Abstract
Although Hodgkin's disease is highly responsive to treatments that cause apoptosis, it remains resistant to the physiological mechanisms intended to cause cell death. Presumably, the Reed-Sternberg cell defies endogenous apoptosis, persists, accumulates, and manifests the malignant disorder seen clinically. The Reed-Sternberg cell expresses several members of the tumor necrosis factor receptor superfamily. This family of receptors is involved in both activation and proliferation of cells, as well as either protection from or initiation of apoptosis in cells expressing these surface proteins. Signals from these receptors affect transcription. We reasoned that the activation state and resistance to apoptosis of Reed-Sternberg cells might be attributable to dysregulation of genes controling these processes. To determine gene expression by Reed-Sternberg cells, we developed a method of micromanipulation, global reverse transcription, and the reverse transcription-polymerase chain reaction and applied it to 51 single Reed-Sternberg cells and their variants from six cases of Hodgkin's disease. This report analyzes the gene expression of bcl-xs,bcl-xl, bax-α,bax-β, fadd, fas, fas ligand (fas L), ice,TNF-α, TNF-β,TNFR1, TNFR2, TRAF1,TRAF2, TRAF3, cIAP2, and tradd at the level of mRNA in the single Reed-Sternberg cells and their variants. The findings here suggest a molecular mechanism for the activated state and in vivo survival occurring in untreated Reed-Sternberg cells of Hodgkin's disease.
THE ORIGIN of the Reed-Sternberg cell and the biological mechanisms of its growth, regulation, and death remain largely unstudied. This is a consequence of the rarity of the Reed-Sternberg (RS) cell in involved tissues and the difficulty in establishing long-term cultures of primary cells. Despite this, Hodgkin's disease (HD) is often treatable by chemotherapy and radiation therapy.1 The effectiveness of therapy is presumed to be mediated by induction of apoptosis, a form of programmed cell death.2-4 Even histologically, a low level of spontaneous apoptosis in RS cells is suggested by certain karyorrhectic “mummified” cells with condensed nuclei seen in tissue sections.5 Their sensitivity to therapy and endogenous cell death argues that neoplastic cells of HD retain the capacity to undergo apoptosis. The physiological apoptosis, however, is not sufficient to keep the disease in check, as HD spreads inexorably when left untreated. Still other cases resist therapy. Here, we examined cellular pathways in primary RS cells to better understand the molecular mechanisms that cause the RS cell to survive.
Patterns of cell growth, survival, and programmed death (apoptosis) are central to the development and homeostasis of the organism; inappropriate downregulation of apoptosis appears to be a common theme in neoplasia, including non-Hodgkin's lymphoma (NHL). One family of apoptosis-regulating proteins, which includes Bcl-2, Bcl-xl, Bcl-xs, Bax, Bak, and Bad, is dysregulated in NHL.6-8 Bcl-2 and Bcl-xl provide protection from cell death, whereas Bcl-xs, Bax, Bak, and Bad promote death of the cell by apoptosis. Ratios of death to anti-death proteins within the cell are critical, as anti-death proteins probably function by binding to and inactivating apoptosis proteins. In HD, immunohistochemical studies have focused on Bcl-2, Bcl-x, and Bax, showing that all three proteins are expressed by more than 50% of the RS cells, thereby suggesting a mechanism for both in vivo survival and responsiveness to therapy.9 10
The tumor necrosis factor receptor (TNFR) superfamily of cell-surface molecules elicits a panel of responses from proliferation to apoptosic cell death in cells of the immune system and are involved in regulating the BCL family proteins. The RS cell expresses many of these proteins including TNFRs 1 and 2, CD30, CD40, and Fas (CD95).11,12This group of receptors may be divided functionally into those that induce apoptosis when bound by their ligands and those that promote survival and proliferation. TNFR1 and Fas signal through a cytoplasmic “death domain” bound by the intracellular molecules TRADD (TNFR-associated death domain) and FADD (Fas-associated death domain), respectively.13,14 This pathway may involve the death-promoting Bax protein. In the case of TNFR2, CD30, and CD40, a TRAF (TNFR-associated factor)-binding domain binds one or more of four TRAFs.15-19 Interestingly, the LMP 1 protein of Ebstein-Barr virus (EBV), whose products are present in the RS cells in more than 70% of HD cases,20 also contains a TRAF domain and uses the TRAFs in some aspects of its signaling.17 CD40 and LMP 1 upregulate the antideath proteins Bcl-xL and Bcl-2, respectively, and may thus promote survival.21 22
Cell-surface signaling of apoptosis promoted by engagement of Fas with its ligand (FasL) may also involve another family of proteins structurally similar to interleukin-1β converting enzyme (ICE), a serine protease homologous to the CED-3 gene in the nematodeCaenorhabditis elegans.23-26 One such enzyme, FLICE,27 is recruited to the death-inducing signal complex upon Fas stimulation. Thus, the substrates for the ICE-like proteases likely mediate Fas-induced apoptosis.28 As yet, these proteins have not been studied in HD.
Loss of function of either Fas or FasL leads to unregulated proliferation of lymphocytes, as evidenced by lpr mice, which lack Fas as a result of a germline mutation of the gene, and gld mice, which lack FasL.29-33 Similarly, a germline defect in Fas in humans can cause an autoimmune lymphoproliferative syndrome (ALS).34 HD is thought to be a lymphoproliferation with a profound immune response and might be a manifestation of an ALS. However, somatic mutations of these genes have not been examined in HD.
Both TNFR1 and TNFR2 and their ligands, the TNFs, are expressed by RS cells.11 Survival and activation promoted by TNFRs may be mediated through the TRAFs. TRAFs 1, 2, and 5 can activate the transcription factor nuclear factor κB (NFκB), an event that is important in both cytokine secretion and resistance to apoptosis.15,19 The mammalian homolog of inhibitor of apoptosis (IAP), a baculoviral protein necessary for preventing apoptosis in an infected insect cell, forms a complex with TRAF2 and may function in the survival-promoting aspect of receptors signaling by TRAF2.35 If TNFR-TRAF signaling is active in RS cells, the cIAP/TRAF relationship may thus be important to their survival and are the subject of this analysis.
The repertoire of expressed genes determines the balance between cell growth, activation state, and apoptotic fate. RS cells may be apoptotic and express members of the TNFR superfamily, but for most of the cell death and TNFR-associated proteins no antibody is suitable for immunohistochemical detection. Here, we specifically examine gene expression at the level of transcription in single RS cells from primary tissues. RS cells appear to have a defective germinal center B-cell phenotype based on their rearranged but nonproductively hypermutated IgH genes36 and a very limited growth fraction, as suggested by DNA content analysis.37 Their persistence in tissue must be partly attributable to regulated cell death. We have begun to test the hypothesis that RS cells and their variants may persist in the untreated state as a result of dysregulated expression of apoptosis-controlling genes. We have used a series of single RS cell 3′ cDNA libraries that we had previously prepared by a unique approach using micromanipulation, reverse transcription (RT), and the polymerase chain reaction (PCR).38 With this technique, essentially unlimited amounts of cDNA were prepared from the mRNA of each of 51 RS cells and their variants obtained from primary cases of HD. cDNAs of single RS cells and their variants were analyzed by PCR for coordinate expression of activation and apoptosis-regulating genes.
MATERIALS AND METHODS
Cells and Tissues
B cells.
Fresh tissue was obtained from tonsillectomy specimens, minced in RPMI medium with penicillin (100 U/mL) and streptomycin (100 μg/mL) and passed through a sterile wire mesh to create a single cell suspension. Mononuclear cells were partially purified by centrifugation over Ficoll. B cells were purified from this population by rosette depletion of T cells with sheep red blood cells as described.39 The B-cell population was 76% pure, as confirmed by the presence of surface antigens CD19 and CD20 and the absence of markers CD2 and CD7 as evaluated by flow cytometry.
T cells.
Leukopheresis-enriched peripheral blood was obtained from the American Red Cross (Baltimore, MD). Mononuclear cells were partially purified by centrifugation over Ficoll. 2 × 109 cells were resuspended in 10 mL sterile RPMI medium and passed over a nylon wool column. After 1-hour incubation at 37°C, nonadherent cells were eluted with 75 mL RPMI at 37°C. The resulting T-cell population was 82% pure, as confirmed by the presence of surface antigens CD2 and CD7 and the absence of CD19 and CD20 as evaluated by flow cytometry.
Cell lines.
KMH2 was kindly provided by Dr Hiroshi Kamesaki (Nagoya, Japan), and L428 was provided by Dr V. Diehl (Cologne, Germany). Jurkat and Raji cell lines were obtained from American Type Culture Collection (ATCC, Rockville, MD). All lines were maintained in RPMI 1640 containing 10% fetal calf serum (FCS) (GIBCO-BRL, Grand Island, NY) and antibiotics in a humidified atmosphere with 5% CO2.
mRNA and cDNA
Messenger RNA was extracted from B, T, Jurkat, and Raji cells by oligo-dT purification (Fast-Track mRNA Isolation kit, San Diego, CA). Total RNA was extracted from KMH2 and L428 cell lines using Trizol (Life Technologies, Gaithersburg, MD) according to the manufacturer's directions. First-strand cDNA was synthesized with random hexamer primers and Superscript RT enzyme (Life Technologies).40Genomic DNA was obtained from human placenta.
Single-Cell RT-PCR
Representative cDNA libraries were the same as described by Trumper et al38 and generated from 51 single RS cells and their variants. For the purposes of this study, 25 μg of each sample was reamplified in a PCR reaction in order to generate sufficient amounts of each cDNA library, for the analysis of multiple genes. The PCR mixture contained 1.0 pmol/μL of the X-only clamp primer,38 1 mmol/L dNTPs, 20 mmol/L Tris-HCl (pH 8.4), 50 mmol/L KCl, 1.5 mmol/L MgCl2, and 2.5 U Taq DNA polymerase in a final volume of 100 μL. Thermal cycling was carried out as follows: one cycle of 94°C for 3 minutes, 65°C for 1 minute, 72°C for 1 minute, followed by 30 cycles of 94°C for 30 seconds, 65°C for 45 seconds, 72°C for 30 seconds, and an additional extension period at 72°C for 5 minutes.
When screening for specific sequences by PCR, 1-μL aliquots of the completed PCR reaction were used in subsequent reactions with appropriate primers. The cDNA was constructed so that their average length was 600 bp, maintaining proportionality of mRNA concentrations; therefore, primers were chosen that would selectively amplify a product within this region at the 3′ end of the target gene. As a positive control for representative gene expression, each expanded cDNA library was tested for β-actin by PCR.
For this study, it was essential to ensure that the single-cell cDNA libraries did not contain genomic DNA. Therefore, each of the cDNA libraries from the previous study38 was tested for genomic DNA by PCR. β2-Microglobulin primers, the upper of which lies within intron 3,41 detected genomic DNA in nine cases, which were subsequently excluded from this study. The reamplified cDNA libraries were then aliquoted and stored in ethanol at −20°C.
Each experimental primer was used at a concentration of 0.5 pmol/μL in a final volume of 50 μL. Both temperature and Mg2+concentration were optimized for each pair of primers using comparable amounts of appropriate positive control cDNA. Positive control cDNAs for each primer pair were as follows: activated T cells forbcl-x; resting B cells for bax; EBV-positive lymphoblastoid cells for traf1, Jurkat for bak,fadd, Fas, FasL, tnfr1, tnfr2,tnf-α, and tradd; KMH2 fortraf2; and Raji for ice and traf3. Other PCR conditions were carried out as in the reamplification reaction described above. A total of 23 μL of each reaction was electrophoresed through a 1.5% agarose gel and visualized with ethidium bromide.
Primers
The following primer pairs were used. For each set, the upper strand (5′) primer is listed first:
actin: 5′-GAA CGG TGA AGG TGA CAG CAG T-3′,5′-TGG GGG ACA AAA AGG GGG AAG G-3′42
bax-α: 5′-TCA GGA TGC GTC CAC CAA GAA GC-3′,5′-TGA GCA CTC CCG CCA CAA AGA TG-3′43
bak-β: 5′-GGT TGT CGC CCT TTT CTA CTT TG-3′,5′-GTG GTG GGG GTG AGG AGG CTT GA-3′43
bak: 5′-TGC ATT TGG CTG AAT CAA GAA CTT-3′,5′-AGT GGG AGA AGG ACT ATC AAC ACC-3′7
bcl-xl: 5′-GGA AAG CGT AGA CAA GGA GAT GC-3′,5′-GGT GGG AGG GTA GAG TGG ATG GT-3′44
bcl-xs: 5′-GGG AGG CAG GCG ACG AGT TTG AA-3′,5′-GGT GGG AGG GTA GAG TGG ATG GT-3′44
fadd: 5′-CTG CCT TGG CAA TTC TGT TAT CAG-3′,5′-TGG CTG GGG TGG GGG TGG GGA GAC-3′14
Fas: 5′-CAC CCC CGA AAA TGT TCA ATA AT-3′,5′-GTT CCA GGT ATC TGT TTC AGT TT-3′45
Fas-L: 5′-GGC AGC ATC TTC ACT TCT AAA TG-3′,5′-CAA AAA TGC ACA TCT ACA TAA AT-3′46
iap: 5′-ATG AAC GAA AAA GAG GTA GCA-3′,5′-GAG AAA TGT GTC CCT GGT GT-3′35
ice: 5′-AAC ATG TTG AAT ACC AAG AAC TGC-3′,5′-AGA GCA GAA AGC GAT AAA ATC CTT-3′23
tnf-α: 5′-CCA GCT CCC TCT ATT TAT GTT TG-3′,5′-TAT TGT TCA GCC TCC GTT TTC AC-3′47
tnf-β: 5′-TTC TCC CCA TTC TGC CTC CAT TC-3′,5′-TGC CCT CTC ACT CTT CAT CTC TT-3′47
tnfr1: 5′-CAA GAG CCT GAG TGG GTG GTT TG-3′,5′-CTG CTT ATG CAC TGT GAA AAA GG-3′48
tnfr2: 5′-AGG CTT GGA AAG CAT CAC CTC A-3′,5′-CCG GCT AAT TTC TGT ATT TTT A-3′49
tradd: 5′-GGT TCC TTC TGC GGC TAT TGC TGA-3′, TGA AAC TGT AAG GGC TGG CTG TAA-3′13
traf1: 5′-GTA ACC CCC ACT GGA CTG ACC TG-3′,5′-TTT ATG CCC CTC TTC TTC TTG AG-3′17
traf2: 5′-CTA GGA ATG CTC CCT TCT CTC CAG-3′,5′-ACC AGC CAG TCC TCA GAT TTC AGA-3′50
traf3: 5′-AAC CGG TCT GTC TTC ACT GAG GTC-3′,5′-AAT CAT GTT TCT TTT GTT TAG CAT-3′17
Immunohistochemistry
Tissue sections were deparaffinized and stained with a monoclonal anti-Bcl-2 antibody (DAKO, Carpinteria, CA), anti-Bax monoclonal antibody (Pharmingen, San Diego, CA) or anti-Bax polyclonal antibody (kindly provided by Dr John C. Reed, La Jolla, CA) using an indirect avidin–biotin–peroxidase complex (ABC).51Positive controls were performed with a single block containing multiple tissues, including placenta, lymph node, spleen, large cell lymphoma, and liver.
RESULTS
Optimization of PCR Conditions
PCR conditions for each set of primers were optimized using an appropriate positive control cDNA known to express the gene of interest (see Materials and Methods). In addition, the positive control cDNA was included in each set of reactions with the single-cell cDNAs to verify reaction conditions. In each round of PCR, the positive control cDNA was consistently positive, and the negative (no template) controls were negative (data not shown), confirming the efficacy and validity of each round of the PCR.
Validation and Quantification of the Expanded Single-Cell cDNA Libraries
To test the relative quality of the reamplified cDNA libraries, each expanded set of single cell cDNAs was analyzed for the expression of a ubiquitously expressed gene and for the absence of genomic DNA. First, each was screened for β-actin using primers that lie within the last exon of the gene. All the cDNA libraries generated for this study contained β-actin titratable out to 1 : 5,000 dilution of the reamplified sample, indicating that the mRNA of a randomly selected, constitutively expressed gene could be consistently detected in all expanded cDNA libraries (Fig 1). To confirm that the cDNA libraries contained only expressed sequences, they were screened for the presence of genomic DNA. For this purpose, a primer was derived from the untranscribed sequence of intron 3 of the β2-microglobulin gene26 and paired with a primer complementary to a sequence in a downstream exon of this gene. A PCR product with this pair of primers would be derived only from unspliced DNA, and thus would indicate genomic contamination of the library. Forty-two of 51 cDNA libraries tested in this manner gave no PCR product with this pair of primers and were included in the analysis.
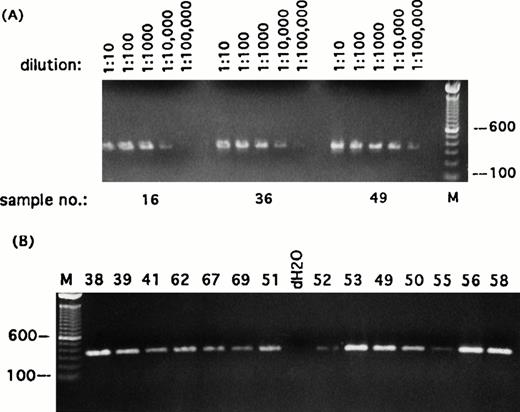
Detection of transcripts for β-actin in single Reed-Sternberg cells and their variants. Representative sampling of PCR results using primers specific for β-actin transcript. (A) Three randomly selected reamplified samples were titrated out to 1 : 100,000 dilution. (B) Each of the 49 reamplified single cell cDNA libraries was then diluted to 1 : 5,000 and screened for expression of β-actin. All single-cell libraries included in this study expressed β-actin titratable to at least 1 : 5,000. PCR products were run on ethidium bromide-stained 1% agarose gels. Positive or negative results were scored on the basis of intensity of ethidium bromide staining. Sample numbers correspond to cell numbers listed in Table 1. Molecular-weight markers (M) are labeled in base pairs.
Detection of transcripts for β-actin in single Reed-Sternberg cells and their variants. Representative sampling of PCR results using primers specific for β-actin transcript. (A) Three randomly selected reamplified samples were titrated out to 1 : 100,000 dilution. (B) Each of the 49 reamplified single cell cDNA libraries was then diluted to 1 : 5,000 and screened for expression of β-actin. All single-cell libraries included in this study expressed β-actin titratable to at least 1 : 5,000. PCR products were run on ethidium bromide-stained 1% agarose gels. Positive or negative results were scored on the basis of intensity of ethidium bromide staining. Sample numbers correspond to cell numbers listed in Table 1. Molecular-weight markers (M) are labeled in base pairs.
RT-PCR Analysis of Apoptosis-Regulating Genes
Although all the selected apoptosis-associated genes were found to be expressed in single RS cells and their variants, their frequency of detection was highly variable. The transcript for bcl-xl was detected in one or more of the RS cells and their variants in four of six cases of HD studied. The transcripts forbax-α or bax-β, however, were found in one and five, respectively, of the 42 RS cells and their variants analyzed. Another member of the bcl-2 family,bak, was found in 13 of 42 RS cells analyzed (Table1).
Apoptosis-Related Gene Expression
| Case . | Cell No. . | bcl-xs . | bcl-xl . | bax-α . | bax-β . | bak . | fas . | fasL . | ice . |
|---|---|---|---|---|---|---|---|---|---|
| 1 (LP) | 14 | − | − | − | − | − | +++ | − | +++ |
| 16 | ++ | + | − | − | − | +++ | − | − | |
| 17 | + | + | − | + | − | − | − | − | |
| 18 | ++ | − | − | − | − | − | − | − | |
| 20 | ++ | + | − | − | +++ | − | − | + | |
| 21 | − | ++ | − | − | − | ++ | − | +++ | |
| 22 | − | − | − | − | + | − | − | − | |
| 23 | − | − | − | − | + | + | − | + | |
| 24 | + | + | − | − | − | − | − | ++ | |
| 25 | ++ | + | − | + | − | − | − | ++ | |
| 2 (NS) | 34 | ++ | − | − | + | − | +++ | − | − |
| 36 | ++ | − | − | − | − | +++ | − | − | |
| 37 | − | − | − | − | − | +++ | − | +++ | |
| 38 | + | + | − | − | +++ | − | − | − | |
| 39 | + | − | − | + | + | − | − | − | |
| 41 | + | − | − | − | +++ | − | − | − | |
| 3 (NS) | 51 | − | − | − | − | − | +++ | − | − |
| 52 | − | − | − | − | − | − | − | − | |
| 53 | − | − | − | − | − | − | − | − | |
| 49 | + | − | − | − | +++ | +++ | − | ++ | |
| 50 | − | − | − | − | − | +++ | − | − | |
| 55 | − | − | − | − | − | − | − | − | |
| 56 | − | − | − | − | − | +++ | − | +++ | |
| 58 | − | − | − | − | + | ++ | − | − | |
| 5 (NS) | 62 | − | − | − | − | − | +++ | − | − |
| 67 | + | − | − | − | − | − | − | ++ | |
| 69 | − | − | − | − | +++ | +++ | − | − | |
| 6 (NS) | 75 | − | − | − | − | − | − | − | − |
| 77 | − | − | − | − | − | ++ | − | − | |
| 78 | + | + | − | − | − | ++ | + | − | |
| 79 | − | − | − | − | +++ | − | − | − | |
| 80 | − | − | +++ | − | − | ++ | − | − | |
| 7 (MC) | 89 | − | − | − | − | − | ++ | − | − |
| 90 | − | − | − | − | − | − | − | − | |
| 93 | − | − | − | − | − | + | − | − | |
| 91 | + | ++ | − | − | +++ | − | − | − | |
| 94 | − | ++ | − | − | +++ | − | − | +++ | |
| 95 | − | + | − | − | + | − | + | − | |
| 96 | − | + | − | − | − | ++ | − | − | |
| 97 | − | − | − | − | − | +++ | − | − | |
| 98 | − | − | − | − | − | ++ | − | +++ |
| Case . | Cell No. . | bcl-xs . | bcl-xl . | bax-α . | bax-β . | bak . | fas . | fasL . | ice . |
|---|---|---|---|---|---|---|---|---|---|
| 1 (LP) | 14 | − | − | − | − | − | +++ | − | +++ |
| 16 | ++ | + | − | − | − | +++ | − | − | |
| 17 | + | + | − | + | − | − | − | − | |
| 18 | ++ | − | − | − | − | − | − | − | |
| 20 | ++ | + | − | − | +++ | − | − | + | |
| 21 | − | ++ | − | − | − | ++ | − | +++ | |
| 22 | − | − | − | − | + | − | − | − | |
| 23 | − | − | − | − | + | + | − | + | |
| 24 | + | + | − | − | − | − | − | ++ | |
| 25 | ++ | + | − | + | − | − | − | ++ | |
| 2 (NS) | 34 | ++ | − | − | + | − | +++ | − | − |
| 36 | ++ | − | − | − | − | +++ | − | − | |
| 37 | − | − | − | − | − | +++ | − | +++ | |
| 38 | + | + | − | − | +++ | − | − | − | |
| 39 | + | − | − | + | + | − | − | − | |
| 41 | + | − | − | − | +++ | − | − | − | |
| 3 (NS) | 51 | − | − | − | − | − | +++ | − | − |
| 52 | − | − | − | − | − | − | − | − | |
| 53 | − | − | − | − | − | − | − | − | |
| 49 | + | − | − | − | +++ | +++ | − | ++ | |
| 50 | − | − | − | − | − | +++ | − | − | |
| 55 | − | − | − | − | − | − | − | − | |
| 56 | − | − | − | − | − | +++ | − | +++ | |
| 58 | − | − | − | − | + | ++ | − | − | |
| 5 (NS) | 62 | − | − | − | − | − | +++ | − | − |
| 67 | + | − | − | − | − | − | − | ++ | |
| 69 | − | − | − | − | +++ | +++ | − | − | |
| 6 (NS) | 75 | − | − | − | − | − | − | − | − |
| 77 | − | − | − | − | − | ++ | − | − | |
| 78 | + | + | − | − | − | ++ | + | − | |
| 79 | − | − | − | − | +++ | − | − | − | |
| 80 | − | − | +++ | − | − | ++ | − | − | |
| 7 (MC) | 89 | − | − | − | − | − | ++ | − | − |
| 90 | − | − | − | − | − | − | − | − | |
| 93 | − | − | − | − | − | + | − | − | |
| 91 | + | ++ | − | − | +++ | − | − | − | |
| 94 | − | ++ | − | − | +++ | − | − | +++ | |
| 95 | − | + | − | − | + | − | + | − | |
| 96 | − | + | − | − | − | ++ | − | − | |
| 97 | − | − | − | − | − | +++ | − | − | |
| 98 | − | − | − | − | − | ++ | − | +++ |
Results of single-cell RT-PCR. −, no band; +, weak; ++, intermediate; +++, strong.
Abbreviations: LP, lymphocyte predominant type HD; NS, nodular sclerosing; MC, mixed cellularity.
RT-PCR Analysis of TNFR Signaling Proteins
Ligands.
TNF-α and TNF-β were expressed by 17 and 12 RS cells, respectively. Two of the cases expressed exclusively TNF-α (Table2).
TNFR-Related Gene Expression
| Case . | Cell No. . | TNF-β . | TNF-α . | TNFR1 . | TNFR2 . | TRAF1 . | TRAF2 . | TRAF3 . | cIAP2 . | TRADD . | FADD . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (LP) | 14 | − | − | − | ++ | ++ | + | − | +++ | +++ | + |
| 16 | − | − | ++ | − | − | ++ | ++ | +++ | − | + | |
| 17 | ++ | ++ | − | − | − | +++ | + | − | − | + | |
| 18 | − | − | − | − | − | ++ | − | − | − | − | |
| 20 | + | ++ | ++ | − | − | + | + | ++ | − | ++ | |
| 21 | + | − | − | − | − | − | − | − | − | ++ | |
| 22 | + | − | − | − | − | − | − | − | − | − | |
| 23 | − | ++ | ++ | − | − | ++ | + | − | +++ | + | |
| 24 | − | − | + | − | − | ++ | + | − | +++ | − | |
| 25 | − | ++ | − | − | − | ++ | + | + | − | + | |
| 2 (NS) | 34 | +++ | − | − | − | + | − | − | +++ | +++ | + |
| 36 | − | + | ++ | − | +++ | − | − | ++ | − | − | |
| 37 | − | − | − | − | ++ | − | +++ | +++ | − | − | |
| 38 | − | ++ | − | − | − | ++ | − | +++ | − | − | |
| 39 | − | ++ | − | − | − | − | − | − | − | − | |
| 41 | + | ++ | − | + | − | − | − | +++ | + | + | |
| 3 (NS) | 51 | − | − | +++ | ++ | − | − | ++ | − | − | − |
| 52 | − | + | − | − | − | − | − | +++ | − | − | |
| 53 | − | ++ | − | − | − | − | − | − | − | − | |
| 49 | − | ++ | − | − | − | − | − | − | − | − | |
| 50 | − | +++ | − | − | − | +++ | − | − | − | − | |
| 55 | − | − | − | − | − | − | − | − | − | − | |
| 56 | − | − | − | − | − | + | + | +++ | +++ | − | |
| 58 | − | − | +++ | − | − | ++ | + | − | − | + | |
| 5 (NS) | 62 | − | +++ | − | − | + | − | +++ | +++ | − | − |
| 67 | − | − | − | ++ | − | − | − | +++ | − | − | |
| 69 | +++ | − | − | − | +++ | + | +++ | +++ | +++ | − | |
| 6 (NS) | 75 | − | − | ++ | + | + | − | − | − | − | − |
| 77 | − | − | +++ | − | + | − | − | +++ | − | − | |
| 78 | − | ++ | − | − | + | + | − | − | − | − | |
| 79 | − | − | − | − | +++ | − | − | +++ | − | − | |
| 80 | − | ++ | − | − | − | − | +++ | − | +++ | − | |
| 7 (MC) | 89 | + | − | − | − | − | − | − | − | − | + |
| 90 | − | − | − | − | − | + | + | +++ | − | ++ | |
| 93 | − | − | − | − | − | − | − | ++ | − | − | |
| 91 | + | − | − | − | +++ | + | − | +++ | − | − | |
| 94 | + | ++ | − | − | +++ | − | − | +++ | − | − | |
| 95 | + | − | − | − | − | − | − | +++ | − | − | |
| 96 | − | − | − | − | +++ | + | ++ | +++ | ++ | − | |
| 97 | − | ++ | +++ | − | − | − | − | +++ | − | − | |
| 98 | +++ | − | − | − | +++ | − | − | +++ | + | − |
| Case . | Cell No. . | TNF-β . | TNF-α . | TNFR1 . | TNFR2 . | TRAF1 . | TRAF2 . | TRAF3 . | cIAP2 . | TRADD . | FADD . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (LP) | 14 | − | − | − | ++ | ++ | + | − | +++ | +++ | + |
| 16 | − | − | ++ | − | − | ++ | ++ | +++ | − | + | |
| 17 | ++ | ++ | − | − | − | +++ | + | − | − | + | |
| 18 | − | − | − | − | − | ++ | − | − | − | − | |
| 20 | + | ++ | ++ | − | − | + | + | ++ | − | ++ | |
| 21 | + | − | − | − | − | − | − | − | − | ++ | |
| 22 | + | − | − | − | − | − | − | − | − | − | |
| 23 | − | ++ | ++ | − | − | ++ | + | − | +++ | + | |
| 24 | − | − | + | − | − | ++ | + | − | +++ | − | |
| 25 | − | ++ | − | − | − | ++ | + | + | − | + | |
| 2 (NS) | 34 | +++ | − | − | − | + | − | − | +++ | +++ | + |
| 36 | − | + | ++ | − | +++ | − | − | ++ | − | − | |
| 37 | − | − | − | − | ++ | − | +++ | +++ | − | − | |
| 38 | − | ++ | − | − | − | ++ | − | +++ | − | − | |
| 39 | − | ++ | − | − | − | − | − | − | − | − | |
| 41 | + | ++ | − | + | − | − | − | +++ | + | + | |
| 3 (NS) | 51 | − | − | +++ | ++ | − | − | ++ | − | − | − |
| 52 | − | + | − | − | − | − | − | +++ | − | − | |
| 53 | − | ++ | − | − | − | − | − | − | − | − | |
| 49 | − | ++ | − | − | − | − | − | − | − | − | |
| 50 | − | +++ | − | − | − | +++ | − | − | − | − | |
| 55 | − | − | − | − | − | − | − | − | − | − | |
| 56 | − | − | − | − | − | + | + | +++ | +++ | − | |
| 58 | − | − | +++ | − | − | ++ | + | − | − | + | |
| 5 (NS) | 62 | − | +++ | − | − | + | − | +++ | +++ | − | − |
| 67 | − | − | − | ++ | − | − | − | +++ | − | − | |
| 69 | +++ | − | − | − | +++ | + | +++ | +++ | +++ | − | |
| 6 (NS) | 75 | − | − | ++ | + | + | − | − | − | − | − |
| 77 | − | − | +++ | − | + | − | − | +++ | − | − | |
| 78 | − | ++ | − | − | + | + | − | − | − | − | |
| 79 | − | − | − | − | +++ | − | − | +++ | − | − | |
| 80 | − | ++ | − | − | − | − | +++ | − | +++ | − | |
| 7 (MC) | 89 | + | − | − | − | − | − | − | − | − | + |
| 90 | − | − | − | − | − | + | + | +++ | − | ++ | |
| 93 | − | − | − | − | − | − | − | ++ | − | − | |
| 91 | + | − | − | − | +++ | + | − | +++ | − | − | |
| 94 | + | ++ | − | − | +++ | − | − | +++ | − | − | |
| 95 | + | − | − | − | − | − | − | +++ | − | − | |
| 96 | − | − | − | − | +++ | + | ++ | +++ | ++ | − | |
| 97 | − | ++ | +++ | − | − | − | − | +++ | − | − | |
| 98 | +++ | − | − | − | +++ | − | − | +++ | + | − |
Results of single cell RT-PCR. −, no band; +, weak; ++, intermediate; +++, strong.
Abbreviations: LP, lymphocyte predominant; NS, nodular sclerosing; MC, mixed cellularity.
Receptors.
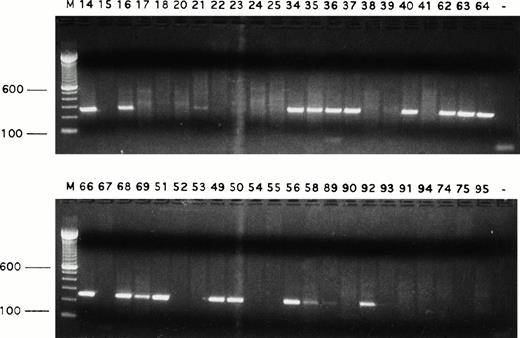
Both TNFR1 and TNFR2 were detected in all cases of HD but at low (10 cells and 5 cells, respectively) frequency. Fas (CD95) gene expression was frequent (24 of 42 RS cells) (Fig 2; Table1), but FasL mRNA was rarely detected.
Detection of Fas transcripts in single Reed-Sternberg cells and their variants. Fas transcript was expressed by many single Reed-Sternberg cells and their variants. PCR products were run on ethidium bromide-stained 1.5% agarose gels. Positive or negative results were scored on the basis of intensity of ethidium bromide staining. Sample numbers correspond to cell numbers listed in Table 1. Molecular-weight markers (M) are labeled in base pairs.
Detection of Fas transcripts in single Reed-Sternberg cells and their variants. Fas transcript was expressed by many single Reed-Sternberg cells and their variants. PCR products were run on ethidium bromide-stained 1.5% agarose gels. Positive or negative results were scored on the basis of intensity of ethidium bromide staining. Sample numbers correspond to cell numbers listed in Table 1. Molecular-weight markers (M) are labeled in base pairs.
Cytoplasmic mediators.
TRAF1, 2, and 3 were detected in all cases of HD examined. TRAF2 and 3 were present evenly throughout the cases, while TRAF1 appeared to be expressed either highly (more than 50% of cells) in four of six or rarely or not at all (two of six cases). cIAP2 transcript was found in 57% of RS cells, more than twice the frequency of TRADD transcript. FADD transcript was detected very frequently (79% of cells) in the case of LP HD but was rare to absent in the NS and MC cases analyzed.ICE transcript was detected in one or more cells in every case of HD analyzed (Table 1).
Immunohistochemical Detection of Bcl-2 and Bax Protein in Selected Cases
Cases 1, 3, 5, and 7 were analyzed by immunohistochemistry for Bcl-2 protein expression. Bcl-2 protein was found to be expressed strongly in cases 3 and 7, moderately in case 5, and weakly in case 1. Case 2, 6, and 7 were analyzed with both a polyclonal antibody and a monoclonal antibody directed against the Bax protein. All cases were negative for Bax, using the monoclonal antibody, whereas the polyclonal antibody reacted with RS cells strongly in case 6, moderately in case 7, and faintly in case 2. Lymophocytes displayed negative staining in all cases with both antibodies (data not shown).
Clinical Data
All patients from whom cells were used in this study achieved complete remission for at least three years (Table3). Patient number 7 relapsed at 3 years and subsequently demised despite treatment.
Clinical Data on Hodgkin's Patients
| Case . | Class . | Stage . | Status at Study . | Survival (yr) . | Notes . |
|---|---|---|---|---|---|
| 1 | LP | IIIA | 1st relapse | 16+ | Presented @ age 31 → MOPP/DXT → CR → relapse |
| 2 | NS | IIIA | Presentation | 2+ | LTF |
| 3 | NS | IIB | Presentation | 3+ | DXT → CR |
| 5 | NS | IIIB | Presentation | 3+ | ABVD → CR |
| 6 | NS | IIA | Presentation | 3+ | DXT → CR |
| 7 | MC | IIA | 2nd relapse | 7 | MOPP → CR → relapse @ 3 yrs → MOPP/ABVD → demised post ABMT |
| Case . | Class . | Stage . | Status at Study . | Survival (yr) . | Notes . |
|---|---|---|---|---|---|
| 1 | LP | IIIA | 1st relapse | 16+ | Presented @ age 31 → MOPP/DXT → CR → relapse |
| 2 | NS | IIIA | Presentation | 2+ | LTF |
| 3 | NS | IIB | Presentation | 3+ | DXT → CR |
| 5 | NS | IIIB | Presentation | 3+ | ABVD → CR |
| 6 | NS | IIA | Presentation | 3+ | DXT → CR |
| 7 | MC | IIA | 2nd relapse | 7 | MOPP → CR → relapse @ 3 yrs → MOPP/ABVD → demised post ABMT |
Clinical outcomes of patients involved in this study.
Abbreviations: MOPP, mechlorethamine-oncovin (vincristine)-procarbazine-prednisone; ABVD, Adriamycin-bleomycin-vinblastine-dacarbazine; DXT, deep X-ray therapy; ABMT, autologous bone marrow transplant; CR, complete remission; LTF, lost to follow-up.
HD Cell Lines
The Hodgin-derived cell lines KMH2 and L428 were tested by RT-PCR for the expression of each gene that was assayed in single, primary RS cells (data not shown). KMH2 was found to express all the genes in this study except FasL. L428 expressed transcripts for all except FasL and TNFR1.
DISCUSSION
The single-cell strategy applied here overcomes the previous obstacles in gene expression analysis of RS cells by examining the mRNA of isolated living cells, uncontaminated by surrounding host cells. Several single cells in each case were evaluated to provide a more accurate picture of gene expression because it is likely that expression of genes, even those constitutively expressed, may vary among individual cells. Because of this, the level of expression may be extrapolated from the number of positive or negative cells in a population. The data from these experiments were evaluated on this basis.
TNFR Family Signaling May Favor Survival
RS cells and their variants are known to express members of the TNFR superfamily. For example, CD30 and CD40 are abundantly expressed by RS cells in most HD cases, as shown by immunohistochemistry.11TNFRs 1 and 2 were shown previously to be infrequently detected by immunohistochemistry, and this was corroborated in this study by RT-PCR. In the present study, RS cells were found to express mRNA of TNF-α and -β, the ligands of these receptors, which can act in an autocrine, paracrine, or endocrine manner in HD. The intracellular pathways initiated by these four TNFRs may be directed toward cell survival. The downstream signals of these receptors, however, have not been previously examined in primary RS cells, and appropriate immunohistochemical reagents are not available. Here we have specifically looked at cytoplasmic mediators of the TNFR family by single-cell cDNA analysis.
The TRAFs associate with cytoplasmic domains of members of the TNFR family of receptors.15-19 In this study, TRAF1 was detected at high levels in some of the cases of HD. The expression of this protein can be upregulated by EBV infection.17 TRAF1 is important in LMP1-mediated NFκB activation, independent of TRAF2.52 TRAF1 also binds CD30, and therefore may signal directly from this receptor, which also leads to NFκB activation.53
All the HD cases studied expressed TRAF2 at moderate levels. This protein mediates signals from both TNFR1 and TNFR2,15,54and its relevance to cell survival and activation may be determined by the presence of other factors known to act downstream of these receptors. TNFR2 binds directly to TRAF2. In most situations, TNFR2 ligation activates NFκB, a transcription factor that induces proliferation and cytokine secretion. TRAF2 is necessary for the activation of NFκB,15 a factor necessary for cell survival in the presence of many different noxious stimuli.55-57 TRAF2 also binds to cIAP, which is thought to promote cell survival on the basis of the actions of its viral homolog vIAP.35 TNFR1, on the other hand, can transduce signals for survival or death via the cytoplasmic molecule TRADD. TRADD may cause the activation of NFκB when bound to TRAF2, or it can initiate cell death if it subsequently binds to FADD.54 In the 6 cases of HD studied, five expressed TRAF2 at frequencies equal to or higher than FADD, indicating that any signal through TRADD is shunted towards survival rather than death. In case 2 where FADD was more frequent, cIAP was detected at a much higher frequency than FADD, perhaps providing protection from death signals.
TRAF3 binds CD40.16,18 Importantly for HD, CD40 is nearly always expressed (at the protein level) by RS cells,11 and in the present study, TRAF3 was detected in the single RS cells of every case of HD studied, but not in the 14 normal single lymphocytes analyzed (data not shown). Although a distinct function has not yet been assigned to TRAF3, it is implicated in downregulation of CD40-mediated NFκB activation, and in lymphotoxin-β-receptor-mediated cell death.58 The role of TRAF3 in RS cells may be to modulate the signals of CD40 so that the cell remains in an activated state but does not progress to activation-induced apoptosis.
Relative Levels of Apoptosis Family Members
The findings presented in this report suggest that Reed-Sternberg cells and their variants suppress apoptosis by the regulation of genes involved in a cell death pathway. For example, Bax promotes cell death in mammalian cells when its level is high relative to that of Bcl-2 or Bcl-xl,43 and there was infrequent expression in single RS cells of both bax- α andbax-β, which are known to induce apoptosis when abundant. Interestingly, the transcript for the anti-death genebcl-xl was detected more frequently in RS cells than those for the death promoting genes bax-α orbax-β, suggesting that the cells may be protected from death by Bcl-xl, at amounts capable of binding all available Bax. Bcl-xs may function by heterodimerizing with Bcl-xl, thereby displacing Bax and promoting cell death44 and its expression may play a role in the responsiveness of HD to chemo- and radiation therapy. CD40 upregulates the expression of Bcl-xl and, therefore, CD40 signaling may be responsible for protecting the RS cell by this pathway. The anti-apoptosis protein Bcl-2 was detected by IHC in all cases tested and may function in a similar manner.
Bak was detected in 13 of 42 single RS cells and their variants. Overexpression of Bak has been shown to induce apoptosis in cells protected by Bcl-2 or Bcl-xl perhaps by its association with the apoptosis-protecting proteins.7 Alternatively, Bak itself may be modulated: Bak inhibits cell death in an EBV-transformed cell line,59 raising the possibility of a similar role in HD since EBV is frequently present and expressed in RS cells.20
In general, the mRNA levels of a gene correspond to protein levels in a cell. This correlation held true in this study of the TNFR family in cases in which protein studies are available, such as Fas and TNFRs 1 and 2. The relative level of reactivity with the Bax polyclonal antibody did correspond with our RT-PCR findings, as case 6 was the most strongly positive with both methods of detection. In the analysis of Bcl-2, Bcl-x, and Bax, however, overall detection was unexpectedly low based on immunohistochemical studies with the few available antibodies.10 Anti-bax polyclonal, but not monoclonal, antibodies reacted with RS cells. This raises a question about the specificity of the two antibody types and, perhaps, cross-reactivity of the polyclonal with some other proteins, a problem which is broadly encountered with polyclonal antisera. In normal tissue, the polyclonal antibody reacted with more cell types than monoclonal, including germinal center cells, lymph node mantle cells, and placenta/decidua. Thus, the relationship of bax mRNA and protein levels is unclear. A high rate of translation versus transcription or a relatively long half-life of the protein as compared to the mRNA in the cell would affect the protein-mRNA relationship. In addition, Bcl-2 has at least three splice variants and is difficult to detect using 3′ RT-PCR. In fact, Bcl-2 protein was detected in four of the cases, but only two of the cells had detectable levels of the most common variant of bcl-2 transcript (data not shown). These factors have made 3′ RT-PCR analysis of the Bcl family of genes difficult to perform. For this reason, further immunohistochemical studies of all molecules discussed here are warranted, once suitable antibodies are developed.
Receptors for T-Lymphocyte–Mediated Cell Death Are Present in RS Cells
Fas, known to be expressed on the surface of RS cells by IHC,12 was detected here at high frequency by RT-PCR. RS cells are often surrounded in vivo by adherent, activated T cells capable of expressing the ligand for Fas. When activated by its ligand, Fas can induce apoptosis in lymphocytes.60-62 Cytotoxic T lymphocytes are known to express FasL and are thought to mediate killing through activation of Fas on target cells63 by activation of downstream ICE-like cysteine proteases or “caspases.”28 Overexpression of ICE itself is capable of inducing apoptosis.24 Both Fas and ICE were detected at the mRNA level in RS cells. However, Bcl-2 and Bcl-xl prevent a step in the Fas pathway that occurs before caspase activation. Since RS cells and their variants survive in vivo, the caspases in these cells may be in an inactive form. The Fas/FasL pathway may be active in some RS cells, because occasional evidence of presumably apoptotic cells with karyorrhectic nuclei is seen histologically. This mode of killing, however, must be insufficient to cause the complete elimination of RS cells and to prevent the spread of disease seen in untreated patients.
Another explanation for resistance to Fas-mediated apoptosis may be found in the cell-cycle phase of Reed-Sternberg cells. Death by Fas occurs only in cells induced to activation or proliferation. Apoptosis by Fas activation occurs only during late G1 phase or G1/S transition, whereas cells arrested early in G1 are resistant to Fas-induced apoptosis.64Reed-Sternberg cells in vivo may have a dysregulated cell cycle and are not in a highly proliferative state. Their doubling time is relatively long in comparison to other malignant lymphomas, yet both the S phase and the G1 phase are short.37 This indicates that most cells are not in a phase of the cell cycle where they would be susceptible to Fas-mediated apoptosis.
Apoptosis Genes Allow Alternate Pathways to Cell Death
Because HD is very sensitive to treatment, alternative pathways of apoptosis may be intact. In fact, all patients from whom cells were used in this study achieved complete remission for at least 3 years (Table 3). The presence of bax, bak, and ICEtranscripts in RS cells leaves the opportunity for apoptosis signaling through these pathways. These cytoplasmic effectors may be upregulated or activated in RS cells exposed to chemotherapeutic agents or irradiation, which presumably act by inducing apoptosis in the tumor cells. Chemotherapy with Adriamycin upregulates FasL expression on certain T-cell leukemia cells and kills the leukemia by inducing Fas-mediated apoptosis.65 This mechanism may also play a role in Adriamycin-treated HD, enhancing the ability of the T cells to kill the RS cell through the Fas pathway.53 It would be intriguing to determine the effects of treatment on expression of these and other apoptosis-regulating genes in RS cells.
In conclusion, HD has been an exceptionally challenging lymphoma to study because of the difficulty in obtaining RS cells. In this study, a first glimpse has been observed of the expression of genes involved in apoptosis and lymphocyte activation at the transcriptional level in HD. It is hoped that this preliminary observation will prompt further investigation into the role of these intracellular mechanisms in the pathogenesis and treatment of this peculiar form of lymphoma.
ACKNOWLEDGMENT
We thank Dr John C. Reed for his generous gift of the anti-Bax polyclonal antibody. We thank Celeste Riley, MD, Winnie Robinson, and Eileen Stein for laboratory help and Maria Ferguson for technical advice. We thank Loesje Troglia for technical help with manuscript preparation and Metin Ozdemirli, MD, for valuable discussions.
Supported by American Cancer Society Grant No. DHP112 (JC) and by a Special Fellowship of the Leukemia Society of America (A.B.).
Address correspondence to Jeffrey Cossman, MD, Georgetown University, 3900 Reservoir Rd, NW, Washington, DC 20007.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal