shear stress. vWF does not bind spontaneously to GpIb. Conformational changes are thought to expose the vWF-A1 domain after the binding of other regions of vWF to collagen on the subendothelium. In vitro tests show that vWF can be stimulated to bind to platelets by the antibiotic ristocetin (molecular weight of several thousands) and two forms of the snake venom derivative botrocetin (molecular weight 25,000 and 26,500), although these two compounds involve different residues in the vWF-A1 domain.5 The vWF-A1 domain also contains distinct binding sites for heparin and sulphatides and a further site for collagen.2,4 The binding of vWF to collagen permits its interaction with GpIb, whereas vWF binding to heparin inhibits it. The vWF-A2 domain spans residues 717 to 909 and does not contain a disulfide bridge between its N- and C-termini (although there is a bridge between Cys906 and Cys907). It contains a physiological cleavage site between Tyr842 and Met843 that is associated with a metalloprotease in normal circulation.6,7 The vWF-A3 domain spans residues 910 to 1111 in which its N- and C-termini are joined by a disulfide bridge between Cys923 and Cys1109.8 This contains the major binding site for fibrillar collagens.9 10
von Willebrand's Disease (vWD) is the most common inherited bleeding disorder, in which type 2 vWD corresponds to functional abnormalities in vWF (as opposed to a deficiency or absence of vWF in types 1 and 3 vWD, respectively).11-13 A large number of mutated residues that give rise to three variant types 2A, 2B, and 2M vWD have been found in the vWF-A1 and vWF-A2 domains. Type 2B vWD is characterized by the loss of high molecular weight multimers of vWF as a result of an increased affinity of vWF for GpIb, and shown in vitro by increased ristocetin-induced binding of vWF to platelets. Most type 2B vWD mutations involve a short segment in the vWF-A1 domain.11,14,15 Type 2M vWD is characterized by a normal pattern of vWF multimers but a decreased binding of vWF to GpIb and a decreased ristocetin-induced binding of vWF to platelets. Mutations causing type 2M vWD are also located in the vWF-A1 domain.16-19 Type 2A vWD results from the loss of high and intermediate vWF multimers, either through the impaired intracellular transport of vWF multimers (group I mutations) or the abnormal proteolytic degradation of vWF (group II mutations).20 Type 2A mutations arise mainly within the vWF-A2 domain.11 21-23
Three-dimensional structures are required to provide a molecular interpretation of vWF function and a molecular explanation of the mutations involved in type 2 vWD. The vWF-A domains belong to a large superfamily that include other plasma proteins, collagens, integrins, and other extracellular proteins that mediate cell-cell, cell-matrix, and matrix-matrix interactions.24,25 The vWF-A protein fold was first identified by our structure prediction analysis, and this included an initial consideration of vWF mutation sites.26,27 Crystal structures of the vWF-A domains in human complement receptor type 3 (CR3) and the leukocyte integrin lymphocyte function-associated antigen-1 (LFA-1) have since been determined to confirm the prediction.28-31 Since then, vWF sequences from 28 different mammalian species from 15 placental orders are now available.25,32 The combination of our vWF-A predictions with these crystal structures and new sequences now make it possible to create homology models for the human vWF-A1, -A2, and -A3 domains and assess them. We show that major functional sites in the A1 and A2 domains correspond to the classic active site region of this protein fold. Recent crystal structures of protein-heparin complexes33 permit a set of conserved residues in vWF-A1 to be identified as a heparin binding site. The type 2B vWD mutations are shown to be located between the heparin and GpIb binding sites and this provides an explanation of how vWF binding is upregulated, whereas those involved in type 2M vWD are shown to be directly associated with the GpIb binding site and explain how vWF binding is downregulated. The vWF-A2 mutations that lead to type 2A vWD are shown to correspond to buried conserved residues, and these are discussed also.
MATERIALS AND METHODS
Crystal structures in the vWF-A superfamily.
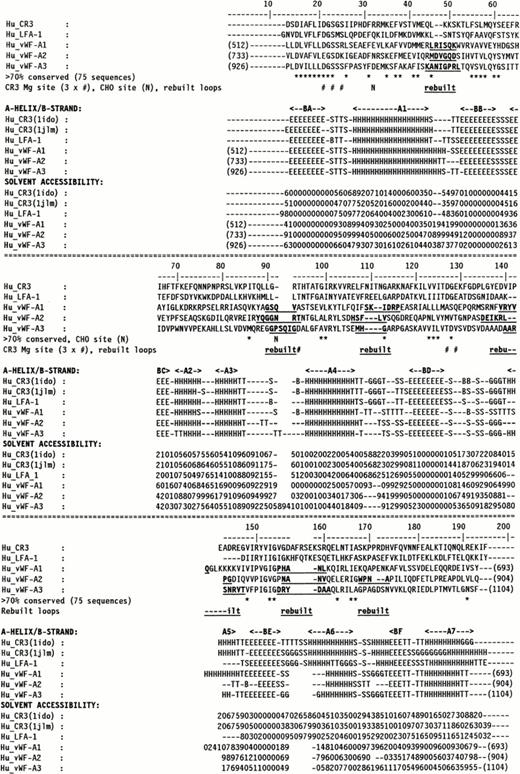
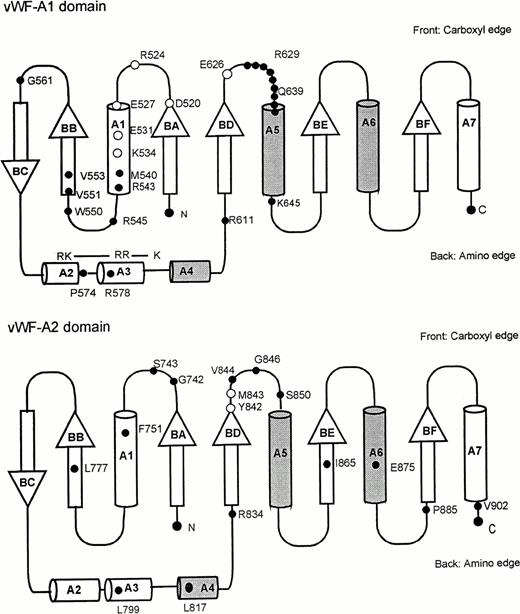
Molecular graphics models of the three vWF-A1, -A2, and -A3 domains in human vWF were constructed as follows. The multiple sequence alignment of 75 vWF-A sequences25 was used to align the three vWF-A sequences to be modeled with the crystal structures of CR3 (CD11b/CD18) and LFA-1 (CD11a/CD18), and this is summarized in Fig 1 using the same sequence numbering as in Perkins et al.25 Residues L512-Y693 of the vWF-A1 domain (out of residues 479 to 716), V733-Q904 of the vWF-A2 domain (out of residues 717 to 909), and P926-F1104 of the vWF-A3 domain (out of residues 910 to 1111) could be modeled. All three models were based on the crystal structure of the Mg-bound CR3 vWF-A domain28 (Brookhaven code 1ido). As a control of this structure, the 1ido coordinates were compared with those for Mn-bound CR329 (code 1jlm) and for LFA-1,30,31 which correspond to metal-free LFA-1 (code 1zon), Mg-bound LFA-1 (code 1zoo), and Mn-bound LFA-1 (code 1zop) in Fig 1. A conformational change has been reported between Mg-bound and Mn-bound CR3 that involves the metal site, the α-helix A7, and three of the five loops on the carboxyl-edge of the β-sheet.29 Only α-helix A7 moves in metal-free LFA-1, which has a structure otherwise identical to Mg-bound and Mn-bound LFA-1, the latter pair having identical structures.31 The DSSP program34was used to assign secondary structure in the crystal structures. Side-chain solvent accessibilities were calculated by the method of Lee and Richards using a probe diameter of 1.4 Å in the COMPARER program.35 36 The secondary structures and accessibilities of 1zon, 1zoo, and 1zop were indistinguishable, and their mean is shown in Fig 1. Figure 1 shows that the secondary structures of LFA-1 and both forms of CR3 are very similar with the exception of changes in the 310-helix (G) and α-helix (H) content of A7 and minor ones in A6. Likewise, if the sidechain accessibilities were classified as exposed (values of 2 to 9) or inaccessible (0 to 1), Fig 1 shows that the three crystal structures are very similar except for small changes at two carboxyl-edge loops connecting BD with A5 and BE with A6, and the C-terminal β-strand BF and α-helix A7. These differences are negligible in terms of the analyses of the ristocetin binding site and the types 2B, 2M, and 2A vWD mutations below.
Sequence alignment for human CR3 and LFA-1 and the vWF-A1, -A2, and -A3 domains. The vWF sequence numbering is indicated at the start and end of each vWF-A sequence. The α-helix and β-strand secondary structures in the CR3 and LFA-1 crystal structures and the three vWF-A models are denoted by A1 to A7 and BA to BF, respectively. Residues that were rebuilt as new loops are denoted in bold and underlined. The DSSP analysis is represented as follows: E, β-strand; B, single residue β-ladders; T, turn; G, 310helix; and H, α-helix. The COMPARER sidechain solvent accessibilities are on a scale of 0 to 9 for each residue, where 0 corresponds to 0% to 9% solvent exposure, 1 to 11% to 19% solvent exposure, and so on, and buried residues have accessibilities of 0 or 1.
Sequence alignment for human CR3 and LFA-1 and the vWF-A1, -A2, and -A3 domains. The vWF sequence numbering is indicated at the start and end of each vWF-A sequence. The α-helix and β-strand secondary structures in the CR3 and LFA-1 crystal structures and the three vWF-A models are denoted by A1 to A7 and BA to BF, respectively. Residues that were rebuilt as new loops are denoted in bold and underlined. The DSSP analysis is represented as follows: E, β-strand; B, single residue β-ladders; T, turn; G, 310helix; and H, α-helix. The COMPARER sidechain solvent accessibilities are on a scale of 0 to 9 for each residue, where 0 corresponds to 0% to 9% solvent exposure, 1 to 11% to 19% solvent exposure, and so on, and buried residues have accessibilities of 0 or 1.
Construction of vWF-A homology models.
Protein structures were visualized and modeled using INSIGHT II 95.0 software with HOMOLOGY and DISCOVER modules (Biosym/MSI, San Diego, CA) on INDY Workstations (Silicon Graphics, Reading, UK). The rigid body fragment assembly method used in HOMOLOGY was used to construct the three models. A conserved core of 138 to 156 α-helix, β-strand and loop residues was defined based on the 1ido crystal structure. In the vWF-A1 model, the remaining 26 residues at five surface loop regions were modeled from a database of protein fragments using the Brookhaven loop database of INSIGHT II. These were between L544-K549, G588-V591, S607-P612, V635-Q639, and P655-L659 where sequence insertions or deletions occurred (Fig 1). In the vWF-A2 model, 34 residues in six loops were modeled at M765-D770, Y807-T813, H825-V829, D851-G858, N870-V873, and W881-A884. In the vWF-A3 model, 29 residues in five loops were modeled at K957-L964, G1003-G1008, M1022-G1024, A1051-T1056, and D1066-A1071. All the sidechains were replaced according to the human vWF-A sequences. Energy refinements using DISCOVER were performed at the loop splice junctions, then on the loop regions, and finally the mutated core residues. The core residues were tethered to their original positions with greater energy terms than the reconstructed loops. The refinements were based on the consistent valence forcefield, and iterations were made using a combination of the steepest descent and conjugate algorithms. These improved the connectivity of the model and reduced the proportion of bad contacts or stereochemistry as confirmed by the use of the PROCHECK program.37 The vWF mutation residues in the vWF-A1 and vWF-A2 domains are summarized in Table 1. The site of each one was identified by a sphere at the α-carbon atom in the vWF-A models of Fig 6.
Mutations in the vWF-A1 and vWF-A2 Domains of vWF Giving Rise to vWD
| vWD Type . | Mutation Site in Mature Polypeptide . | Database Notation* . | Location of Mutation in Model (Fig 3) . | Reference . |
|---|---|---|---|---|
| vWF-A1 domain | ||||
| 2B | 540insM | In A1 | 11 | |
| 2B | R543W | R1306W/Q/C | In A1 | 11 |
| 2B | R545C | R1308C/P | Between A1 and BB | 11 |
| 2B | W550C | W1313C | Between A1 and BB | 11 |
| 2B | V551L/F | V1314L/F | In BB | 11, 14, 15 |
| 2B | V553M | V1316M | In BB | 11 |
| 2B | P574L | P1337L | Between A2 and A3 | 11 |
| 2B | R578Q | R1341Q/P | In A3 | 11 |
| 2M | G561S | G1324S | Between BB and BC | 16 |
| 2M | R611H/C | Between A4 and BD | 17 | |
| 2M | K645del | K1408del | Between A5 and BE | 18 |
| 2M | R629-Q639del | Between BD and into A5 | 19 | |
| vWF-A2 domain | ||||
| 2A | G742E/R | G1505E/R | Between BA and A1 | 11 |
| 2A | S743L | S1506L | Between BA and A1 | 11 |
| 2A | F751C | F1514C | In A1 | 11 |
| 2A | L777P | L1540P | In BB | 11 |
| 2A | L799P | L1562P | In A3 | 11 |
| 2A | L817P | L1580P | In A4 | 21 |
| 2A | R834W/Q | R1597W/Q/G | Between A4 and BD | 11 |
| 2A | V844D | V1607D | Between BD and A5 | 11 |
| 2A | G846R | G1609R | Between BD and A5 | 22 |
| 2A | S850P | S1613P | Between BD and A5 | 11 |
| 2A | I865T | I1628T | In BE | 11 |
| 2A | E875K | E1638K | In A6 | 11 |
| 2A | P885S | P1648S | In BF | 23 |
| 2A | V902E | V1665E | Post A7 | 22 |
| vWD Type . | Mutation Site in Mature Polypeptide . | Database Notation* . | Location of Mutation in Model (Fig 3) . | Reference . |
|---|---|---|---|---|
| vWF-A1 domain | ||||
| 2B | 540insM | In A1 | 11 | |
| 2B | R543W | R1306W/Q/C | In A1 | 11 |
| 2B | R545C | R1308C/P | Between A1 and BB | 11 |
| 2B | W550C | W1313C | Between A1 and BB | 11 |
| 2B | V551L/F | V1314L/F | In BB | 11, 14, 15 |
| 2B | V553M | V1316M | In BB | 11 |
| 2B | P574L | P1337L | Between A2 and A3 | 11 |
| 2B | R578Q | R1341Q/P | In A3 | 11 |
| 2M | G561S | G1324S | Between BB and BC | 16 |
| 2M | R611H/C | Between A4 and BD | 17 | |
| 2M | K645del | K1408del | Between A5 and BE | 18 |
| 2M | R629-Q639del | Between BD and into A5 | 19 | |
| vWF-A2 domain | ||||
| 2A | G742E/R | G1505E/R | Between BA and A1 | 11 |
| 2A | S743L | S1506L | Between BA and A1 | 11 |
| 2A | F751C | F1514C | In A1 | 11 |
| 2A | L777P | L1540P | In BB | 11 |
| 2A | L799P | L1562P | In A3 | 11 |
| 2A | L817P | L1580P | In A4 | 21 |
| 2A | R834W/Q | R1597W/Q/G | Between A4 and BD | 11 |
| 2A | V844D | V1607D | Between BD and A5 | 11 |
| 2A | G846R | G1609R | Between BD and A5 | 22 |
| 2A | S850P | S1613P | Between BD and A5 | 11 |
| 2A | I865T | I1628T | In BE | 11 |
| 2A | E875K | E1638K | In A6 | 11 |
| 2A | P885S | P1648S | In BF | 23 |
| 2A | V902E | V1665E | Post A7 | 22 |
*Database at http://mmg2im.med.umich.edu/vWF/mut-list.cgi. Subtract 763 to obtain the mature polypeptide residue number.
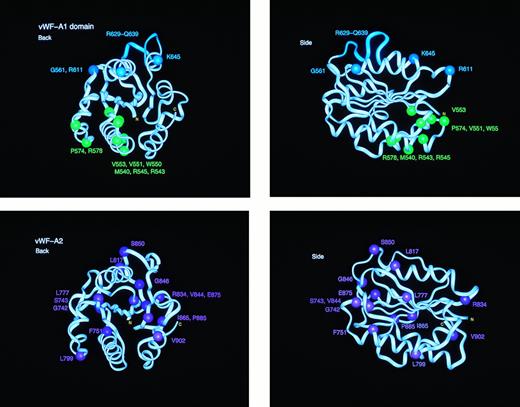
Location of the types 2B, 2M, and 2A mutation sites in homology models of the vWF-A1 and vWF-A2 domains. Each of the vWF-A1 and vWF-A2 domains is shown in two orthogonal perspectives rotated by 90° along the vertical axis. The central β-sheet is seen edge-on in both views. The mutation residues are identified by spheres at their α-carbon atom. The protein backbone is represented by a ribbon. The eleven residues deleted in the type 2M “Milwaukee” mutation are shown as the cyan ribbon in the vWF-A1 model.
Location of the types 2B, 2M, and 2A mutation sites in homology models of the vWF-A1 and vWF-A2 domains. Each of the vWF-A1 and vWF-A2 domains is shown in two orthogonal perspectives rotated by 90° along the vertical axis. The central β-sheet is seen edge-on in both views. The mutation residues are identified by spheres at their α-carbon atom. The protein backbone is represented by a ribbon. The eleven residues deleted in the type 2M “Milwaukee” mutation are shown as the cyan ribbon in the vWF-A1 model.
Electrostatic determination of the surfaces was performed using DELPHI (Biosym/MSI).38 The total electrostatic energy of the system was calculated using a full coulombic boundary condition, with the interior and exterior dielectric constants set as 2 and 80, respectively, and assuming an ionic strength corresponding to a 0.145 mol/L salt solution. The solvent accessible surface of the molecule was displayed using the Connolly algorithm with a solvent probe radius of 0.14 nm. This surface was colored red for potentials less than −4 kT (acidic), blue for potentials greater than 4 kT (basic), and white for neutral potentials of 0 kT. Linear interpolation was used to produce the colors between these values.
RESULTS
Homology models for the vWF-A1, -A2, and -A3 domains.
The homology modeling of the vWF-A1, -A2, and -A3 structures on the basis of five related crystal structures28-31 was straightforward (Fig 1). The use of a multiple sequence alignment for the vWF-A superfamily significantly enhanced the reliability of aligning the vWF-A1, -A2, and -A3 sequences with those of the crystal structures.25 The three vWF-A structures containing 172 to 182 residues were modeled starting from the CR3 crystal structure using five to six short remodeled loops with lengths of three to eight residues that totalled 26 to 34 residues. Regions of sequence insertion and deletion corresponded to surface locations in the structure between α-helices and/or β-strands, including the partial exceptions of α-helices A5 and A6. An additional three to four residues were added at position 142/143 in the vWF-A1 domain and position 92/93 in the vWF-A3 domain. Two segments of 3/4 residues and 6 residues were deleted at positions 109 to 112 and 157/158, respectively, in the models. These changes showed that only minimal perturbations of secondary structures were required in the three modelings, although a one-to-one correspondence of residues cannot be assumed in the loop regions that were rebuilt.
Support for a satisfactory outcome of the modelings was provided by DSSP analysis of the secondary structure to show that all 13 α-helices and β-strands seen in the crystal structures were retained after energy refinement, except for the truncated A5 α-helix in the vWF-A2 domain (Fig 1). Likewise the COMPARER analysis of sidechain accessibilities in the three models showed that these agreed with the crystallographic values (Fig 1). Two glycosylation sites in the vWF-A2 domain at N751 on α-helix A1 and N811 between α-helices A3 and A4 were exposed to solvent as required. All three vWF-A models also satisfied stereochemical verification by PROCHECK.
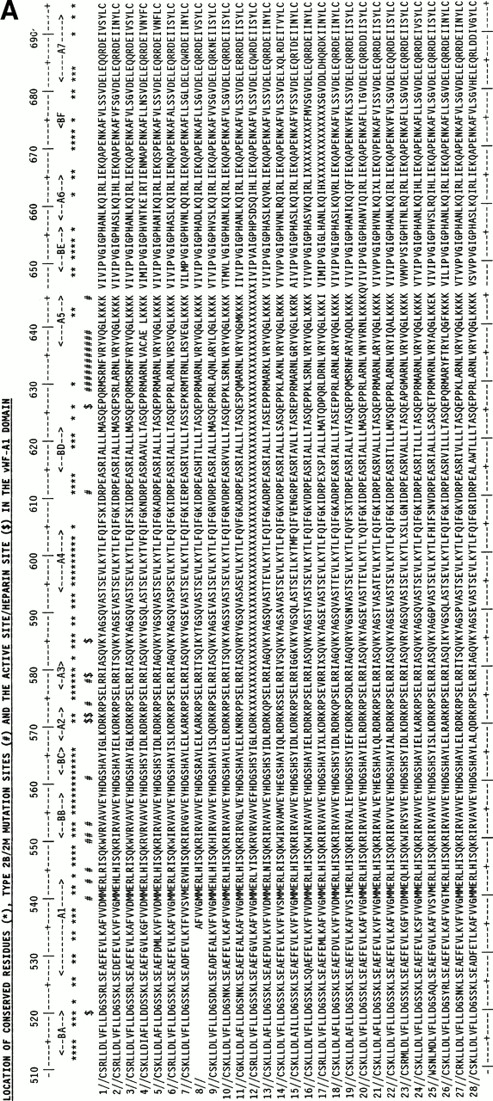
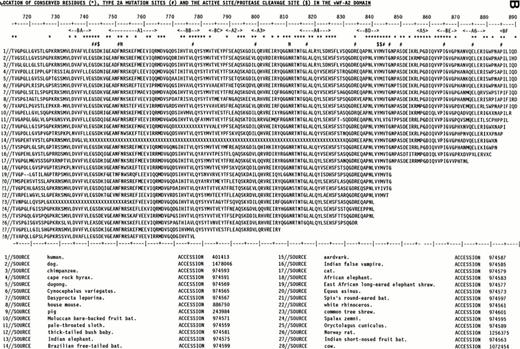
The use of the vWF-A1 and -A2 models was enhanced by the use of 28 mammalian vWF-A1 and -A2 sequences representing at least 15 placental orders and mostly derived from the recent sequencing of exon 28 in the vWF gene.32 Their alignment (Fig 2) showed only a single one-residue insertion in the A1 sequences and none in the A2 sequences, and no gaps. On the basis of Fig 2, residues that are seen to be 100% conserved in all 28 sequences may be essential for function or structural folding in these two domains.
Alignment of vWF-A1and vWF-A2 sequences from 28 mammalian species. The sequences are identified by accession numbers. The numbering is taken from human vWF, and the location of α-helices and β-strands follows that of Fig 1. Residues that are 100% absolutely or conservatively conserved are indicated by an asterisk. (Conservative: G=A=S, D=E, I=L=V=M; S=T, R=K=H, F=Y=W=H; X is disregarded). (A) The positions of D520 and E626 at the active site and the proposed heparin site RK-RR-K (residues 571-585) are marked by a dollar sign ($). The positions of the type 2B and 2M mutations are marked by a number sign (#). (B) The position of D744 at the active site and the protease cleavage site at Y842-M843 are marked by $. The positions of the type 2A mutations are marked by #. The positions of two conserved putative N-linked glycosylation sites are marked by N.
Alignment of vWF-A1and vWF-A2 sequences from 28 mammalian species. The sequences are identified by accession numbers. The numbering is taken from human vWF, and the location of α-helices and β-strands follows that of Fig 1. Residues that are 100% absolutely or conservatively conserved are indicated by an asterisk. (Conservative: G=A=S, D=E, I=L=V=M; S=T, R=K=H, F=Y=W=H; X is disregarded). (A) The positions of D520 and E626 at the active site and the proposed heparin site RK-RR-K (residues 571-585) are marked by a dollar sign ($). The positions of the type 2B and 2M mutations are marked by a number sign (#). (B) The position of D744 at the active site and the protease cleavage site at Y842-M843 are marked by $. The positions of the type 2A mutations are marked by #. The positions of two conserved putative N-linked glycosylation sites are marked by N.
Structure and function of the vWF-A1 domain.
The vWF-A protein fold is closely related in topology to the doubly-wound α/β fold found in intracellular nucleotide binding proteins such as flavodoxin and the GTP-binding domain of ras-p21.26,27 In the latter, the nucleotide binding site occurs on the carboxy-edge of the parallel β-sheet found at the center of the fold, close to the switch point when the β-strands wind in opposite directions, and this creates a crevice that is where the active site is located39 (Fig3). The α-helices A4, A5, and A6 are above the plane of the β-sheet to the right as visualized in Fig 3, whereas the α-helices A1, A2, and A3 to the left are below this plane. In many vWF-A domains, two buried conserved Asp residues at positions 21 and 127 in Fig 1 are located at this switch point, which serves as an active site and binds metal.25,26 The CR3 and LFA-1 crystal structures show bound metal at this location.28-31
Supersecondary structure topology for the doubly wound open α/β fold of the vWf-A domain. α-Helices are represented as cylinders, for which those above the central β-sheet are shaded and those below are unshaded. The β-strands are shown as arrows and labeled to follow Fig 1. N and C denote the N- and C-terminus, respectively. The positions of mutations are shown on the appropriate topology diagram as filled circles. Residues important in ristocetin binding are shown as clear circles in the vWF-A1 domain. Residues important in the protease cleavage site Y842-M843 are likewise shown as clear circles in the vWF-A2 domain.
Supersecondary structure topology for the doubly wound open α/β fold of the vWf-A domain. α-Helices are represented as cylinders, for which those above the central β-sheet are shaded and those below are unshaded. The β-strands are shown as arrows and labeled to follow Fig 1. N and C denote the N- and C-terminus, respectively. The positions of mutations are shown on the appropriate topology diagram as filled circles. Residues important in ristocetin binding are shown as clear circles in the vWF-A1 domain. Residues important in the protease cleavage site Y842-M843 are likewise shown as clear circles in the vWF-A2 domain.
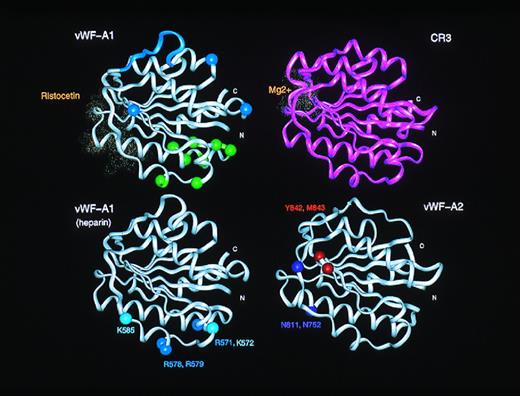
Inspection of the vWF-A1 domain shows that D520 is located in this active site crevice and is fully conserved in CR3, LFA-1, and other vWF-A sequences (Fig 1) and in all the 28 vWF-A1 sequences (Fig 2). However, the second Asp residue at position 127 in CR3 and LFA-1 (Fig1) is missing from all 28 vWF-A1 sequences. Although the conserved vWF-A1 residue E626 is close to this position, this omission implies that no metal binding occurs at this location in vWF-A1. Residues important for the ristocetin-induced binding of vWF to platelets have been shown to be D520-R524, E527-E531, K534, and E626 in vWF-A1 by scanning alanine mutagenesis.40 Figure 2 shows that D520, S522, E529, F530, K534, and E626 are conserved in vWF from 28 species. As ristocetin is cationic, the involvement of D520, E529, and E626 is of great interest, and the similar location of the ristocetin and Mg2+ coordination sites in the vWF-A structure is shown in Fig 4. Because Fig 3 shows that these three acidic residues are close to the active site, it is concluded that the crevice is of functional significance in the vWF-A1 domain.
Comparison of the active sites in the vWF-A1 and vWF-A2 domains. The upper panels compare the ristocetin-binding residues in the vWF-A1 domain with the Mg2+ binding site in the CR3 vWF-A domain. These residues are shown as dot representation of the van der Waal radii of the residues in question. For comparison with Fig 6, the type 2M and 2B mutation sites in the vWF-A1 domain are indicated using spheres at the α-carbon positions. The lower panels show the proposed heparin binding site on the α-helices A3 and A4 in the vWF-A1 domain, and the protease cleavage site at Y842-M843 on the vWF-A2 domain, also using α-carbon positions.
Comparison of the active sites in the vWF-A1 and vWF-A2 domains. The upper panels compare the ristocetin-binding residues in the vWF-A1 domain with the Mg2+ binding site in the CR3 vWF-A domain. These residues are shown as dot representation of the van der Waal radii of the residues in question. For comparison with Fig 6, the type 2M and 2B mutation sites in the vWF-A1 domain are indicated using spheres at the α-carbon positions. The lower panels show the proposed heparin binding site on the α-helices A3 and A4 in the vWF-A1 domain, and the protease cleavage site at Y842-M843 on the vWF-A2 domain, also using α-carbon positions.
The heparin binding site in the vWF-A1 domain can be predicted from the model. This has previously been mapped to residues 565 to 587, and the binding of sulphatides has likewise been mapped to residues 569 to 584.2,41 Both ligands are negatively charged and are complementary in charge to the positively charged surface seen in electrostatic maps at this region of the vWF-A1 model (Fig 5). Within this sequence, Fig 2 shows that the R571-K572 pair, the R578-R579 pair, and K585 are fully conserved in 28 species and that R573 is conserved in 24 species. The significance of this observation is underscored by noting that this pattern of three sets of conserved basic residues is not found in any of the other 72 vWF-A domains in the sequence alignment for this superfamily.25 The vWF-A1 model shows that these five basic residues are located on the α-helices A2 and A3 in an extended linear structure (Fig 4). The K572-R573 pair, the R578-R579 pair, and K585 show 60% to 100% sidechain accessibility to solvent and to heparin (Fig 1). In addition, the α-carbon distances between adjacent sets of these residues are between 10 to 14 Å and are maximally 22 Å between the first and last ones. These dimensions are comparable with those seen for two known heparin binding sites. In the crystal structure of antithrombin III (Brookhaven code 1ant), a heparin binding site has been located on an extended region that involves five basic residues R24, K114, R47, K125, and R129.42 The distances between adjacent pairs of these α-carbon atoms range between 6 to 10 Å, and the distance between the first and fifth α-carbon atoms spans 26 Å. In the crystal structure of the basic fibroblast growth factor-heparin complex (codes 1bfb and 1bfc), heparin makes contact with the basic residues K27, R121, K126, and K136 (whose α-carbon atoms are arranged as a near-rectangle on loops between β-strands at one end of the protein), and no conformational changes were observed between the heparin-free and heparin-bound structures.33 The distances between adjacent pairs of α-carbon atoms of these four basic residues are between 6 to 9 Å and are maximally 14 Å between any pair. Although it is not clear whether all three residues within the set R571-K572-R573 are involved in heparin binding in vWF-A1, it is concluded that the accessibility and separation of these three sets of basic residues in vWF-A1 is comparable with the heparin-binding residues in antithrombin III and basic fibroblast growth factor.
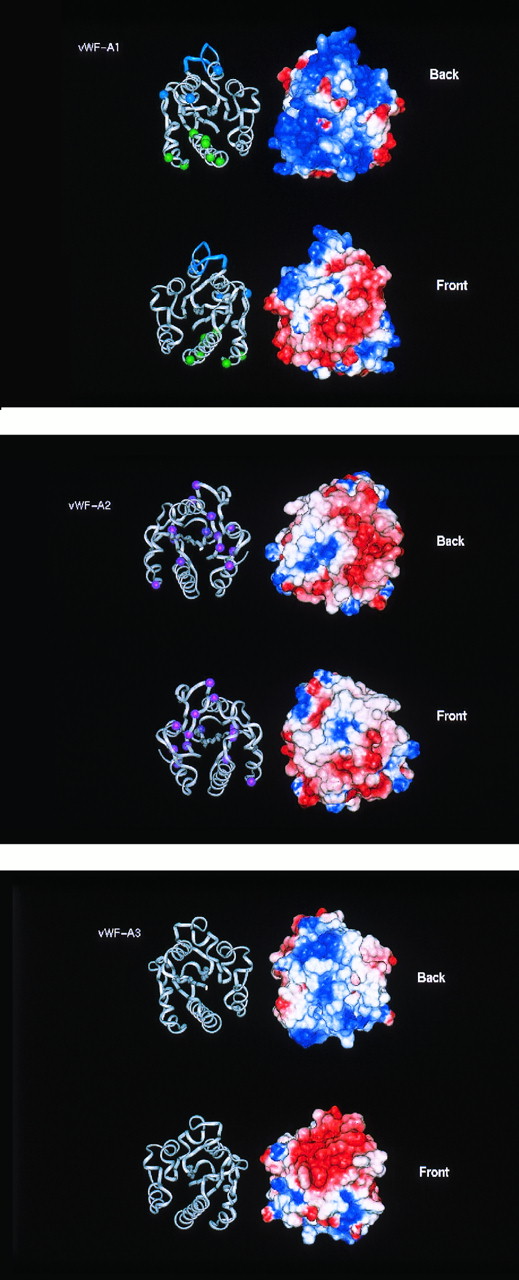
Electrostatic maps of the vWF-A domains in human vWF. The electostatic maps show the Connolly surface of the domains with a color range of red representing a potential of less than −4 kT, blue a potential of more than +4kT (basic) and white as 0kT (neutral). Linear interpolation of the colors represents potentials between −4 kT and +4 kT. The electrostatic surfaces are shown alongside ribbon diagrams of the protein backbone of the respective domains. The domains are shown in two perspectives, rotated along the vertical axis by 180°. The ribbon views permit comparison with Figs 4 and 6.
Electrostatic maps of the vWF-A domains in human vWF. The electostatic maps show the Connolly surface of the domains with a color range of red representing a potential of less than −4 kT, blue a potential of more than +4kT (basic) and white as 0kT (neutral). Linear interpolation of the colors represents potentials between −4 kT and +4 kT. The electrostatic surfaces are shown alongside ribbon diagrams of the protein backbone of the respective domains. The domains are shown in two perspectives, rotated along the vertical axis by 180°. The ribbon views permit comparison with Figs 4 and 6.
Heparin corresponds to a disaccharide repeat of L-iduronic acid and D-glucosamine with three types of sulphate groups connected to the sugar rings via an O-link, a CH2-link, and an N-link. The sulphate and carboxyl groups are positioned on the surface of a central ribbon of sugar rings.33 43 The distances between neighboring pairs of the same type of sulphate group in heparin is 12.5 Å from one side of the ribbon to the opposite and 17.5 Å on the same side of the ribbon (codes 1bfb, 1bfc, and 1hpn). The shortest distances between different types of sulphate groups on the same side of the ribbon are 6 to 8 Å. In relation to the predicted heparin binding site, the separations of anionic groups in heparin are compatible with those found for the conserved residues R571-K572, R578-R579, and K585 in the vWF-A1 domain.
The binding of the snake venom botrocetin to vWF-A1 occurs at different locations to that of ristocetin.5 Peptide studies have shown that residues 539-553, 569-583, and 629-643 are critical in mediating botrocetin-induced vWF binding to platelets. The first two of these correspond to the heparin binding region, whereas the third is close to the ristocetin binding residues on the opposite face of the vWF-A1 structure (Fig 3). As botrocetin is a large protein, its size may either permit it to mask a large surface of the vWF-A1 domain or to induce conformational changes that are transmitted through the vWF-A1 domain to affect both sides of the domain.
Mapping of type 2B and 2M vWD mutation sites in the vWF-A1 domain.
The mapping of eight type 2B vWD mutation sites in the vWF-A1 domain (Table 1) showed that these are located in one spatial region of the structure (Figs 3 and 6). This is at the amino-edge of the β-sheet, and all are located below the plane of this β-sheet. Four mutations (R543W, R545C, W550C, and P574L) correspond to replacements of surface-exposed loop residues that are moderately conserved in vWF from 28 species. Two mutations, V551L/F and V553M, correspond to the replacement of two fully conserved smaller hydrophobic residues by larger ones in the buried β-strand BB (Fig2a). Both these are presumed to disrupt the packing of the local protein structure. The insertion of a third methionine at 540insM (where M540 and M451 are both buried) will alter the conformation and packing of α-helix A1. One mutation involves the replacement of the predicted heparin-binding residue R578Q with a neutral residue. The location of these mutations in the vWF-A1 model corresponds well to a region that is situated between the heparin site at α-helices A2 and A3 and the GpIb binding site between residues 514-542 that encompasses β-strand BA and α-helix A1. As the effect of heparin binding is to inhibit vWF,41 it is concluded that the type 2B mutations (while not necessarily preventing heparin binding) will perturb the local protein structure involved in heparin binding and the transmission of an allosteric signal from the heparin site to the GpIb site. This would explain why these modifications upregulate vWF binding to GpIb.
The mapping of the type 2M mutation sites showed that these are distributed on the carboxyl-edge and amino-edge of the β-sheet, and all are above the plane of the β-sheet (Fig 3). This occurred in a different region of vWF-A1 from that corresponding to the type 2B mutations. The three single-site 2M mutations (G561S, R611H/C, and K645del) involve high solvent accessibilities and are located on loops. As two of these three residues are fully conserved in all species, whereas the third K645del corresponds to a deletion of one of four lysines of which K642 and K643 are fully conserved, this implies that loop structures have been perturbed. Because type 2M vWD corresponds to a downregulation of vWF binding to GpIb, it is possible that the GpIb binding site is directly affected. This could be confirmed in the case of R611, because R611 is close to D514 at the N-terminus of the vWF-A1 domain and D514 has been identified to be important for the interaction of vWF with platelets via the GpIb receptor.44 Both R611 and D514 are fully conserved in all 28 vWF-A1 sequences (Fig 2), and D514 is unusual in that it is buried (Fig 1). Molecular graphic views show that these form a well-defined salt bridge in the crystal structure of CR3 (Fig 3). The multiple-residue 2M Milwaukee deletion R629-Q639 is at the loop between BD and A5 (Fig 2). The construction of a vWF-A1 model with this deletion was stereochemically feasible, and no loss of secondary structure was found apart from a shortening of the α-helix A5 by four residues. The effect of this mutation is nonetheless sizeable, and would be expected to alter the protein structure at its GpIb binding site between residues 514 and 542 (Fig3). Although there is no simple explanation for the effect of the remaining mutations at G561 and K645 at present, it is conceivable that they, too, can influence GpIb binding.
Structure and function of the vWF-A2 domain.
The main structural difference between the vWF-A2 and -A1 models is that the former has a more even distribution of positive and negative charges on its surface than the predominantly positively charged surface of the latter (Fig 5). The vWF-A2 domain is the only one of the three in vWF that contains two putative glycosylation sites at N752 and N811. Figure 2 shows that these are fully conserved in all 28 vWF-A2 sequences, suggesting that this may be important.
A metalloprotease that is involved in the physiological cleavage of large vWF multimers acts on the vWF-A2 domain.6,7 In the presence of divalent cations with Ca2+ as the activator,45 it cleaves the peptide bond between Y842 and M843. Both Y842 and M843 are fully conserved in the 28 vWF-A2 sequences. In addition, M843 corresponds precisely in position to the absent second conserved Asp residue in CR3 and LFA-1 that binds Mg2+ in most members of the vWF-A superfamily and, therefore, is located centrally in the active site of the doubly-wound α/β fold.39 The vWF-A2 model also shows that the two conserved glycosylation sites N752 and N811 are close to the active site but on the opposite side of this crevice from Y842 and M843 (Fig4). It is possible that the glycosylation of these sites may define access to this region to direct protease activity specifically towards Y842 and M843.
Whereas not all type 2A mutations are localized to the vWF-A2 domain, those within this domain (Table 1) are found in diverse locations (Figs3 and 6). All 13 mutations correspond to fully conserved residues in vWF from 28 species, except that the L817P mutation corresponds to L817, which is conserved in 27 of the 28 species (Fig 2b). Interestingly, the sidechain solvent accessibilities of the 13 mutation sites (Fig 1) showed that nine correspond to buried locations, unlike the predominantly surface-exposed mutation sites in types 2B and 2M vWD. These are G742E/R, S743L, F751C, L777P, L817P, R834W/Q, V844D, I865T, and E875K. Three more surface-exposed 2A mutations (L799P, S850P, and P885S) involved mutations to or from Pro residues, which are sterically constrained in their sidechain conformations, and one (G846R) involved a mutation from a small residue to a larger one.
There are two groups of type 2A mutations.11 20 The group I mutations are characterized by abnormal intracellular transport resulting in a decrease in vWF secretion and a selective loss of the larger vWF multimers, and this may be consistent with defective protein folding. The group I mutation sites known to date (G742E/R, S743L, L777P, L817P, and V844D) are not spatially located to one region in vWF-A2 (Figs 2 and 3). The group II mutation sites (G742E/R, L799P, I865T, and R834W/Q) are characterized by the secretion of normal vWF multimers but are sensitive to protease activity. These sites are also not located spatially in vWF-A2. One site at F751C is adjacent to the conserved glycosylation site at N752. Because all these mutations correspond to conserved buried or sterically constrained residues, this implies that type 2A vWD is associated with alterations in protein folding, which, although minor in scope, are sufficient to disable its secretion or increase its susceptibility to proteolysis.
Structural properties of the vWF-A3 domain.
Although the protein model of the vWF-A3 domain is similar to those of the -A1 and -A2 domains, its electrostatic map shows that there is a large negatively charged area on its front surface that is not seen in the -A1 and -A2 domains, and this corresponds to the crevice of the doubly-wound α/β fold. No large positively charged areas were found that correspond to those seen in the vWF-A1 domain. The negatively charged surface of the A3 domain resembles one calculated for the corresponding area of CR3 (not shown) where Mg2+ is coordinated. The DxSxS motif of half the metal binding site of CR3 at positions 21 to 25 in Fig 1 is conserved in the vWF-A3 domain, unlike the vWF-A1 and -A2 domains. Although two Asp residues are found at positions 94 and 128 in the vWF-A3 domain that correspond to the other half of the CR3 metal-binding motif at residues 94, 127 ,and 129, the latter three metal binding residues are not conserved in vWF-A3. It is unlikely that the active site of this domain interacts with cations.
The vWF-A3 domain is critical for the binding of vWF to collagen,9,10 and residues 948 to 998 have been implicated in this.2 These residues are mapped to the N-terminal region of the domain including the α-helices A1 to A3 below the central β-sheet. The molecular mechanism of the interaction with collagen is unknown. The binding of some collagen groups to platelets is often mediated and dependent via divalent cations, in which the binding of collagen types I, III, and IV to platelet receptors at high shear stress rates is particularly sensitive to variations in Mg2+ concentration.46 Possibly the binding of vWF-A3 to collagen acts by a similar mechanism.
DISCUSSION
Past experience with homology modeling47,48 shows that, although the accuracy of the models will be better in the protein core where it is aligned with the 13 α-helices and β-strands and will be reduced in the rebuilt loop regions (Fig 1), the use of a good alignment from multiple sequences25 together with known homologous crystal structures28-31 is sufficiently accurate to assess vWF function.
Functional sites in the three vWF-A domains.
Residues at the switch point between the two halves of the β-sheet in the vWF-A domains have functional significance because these occur in a crevice. The conserved DxSxS motif and the Thr, Asp, and Glu residues in the metal binding site of CR3 is conserved in many vWF type-A domains. Despite that, this is only partially conserved in vWF-A3, and is replaced in vWF-A1 by DxSxR and in vWF-A2 by ExSxK. Metal binding is not predicted for the vWF-A domains of vWF. Despite these sequence differences, the present modeling has shown that this active site region in vWF-A is involved in ristocetin binding in vWF-A1 and contains the physiologically relevant protease cleavage site in vWF-A2. In distinction to these active sites, the predicted heparin binding site R571-K572-R578-R579-K585 occurs on the reverse side of the vWF-A1 structure to that of the active site. An analogous location was identified for the heparin binding site in antithrombin III, which is also distant from the physiological active site in this protein.42
Mutation sites on the vWF-A1 domain.
Whereas earlier analyses of the type 2B mutation sites on the vWF-A1 domain had indicated some degree of spatial clustering,26,30 the present modeling now showed that all the currently known type 2B mutations are grouped at the amino edge of the β-sheet in the vWF-A1 domain below the β-sheet and close to a predicted heparin binding site. In distinction, the type 2M mutations correspond to a spatially different set of residues that are located on the carboxyl-edge of the β-sheet above the β-sheet (Fig 6). It is likely that the separate location of these mutation types in vWF-A1 are related to the upregulation (type 2B) or downregulation (type 2M) of GpIb binding by vWF. It is known that heparin binding inhibits the vWF-GpIb interaction.49 The simplest molecular explanation of these mutations is that type 2B vWD involves the decoupling of the inhibitory effects of heparin binding on the vWF-GpIb interaction, leading to upregulation, whereas type 2M involves the disabling of GpIb binding to vWF, leading to downregulation. Both these explanations require the involvement of two vWF-A1 conformations that are transmitted through its structure. This is consistent with a recent proposal that the vWF-A1 domain exists in either the “on” or “off” states,15 and that the type 2B mutations switch it to “on” to facilitate GpIb binding. The ristocetin-binding site would be left exposed to facilitate vWF binding to GpIb. Evidence that other members of the vWF-A superfamily can transmit such signals has been observed in LFA-1. Mutations at residues IKGN at the N-terminus of LFA-1 (Fig 1), which is at the opposite end of the vWF-A structure to the metal-binding site (Fig 3), disrupt the interactions of LFA-1 with its ligand intracellular adhesion molecule-3 (ICAM-3) without affecting the interactions of LFA-1 with ICAM-1.31,50 Structural studies on the LFA-1 vWF-A domain show that there are significant alterations in the metal binding site in the bound and free states.28 In distinction to this, but consistent with this, the structure of the CR3 vWF-A domain has been seen to switch between two conformations depending on whether Mg2+ or Mn2+ is bound.28 29
The type 2M mutations are characterized by decreased vWF-A1 binding to GpIb, as detected by reduced vWF binding to platelets in the presence of ristocetin, as well as from binding studies of recombinant wild-type and mutated vWF-A1 to GpIb. This is often accompanied by a decreased vWF binding to platelets in the presence of botrocetin. These observations imply that the molecular effect of the type 2M mutations would involve the direct perturbation of the ristocetin or GpIb binding sites. Evidence for this was found from the analysis of the type 2M mutation involving the loss of R611, which in turn causes the salt bridge with D514 in the GpIb binding site to be lost. Likewise the large deletion R629-Q639 would be expected to perturb GpIb binding directly.
Mutation sites on the vWF-A2 domain.
Defective mutations in the vWF-A2 domain do not involve direct functional interactions with ligands. Unlike the type 2B and 2M mutations, all the type 2A mutations were found to involve sterically-significant residues that were fully conserved at buried sites in vWF sequences from 28 species. This implies that the mutations may correspond to a modified folding of the vWF-A2 protein structure, which would affect its physiological function directly or indirectly through a signal transmission mechanism. Several of the type 2A mutations are close to the protease cleavage site at Y842-M843 or to the nearby glycosylation site at N752. Evidence that makes this hypothesis likely is found from the nature of groups I and II of type 2A vWD, which are implicated in the secretion of vWF and its stability to proteolytic attack. An altered folding of the vWF-A2 domain may hinder vWF secretion or alter the likelihood of physiological cleavage.
CONCLUSION
In this study, the comparative homology modeling of the vWF-A1, -A2, and -A3 domains has been successful in correlating known biochemical properties of these three domains with the basic molecular features of the α/β fold in these structures. Although the models will not substitute for the atomic detail available in crystal structures for each domain, recent reports of forthcoming crystal structures for the vWF-A1 and -A3 domains support the use of homology modeling for these structures.51,52 In the present study, the active site crevices in the vWF-A1 and -A2 domains were shown to contain functionally important residues, and a separate heparin binding site in the vWF-A1 domain was predicted. The models have also enabled a molecular mechanism for the vWD type 2B, 2M, and 2A mutation sites to be assessed. This explanation of the molecular defects arising from these mutations will facilitate the rational planning of experiments to explore the molecular pathology of vWD. In addition, the sequence conservation analysis of Fig 2 together with the views in Figs 3 and 6will provide a rational basis for the assessment of further mutation sites that may be discovered. This is exemplified by our recent characterization of the K645del mutation and its identification as a type 2M defect.18 53
ACKNOWLEDGMENT
P.V.J. thanks members of the Protein Structure Unit in the Department of Biochemistry and Molecular Biology for their assistance and helpful advice. S.J.P. thanks the Wellcome Trust for financial support.
Address reprint requests to Stephen J. Perkins, DPhil, Department of Biochemistry and Molecular Biology, Royal Free Hospital School of Medicine, Rowland Hill St, London NW3 2PF, UK.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal