Abstract
The mi locus encodes a member of the basic-helix-loop-helix-leucine zipper protein family of transcription factors (hereafter called MITF). We have reported that the expression of several genes was impaired in cultured mast cells (CMCs) ofmi/mi genotype, and demonstrated the involvement of MITF in the transcription of these genes. To obtain new genes whose transcription may be regulated by MITF, we prepared a subtracted cDNA library using +/+ and mi/mi CMCs. We found two clones carrying the granzyme (Gr) B and tryptophan hydroxylase (TPH) cDNAs in the subtracted library. The expression of the Gr B and TPH genes decreased in mi/mi CMCs, and recovered to nearly normal level by the overexpression of normal (+) MITF but not of mutant (mi) MITF. The +-MITF bound three and one CANNTG motifs in the Gr B and TPH promoters, respectively, and transactivated these two genes, indicating the involvement of +-MITF in their expression. Because TPH is the rate-limiting enzyme for serotonin synthesis, we examined the serotonin content of +/+ and mi/mi CMCs. The serotonin content was significantly smaller in mi/mi CMCs than in +/+ CMCs. The introduction of +-MITF but not of mi-MITF normalized the serotonin content in mi/mi CMCs.
THE mi LOCUS of mice encodes a member of the basic-helix-loop-helix-leucine zipper (bHLH-Zip) protein family of transcription factors (hereafter calledmi-transcription factor, MITF).1,2 The MITF encoded by the mutant mi allele deletes 1 of 4 consecutive arginines in the basic domain (hereafter mi-MITF).1,3,4 Themi-MITF is defective in the DNA binding activity and the nuclear localization potential,5,6 and it does not transactivate target genes.6-10 The mi/mi mice show microphthalmia, depletion of pigment in both hair and eyes, osteopetrosis, and decrease in number of mast cells.11-15In addition to the decrease in number, the phenotype of mast cells is abnormal in mi/mi mice.16-19 Mast cells in the skin of normal (+/+) mice were stained with berberine sulfate that bound heparin proteoglycan and expressed the mouse mast cell protease 6 (MMCP-6) and c-kit genes, but mast cells in the skin of mi/mi mice were berberine sulfate-negative and did not express MMCP-6 and c-kit genes.17 20-23
We have shown the involvement of +-MITF in the transcription of c-kit, MMCP-6, MMCP-5, and p75 nerve growth factor (NGF) receptor gene in cultured mast cells (CMCs).7-10 To obtain new genes whose transcription may also be regulated by +-MITF, we elaborated cDNA libraries from +/+ CMCs and mi/mi CMCs and subtracted the latter from the former. The subtraction process consisted of the removal of clones carrying inserts complementary tomi/mi CMC mRNAs from the +/+ CMC cDNA library by hybridization. After repeating the subtraction process several times, we could yield a subtracted cDNA library made up of a high frequency of clones that were expressed in a +/+ CMC-specific manner. Screening 400 clones randomly selected from this library, we isolated several new genes as potential transcriptional targets of MITF, two of which turned out to be the granzyme (Gr) B and tryptophan hydroxylase (TPH) genes.
Gr B is a serine protease essential for cell-mediated immunity. It is most specifically detected in cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells, but it has also been shown to be expressed by CMCs and ABFTL1 mastocytoma cells.24 The murine Gr B gene resides on chromosome 14 and links with a gene complex encoding the other granzymes (Grs) (Grs C, D, E, F, and G), and the mast cell–specific serine proteases (MMCP-1, -2, -4, and -5).25-28 On the other hand, TPH is the rate-limiting enzyme for the synthesis of serotonin,29-33 a chemical mediator of immediate hypersensitivity reaction that is preformed and stored in the basophilic granules of mast cells.34 We examined whether +-MITF directly transactivated the Gr B and TPH genes. The +-MITF specifically bound three CANNTG motifs in the Gr B promoter and one CAGGTG motif in the TPH promoter, and the binding transactivated the luciferase genes under the control of the Gr B or TPH promoter.
MATERIALS AND METHODS
Mice and cells.
The original stock of C57BL/6-mi/+ (mi/+) mice was purchased from the Jackson Laboratory (Bar Harbor, ME) and was maintained in our laboratory by consecutive backcrosses with our own inbred C57BL/6 colony (more than 12 generations at the time of the experiments described here). Female mi/+ mice were crossed with male mi/+ mice, and the resulting mi/mi mice were selected on the basis of their white coat color.11,12Pokeweed mitogen–stimulated spleen cell conditioned medium (PWM-SCM) was prepared according to the method described by Nakahata et al.35 Mice of mi/mi genotype and their normal (+/+) littermates with an age of 2 to 3 weeks were used to obtain CMCs. Mice were killed by decapitation after ether anesthesia and the spleens were removed. Spleen cells derived from mi/mi or +/+ mice were cultured in α-minimal essential medium (α-MEM; ICN Biomedicals, Costa Mesa, CA) supplemented with 10% PWM-SCM and 10% fetal calf serum (FCS; Nippon Biosupp Center, Tokyo, Japan). Half of the medium was replaced every 7 days. Four weeks after initiation of the culture more than 95% of cells were CMCs.22 CMCs overexpressing +-MITF or mi-MITF were obtained as described previously and maintained in α-MEM supplemented with 10% PWM-SCM and 10% FCS.7 8 The NIH/3T3 fibroblast cell line was generously provided by Dr S.A. Aaronson (National Cancer Institute, Bethesda, MD) and maintained in Dulbecco's modification of Eagle's medium (ICN Biomedicals) supplemented with 10% FCS.
Construction of the pAP3neo vector.
The expression vector, pAP3neo, which harbors an SV40 promoter and allows efficient expression of cDNA inserts in mammalian cells, was prepared as follows. First, the 0.4-kb DNA fragment (AflIII/Kpn I cut) of the pBluescript II vector (Stratagene, La Jolla, CA) was replaced by a 0.7-kb DNA fragment (AflIII/Kpn I cut) of the pcDV1 vector.36Then, the 0.6-kb DNA fragment (Pst I/HindIII cut) of the pL1 vector36 carrying the SV40 promoter was inserted into the Pst I/BssHII site of this plasmid DNA using appropriate adapter/linkers. Subsequently, a pair of chemically synthesized oligonucleotides designed to yield a double-stranded multicloning site was inserted into this plasmid DNA via PstI/Kpn I sites. The multicloning site contains the recognition sequences for the following enzymes: 5′-Sse I(PstI)-Sac I-Cpo I-Apa I-Sau I-MluI-T7 RNA polymerase-EcoRI-AatII-XbaI-BglII-AflIII-Asc I-BstXI-BalI-SnaBI-BstEII-DraIII-Nhe I-SceI-Not I-T3 RNA polymerase-Swa I-SplI-Nru I-Pac I-SacII-Kpn I-3′. Finally, a neomycin unit was inserted via the Sfi I site into the middle of the SV40 promoter, and we named this vector pAP3neo.
Preparation of cDNA libraries carrying directional inserts.
Before the purification of poly (A)+ RNA, total RNA was extracted by the guanidine thiocyanate/CsTFA method from +/+ andmi/mi CMCs. The cDNA library was prepared as described by Gubler and Hoffmann37 with some modifications. Briefly, cDNA was synthesized from 2 μg of +/+ or mi/mi CMC poly (A)+ RNA with reverse transcriptase in a reaction mixture including 5Me-dCTP and 1.6 μg of oligo (dT) primer carrying an Not I restriction site. RNase H was then added to the reaction mixture and was followed by DNA polymerase I as described by Okayama and Berg.36 Then, the cDNA was blunt-ended with T4 DNA polymerase, and an unphosphorylatedBglII-Sma I adapter was ligated to the 5′ end. After digestion with Not I, small DNA fragments of less than 300 bp were removed by a CHROMA spin 400 column (Clontech, Palo Alto, CA). The cDNA fragments were directionally inserted between the Not I (dephosphorylated) and BglII sites of the pAP3neo vector.
Dephosphorylation of the Not I site of the vector and the unphosphorylated BglII end of the inserts minimized the ligation of multiple cDNA inserts in the library. The ligation mixture was pretreated as described before38 and electroporated into Escherichia coli MC1061A cells. Before propagation of the cDNA library for preparation of a stock solution, we removed an aliquot and counted the cell number to know how many independent clones the cDNA library contained (the complexity of a library).
Preparation of the subtracted cDNA library.
The detailed process for the preparation of the subtracted cDNA library was described previously.39 To prepare single-stranded plasmid DNA, the plasmid DNA prepared from the +/+ CMC cDNA library was introduced into E coli DH5αF′IQ cells by electroporation. After 1 hour of culture in rich medium (2× YT), transformed cells were infected with R408 helper phages. Then, single-stranded DNA was purified from the supernatant of the 8-hour culture. To prepare biotinylated RNA drivers, total RNA was extracted by the guanidine thiocyanate/CsTFA method from mi/mi CMCs, and poly (A)+ RNA was purified and labeled by photobiotin (Vector Lab, Burlingame, CA). One microgram of single-stranded DNA prepared from the +/+ CMC cDNA library was hybridized with 10 μg of biotinylated RNA at 42°C in 25 μL hybridization buffer containing 40% formamide, 50 mmol/L HEPES (pH 7.5), 1 mmol/L EDTA, 0.1% sodium dodecyl sulfate (SDS), 0.2 mol/L NaCl, and 1 μg of oligo-poly(rA). After hybridization for 42 hours, the mixture was transferred to 400 μL of SB (50 mmol/L HEPES [pH 7.5], 2 mmol/L EDTA, 500 mmol/L NaCl), and 10 μg streptoavidin was subsequently added. The mixture was incubated at room temperature for 5 minutes and extracted with phenol/chloroform/isoamyl alcohol (25:24:1). The organic phase was back-extracted with 100 μL TE (10 mmol/L Tris-HCl [pH 7.5], 1 mmol/L EDTA). The aqueous phases were pooled. Streptoavidin binding and phenol treatment were repeated once more. The recovered single-stranded plasmid DNA was subtracted with biotinylated RNA one more time. After repeating the subtraction process, the recovered single-stranded DNA was converted to double-stranded plasmid DNA by the BcaBEST DNA polymerase (TaKaRa, Otsu, Japan) reaction at 65°C for 30 minutes. After phenol extraction and ethanol precipitation, the DNA was dissolved in 20 μL TE buffer and 3-μL aliquots were introduced intoE coli MC1061A cells by electroporation.
Screening of +/+ CMC-specific clones by Southern blot analysis.
Four hundred cDNA clones were prepared from ampicillin-resistant colonies randomly selected from the subtracted cDNA library by an automated plasmid purification machine (PI-100; KURABO, Osaka, Japan). After digestion with both Sma I and Not I to separate the cDNA insert, the plasmid DNA was electrophoresed in an agarose gel and transferred to nylon membranes. The cDNA probes were synthesized with reverse transcriptase (RT) from +/+ or mi/mi CMC poly (A)+ RNA in a reaction mixture containing [α-32P]dCTP. Duplicate membranes were hybridized with the reverse-transcribed cDNA probes for 15 hours. The filters were washed several times with a final stringency of 0.1× SSC (1× SSC = 0.15 mmol/L NaCl, 15 mmol/L sodium citrate, pH 7.2) and 0.1% SDS at 50°C, and autoradiographed at −80°C with an intensifying screen. Clones that hybridized with higher efficiency to the +/+ cDNA probe compared to the mi/mi cDNA probe were selected as candidates for further investigation.
DNA sequencing.
The plasmid DNA was purified individually from the subtracted cDNA library by PI-100 and directly subjected to DNA sequencing. Dideoxy-chain termination sequencing reactions were performed with T7 dye-labeled primers and thermal cycle sequencing kits purchased from LI-COR (Lincoln, NE). The reaction products were analyzed by a Model 4000L Automated DNA Sequencer (LI-COR).
Northern blot analysis.
Five micrograms of total RNA prepared from +/+ or mi/mi CMCs was loaded per lane, fractionated on 1% agarose-formaldehyde gels, and transferred to nylon membranes by capillary action in 20× SSC. Baked membranes were prehybridized for 3 hours at 42°C in a buffer containing 50% formamide, 5× SSC, 5× Denhardt's solution, and 0.1% SDS. The membranes were hybridized with the [α-32P]dCTP-labeled DNA probes at 42°C for 15 hours in the same buffer. Preparation of the DNA probes was performed according to the random hexamer labeling method. cDNA inserts of some clones isolated from the subtracted cDNA library were used as a template. To prepare MMCP-6, mast cell carboxypeptidase A (MC-CPA), histidine decarboxylase (HDC), and β-actin probes, cDNA fragments of approximately 570 bp, 550 bp, 800 bp, and 700 bp were used, respectively. The MITF probe was prepared from the XhoI-Pst I cDNA fragment of about 750 bp.1 After hybridization, the membranes were washed to a final stringency of 0.1× SSC and 0.1% SDS at 50°C, and autoradiographed at −80°C.
To characterize the subtracted cDNA library, 1 μg of the NotI–digested plasmid DNA prepared from the +/+ CMC, mi/mi CMC, or subtracted cDNA library was used as a template for the T7 RNA polymerase reaction (Stratagene). Two micrograms of synthesized RNA was loaded per lane, fractionated on 1% agarose-formaldehyde gels, and transferred to nylon membranes by capillary action in 20× SSC. The hybridization procedures were the same as described above.
In situ hybridization.
Pellets of +/+ and mi/mi CMCs with or without introduction of +- or mi-MITF cDNA were fixed with 4% paraformaldehyde in phosphate buffer (0.1 mol/L, pH 7.2) overnight, embedded in paraffin, and cut serially to a thickness of 3 μm. Three serial sections were used as follows. The first section was stained with hematoxylin and eosin (H&E), the second section was used for in situ hybridization, and the third section was stained with alcian blue to detect mast cells. Details of the in situ hybridization technique have been described previously.40 For cRNA probe preparation, the clones containing Gr B, MC-CPA, TPH, and HDC cDNA inserts were transcribed with the DIG RNA Labeling Kit (Boehringer Mannheim Biochemica, Mannheim, Germany). The number of cells positive for the probes and that of alcian blue-positive cells were counted in serial sections, and the proportion of the former cells was calculated.
RT-polymerase chain reaction (PCR) analysis.
Two sets of oligonucleotide primers were synthesized: the Gr B sense primer 5′-GATTACCCATCGTCCCTAGAGCT-3′ and antisense primer 5′-CATGCCCAGCTCCAATGCAAAC-3′ (nucleotides [nt] 884-906 and nt 1097-1118, respectively)25; and the Gr C-G sense primer 5′-TCCTGACCCTACTTCTGCCTCT-3′ (nt 94-115, nt 106-127, nt 1-22, nt 80-101, and nt 61-82 of Gr C, D, E, F, and G, respectively)41-43 and antisense primer 5′-CTTTTGCCACAGGGATGATCTGC-3′ (nt 335-357, nt 359-381, nt 242-264, nt 321-343, and nt 302-324 of Gr C, D, E, F, and G, respectively).41-43 The Gr C-G sense primer had one nucleotide mismatch for the Gr F sequence (nt 90; T to A).41 As a positive control, splenocytes of +/+ mice were activated by culturing in α-MEM containing PWM (1:300 dilution) for 2 days before RNA extraction. Various amounts of total RNA (0.5, 0.05, and 0.005 μg) extracted from +/+ and mi/mi CMCs, and +/+ splenocytes were reverse-transcribed in 20 μL of a reaction mixture containing 200 U of Superscript II (GIBCO-BRL, Grand Island, NY) and 0.2 μg of oligo (dT) primer. One microliter of each reaction mixture was amplified in 10 μL of PCR mixture containing 0.5 U of Taq DNA polymerase (TaKaRa) and 12.5 pmol of one set of specific primers. The PCR profile consisted of 24 or 30 cycles of denaturation at 94°C for 30 seconds, reannealing at 63°C for 30 seconds, and polymerization at 72°C for 1 minute, followed by extension at 72°C for 5 minutes. Half of the respective PCR products was fractionated on 2% agarose gels and stained with ethidium bromide.
Concentrations of serotonin and histamine.
The concentrations of serotonin and histamine were measured using high-performance liquid chromatography (HPLC) with electrochemical detection44 and HPLC-fluorometry,45respectively. Briefly, CMCs were collected, washed with phosphate-buffered saline (PBS), counted, and sonicated for 20 seconds in a sonicator (Tomy, Tokyo, Japan) in 1 mL of ice-cold 3% perchloric acid containing of 5 mmol/L EDTA and 1 mmol/L sodium metabissulfite. The homogenate was centrifuged at 10,000g for 15 minutes at 4°C and the supernatant was applied directly to the HPLC column. The concentration of serotonin and histamine per 1.0 × 106cells was calculated.
Serotonin uptake.
The uptake of serotonin into +/+ or mi/mi CMCs was determined according to the method described by Miller and Hoffman46with minor modification. CMCs (1.0 × 105) of +/+ ormi/mi genotype were preincubated with 0.1 mmol/L iproniazid (Sigma, St Louis, MO) for 30 minutes in the uptake buffer (20 mmol/L HEPES, 140 mmol/L NaCl, 5 mmol/L KCl, 1.8 mmol/L CaCl2, 0.8 mmol/L MgSO4, 5 mmol/L glucose, and 10 μmol/L pargyline). The buffer was aspirated and fresh uptake buffer containing 200 nmol/L of [3H] serotonin (30 mCi/μmol; Amersham, Arlington Heights, IL) was added to the cells. Incubation was continued for 15 minutes at 37°C; uptake was terminated by aspiration of the medium. After washing three times, the cells were dissolved in 1% SDS/0.2 mol/L NaOH and the radioactivity was measured with a liquid scintillation counter (Amersham).
Electrophoretic gel mobility shift assay (EGMSA).
The production and purification of GST-MITF fusion protein and anti-GST antibody were described previously.5 The fusion protein was used to measure the in vitro binding of MITF to the consensus sequence in EGMSA. The double-stranded oligonucleotides were labeled by filling 5′-overhangs with Klenow enzyme in the presence of deoxynucleotides containing [α-32P]dCTP. Probes were separated from unincorporated nucleotides by gel electrophoresis. The binding reactions were performed at 4°C for 15 minutes with 50 ng of labeled DNA probe and 3.5 μg of protein in a 20-μL reaction mixture containing 10 mmol/L Tris-HCl (pH 8.0), 1 mmol/L EDTA, 75 mmol/L KCl, 1 mmol/L dithiothreitol (DTT), 4% Ficoll type 400 (Sigma), and 50 ng of poly (dI-dC). For competition experiments, unlabeled double-stranded oligonucleotides at a 100-fold molar excess were added to the binding reaction mixture before the addition of the protein. DNA-protein complexes were separated by electrophoresis on a 5% polyacrylamide gel in 0.25× TBE buffer (1× TBE = 90 mmol/L Tris-base, 64.6 mmol/L boric acid, and 2.5 mmol/L EDTA, pH 8.3) at 14 V/cm. The gels were dried on Whatman 3MM chromatography paper (Whatman, Maidstone, UK) and subjected to autoradiography.
Construction of effector and reporter plasmids, and the transient cotransfection assay.
pEF-BOS expression vector47 was kindly provided by Dr S. Nagata (Osaka University, Osaka, Japan). The expression plasmids containing +-MITF or mi-MITF cDNA was constructed as described previously.7,8 The luciferase gene subcloned into pSP72 (pSPLuc) was generously provided by Dr K. Nakajima (Osaka University Medical School, Osaka, Japan). To construct reporter plasmids, the promoter region of the Gr B (nt −910 to +42)48 and TPH (nt −1052 to +86)31 genes were obtained with PCR and subcloned into the upstream region of the luciferase gene in pSPLuc. The deletion of the TPH promoter was produced by PCR using the appropriate primers. The mutations were introduced by PCR with mismatch primers. Deleted or mutated products were verified by sequencing. NIH/3T3 cells (5 × 105) were plated in a 10-cm dish 1 day before the procedure. Cotransfection of 10 μg of a reporter, 5 μg of an effector, and 5 μg of an expression vector containing the β-galactosidase gene was performed by the calcium phosphate precipitation method. The expression vector containing the β-galactosidase gene was used as an internal control. The cells were obtained 48 hours after transfection and lysed with 0.1 mol/L potassium phosphate buffer (pH 7.4) containing 1% Triton X-100 (Nacali, Kyoto, Japan). Soluble extracts were then assayed for luciferase activity with a luminometer LB96P (Berthold GmbH, Wildbad, Germany) and for β-galactosidase activity. The luciferase activity was normalized using the β-galactosidase activity and total protein concentration was estimated according to the method described by Yasumoto et al.49 The normalized value was divided by the value obtained after cotransfection with the reporter and pEF-BOS, and was expressed as the relative luciferase activity.
RESULTS
Preparation and characterization of a subtracted cDNA library.
The systematic cloning of the genes expressed in a +/+ CMC-specific manner, including the transcriptional target genes for MITF, was conducted according to the following strategy. With the aim of developing a new mammalian expression vector for future experiments, a plasmid vector (pAP3neo) was constructed by fusing the Okayama-Berg vector36 with pBluescript II and inserting a chemically synthesized multicloning site (see Materials and methods). The introduction of the f1 origin into the vector enabled the preparation of the single-stranded cDNA library necessary for the subtraction process. Using this vector, a cDNA library of +/+ CMCs was prepared by a modified Gubler-Hoffman method,37 which we refer to as the Linker-primer method in Materials and Methods. The complexities of the cDNA libraries used in this study were 1.2 × 106colony-forming units (CFU) for +/+ CMCs and 1.6 × 106 CFU for mi/mi CMCs. These numbers indicate that the cDNA libraries contain almost all of the mRNA expressed in these cells without any leakage. Clones carrying inserts complementary to mRNAs ofmi/mi CMCs were subtracted from the +/+ CMC cDNA library after formation of the biotin-avidin complex. Because mi/mi CMCs contain a reduced proportion of transcripts from MITF-upregulated genes, the subtraction process should allow enrichment of +/+ CMC-specific cDNA clones and facilitate the cloning of downstream target genes of MITF.
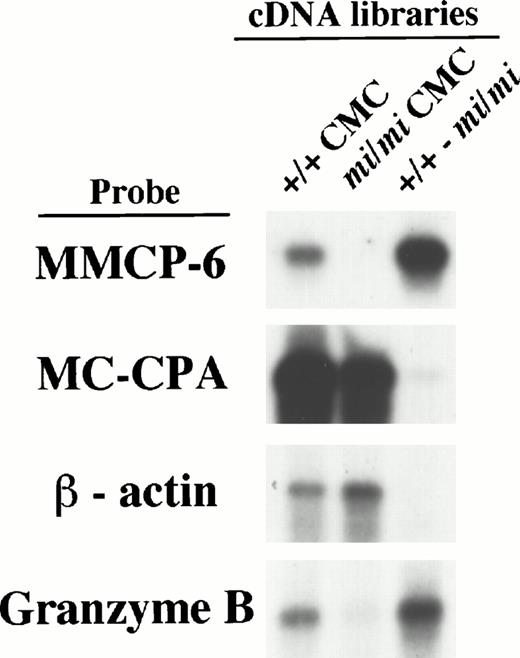
To determine if successful subtraction had been achieved, we examined the subtracted cDNA library by the following experiments. Firstly, the sense RNAs of the cDNA inserts were synthesized by the T7 RNA polymerase reaction using the Not I–digested plasmid DNA of either the +/+ CMC, mi/mi CMC, or subtracted cDNA libraries as templates. They were then fixed to nylon membranes to perform Northern blot analysis (we refer to them as library-Northern blots hereafter) (Fig 1). Both the MC-CPA and β-actin genes have been shown to be expressed equally in +/+ and mi/miCMCs, and the MMCP-6 gene to be expressed only in +/+ CMCs.15 When hybridized with the MC-CPA and β-actin cDNA, the hybridized bands were easily detected with equal intensity in both the RNAs from the +/+ and mi/mi CMC cDNA libraries as well, but were rarely detectable in the RNA from the subtracted cDNA library. On the other hand, when MMCP-6 was used as a probe, the hybridized bands were easily detectable in the RNA from the +/+ CMC cDNA library but no such band was observed in the RNA from the mi/mi CMC cDNA library. In the RNA from the subtracted cDNA library, the intensity of the hybridized band was much stronger than that in the RNA from the +/+ CMC cDNA library. The results of library-Northern blot analysis obtained with the RNAs from +/+ and mi/mi CMC cDNA libraries were consistent with those obtained with the total RNAs of +/+ andmi/mi CMCs, indicating that RNA synthesis from these libraries had been performed without bias. Thus, we concluded that compared with the original +/+ cDNA library, the subtracted cDNA library contained much smaller numbers of cDNA clones from transcripts present at the same level in +/+ and mi/mi CMCs, such as the MC-CPA and β-actin gene transcripts, and much larger numbers of cDNA clones from transcripts present in +/+ CMCs but absent from mi/mi CMCs, such as the MMCP-6 gene transcript.
Characterization of the subtracted cDNA library by Northern blot analysis. Sense RNAs were synthesized by the T7 RNA polymerase reaction using the Not I–digested plasmid DNA of the +/+ CMC, mi/mi CMC, and subtracted cDNA libraries as templates. Two micrograms of synthesized RNA was loaded per lane, electrophoresed, transferred, and fixed onto nylon membranes. Probes were prepared from the cDNAs for MMCP-6, MC-CPA, β-actin, or Gr B using the random hexamer labeling method.
Characterization of the subtracted cDNA library by Northern blot analysis. Sense RNAs were synthesized by the T7 RNA polymerase reaction using the Not I–digested plasmid DNA of the +/+ CMC, mi/mi CMC, and subtracted cDNA libraries as templates. Two micrograms of synthesized RNA was loaded per lane, electrophoresed, transferred, and fixed onto nylon membranes. Probes were prepared from the cDNAs for MMCP-6, MC-CPA, β-actin, or Gr B using the random hexamer labeling method.
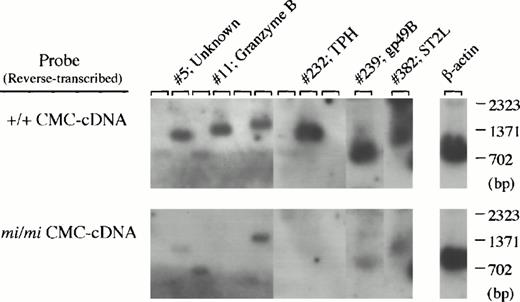
Secondly, 400 cDNA clones were isolated from the subtracted library, and the cDNA insert of each clone was analyzed by Southern blotting using cDNA reverse-transcribed from +/+ or mi/mi CMC poly (A)+ RNA as probes (Fig 2).After adjustment of the two β-actin–specific signals to equal intensity, it may be expected that the clones which hybridize more efficiently with the +/+ cDNA probe than with the mi/mi cDNA probe carry cDNA inserts transcribed specifically in +/+ CMCs. Effectively, the subtracted cDNA library contained such clones (clones no. 5, 11, 232, 239, and 382 in Fig 2) at high frequency (approximately 15%). These two experiments showed that we had achieved successful subtraction and that it was possible to obtain a subtracted cDNA library of sufficient quality to allow an effective search for +/+ CMC-specific cDNA clones.
Screening of +/+ CMC-specific clones by Southern blot analysis. After digestion with both Sma I and Not I to separate the cDNA insert, plasmid DNAs of 400 clones randomly selected from the subtracted cDNA library were electrophoresed in agarose gel and bound to nylon membranes. Duplicate membranes were hybridized with32P-labeled cDNAs synthesized from poly(A)+RNA of +/+ (upper) or mi/mi (lower) CMCs. In the rightmost lanes, an equal amount of the β-actin cDNA fragment was loaded as a control, and we graphically equalized the intensity of their bands for two filters to normalize the intensity of other sets of the bands. The clones which hybridized to a greater degree with +/+ CMC-cDNAs than with mi/mi CMC-cDNAs, such as clones no. 5, 11, 232, 239, and 382, were selected, and subjected to DNA sequencing and a computer-assisted homology search. The identity of these clones is also shown by the clone number.
Screening of +/+ CMC-specific clones by Southern blot analysis. After digestion with both Sma I and Not I to separate the cDNA insert, plasmid DNAs of 400 clones randomly selected from the subtracted cDNA library were electrophoresed in agarose gel and bound to nylon membranes. Duplicate membranes were hybridized with32P-labeled cDNAs synthesized from poly(A)+RNA of +/+ (upper) or mi/mi (lower) CMCs. In the rightmost lanes, an equal amount of the β-actin cDNA fragment was loaded as a control, and we graphically equalized the intensity of their bands for two filters to normalize the intensity of other sets of the bands. The clones which hybridized to a greater degree with +/+ CMC-cDNAs than with mi/mi CMC-cDNAs, such as clones no. 5, 11, 232, 239, and 382, were selected, and subjected to DNA sequencing and a computer-assisted homology search. The identity of these clones is also shown by the clone number.
The subtracted cDNA library had a complexity of 1.0 × 106CFU, as judged by counting the cell number at the final electroporation step (see Materials and Methods). This number indicates that the subtraction process caused almost no leakage of the cDNA species present in the original cDNA library. To know how many independent cDNA clones the subtracted cDNA library contained, 100 clones were randomly selected from the library. A mixture of the cDNA inserts prepared from 20 of these 100 clones by Sma I/Not I digestion were labeled with [α-32P]dCTP. After stripping from the membranes the reverse-transcribed cDNA probes described above, the membranes were rehybridized with this new cDNA probe, and the number of clones showing hybridization signals was counted (data not shown). Five of the 400 clones proved to have cDNA inserts identical to those of the selected 20 clones. The same procedures were performed using the mixture of cDNA inserts from the remaining 80 clones. Out of 400 clones, 20 proved to have identical cDNA inserts to those of the 80 clones. These results indicate that 20 and 80 clones correspond to 5/400 (1.25%) and 20/400 (5%) of the total clone number present in the library, respectively, and that the library contains approximately 20/0.0125 or 80/0.05 clones, ie, approximately 1.6 × 103independent cDNA clones. Considering that the subtracted cDNA library contained +/+-specific clones with a frequency of 15%, we estimated that the number of species of genes transcriptionally upregulated by MITF is, at most, 240 genes.
Identification of the genes transcribed in a +/+ CMC-specific manner.
By screening the cDNA inserts of the subtracted library by Southern blot analysis, 40 clones were picked up and subjected to further analyses. About a 1-kb region from the 5′-end of these cloned cDNAs was sequenced and analyzed with a computer-assisted homology search. Half (16 clones) of them showed no homology to any published sequences. Among the rest of the clones, we repeatedly found cDNAs encoding MMCP-5 (two clones) or MMCP-6 (two clones), both of which are known to be transcribed in a +/+ CMC-specific manner.5,8 9 We also identified cDNAs encoding Gr B (four clones), tryptophan hydroxylase (TPH) (clone no. 232), gp49B50 (clone no. 239), and ST2L (clone no. 382), suggesting that these genes are novel transcriptional targets of MITF.
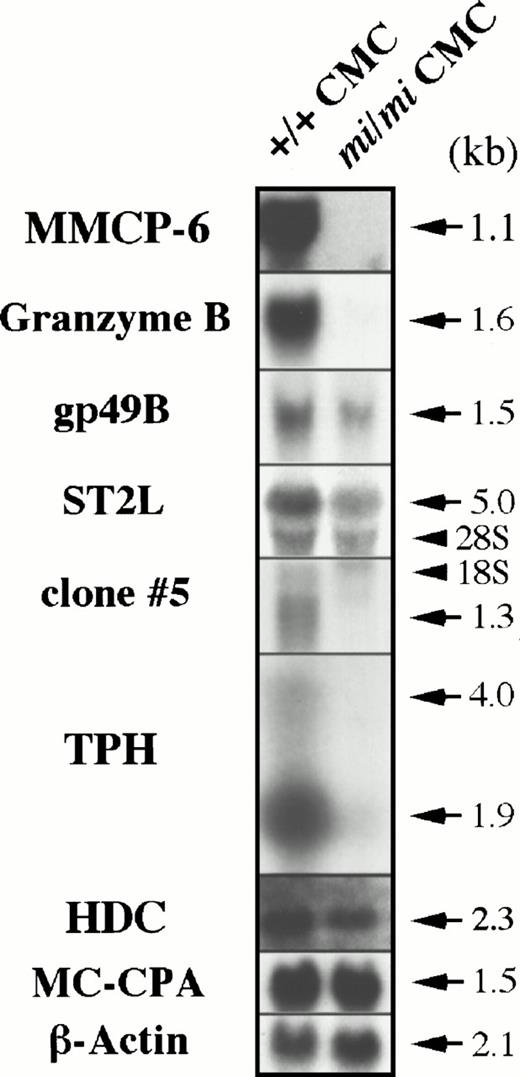
To determine if these cDNAs are actually transcribed exclusively in +/+ CMCs, we performed Northern blot analysis using total RNA obtained from +/+ and mi/mi CMCs. As shown in Fig3, the Gr B and TPH mRNA expression was easily detected in +/+ CMCs but rarely detectable in mi/miCMCs. The gp49B and ST2L genes were also transcribed preferentially in +/+ CMCs. From the clones displaying no homology to any published sequences, clone no. 5 was randomly selected and shown to be expressed in a +/+ CMC-specific manner. These results indicated that the data obtained from Southern and Northern blotting were consistent. To confirm the success of the subtraction process again, the radiolabeled cDNA probe of clone no. 11 insert (Gr B cDNA) was hybridized with the library-Northern blots (Fig 1). As in the case of MMCP-6, the subtracted cDNA library contained much larger numbers of cDNA clones carrying the Gr B cDNA than the original +/+ CMC cDNA library, confirming the efficiency of the subtraction process. Hereafter, we focused on the poor expression of Gr B and TPH mRNA in mi/miCMCs.
Expression of the Gr B, gp49B, ST2L, TPH, and HDC genes in +/+ and mi/mi CMCs. Five micrograms of total RNA prepared from +/+ or mi/mi CMCs was loaded in each lane and fixed onto nylon membranes by capillary action. The membranes were hybridized with specific DNA probes. To prepare the Gr B, gp49B, ST2L, TPH probes, the cDNA inserts of clones no. 11, 232, 239, and 382 were used. Expression of MMCP-6 and MC-CPA mRNA were shown as controls. Arrows indicate the specific signals which are accompanied by their molecular size. Arrowheads indicate the position of 18S and 28S in each panel. Reprobing with the β-actin probe allowed verification that an equal amount of mRNA was loaded per lane.
Expression of the Gr B, gp49B, ST2L, TPH, and HDC genes in +/+ and mi/mi CMCs. Five micrograms of total RNA prepared from +/+ or mi/mi CMCs was loaded in each lane and fixed onto nylon membranes by capillary action. The membranes were hybridized with specific DNA probes. To prepare the Gr B, gp49B, ST2L, TPH probes, the cDNA inserts of clones no. 11, 232, 239, and 382 were used. Expression of MMCP-6 and MC-CPA mRNA were shown as controls. Arrows indicate the specific signals which are accompanied by their molecular size. Arrowheads indicate the position of 18S and 28S in each panel. Reprobing with the β-actin probe allowed verification that an equal amount of mRNA was loaded per lane.
Recovery of Gr B and TPH expression by the introduction of +-MITF cDNA into mi/mi CMCs.
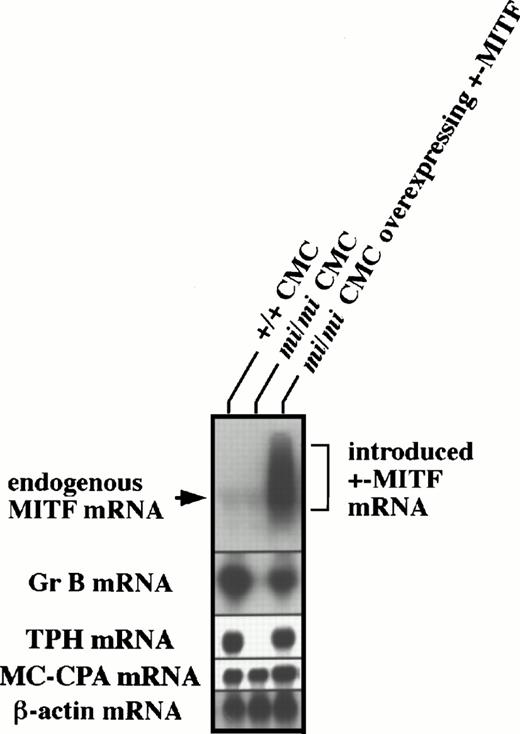
Previously, we were able to successfully introduce +- ormi-MITF cDNA into mi/mi CMCs.7 8 Using these cells, we examined by in situ hybridization whether +-MITF-overexpression in mi/mi CMCs allowed the recovery of Gr B and TPH expression. Approximately 90% and 70% of +/+ CMCs expressed Gr B and TPH mRNA, respectively, whereas only a few mi/mi CMCs were able to do so (Table 1). The introduction of +-MITF cDNA, but not mi-MITF cDNA, increased the proportions of Gr B and TPH mRNA-expressing CMCs up to a level equivalent to those of +/+ CMCs (Table 1). To demonstrate the restoration of Gr B and TPH expression in mi/mi CMCs more quantitatively, total RNAs were extracted from +/+ CMCs, mi/miCMCs, and mi/mi CMCs containing +-MITF cDNA, and subjected to Northern blot analysis. As shown in Fig 4,mi/mi CMCs containing +-MITF cDNA overexpressed the MITF mRNA and recovered the Gr B and TPH expression to the nearly normal levels. After longer exposure, no difference in endogenous MITF gene expression was observed between the original +/+ and the mi/mi CMCs (data not shown).
Recovery of Gr B and TPH mRNA Expression inmi/mi CMCs After Introduction of +-MITF cDNA
| Cells . | Proportion to Alcian Blue+ Cells (%)* . | |||
|---|---|---|---|---|
| Gr B mRNA+ Cells . | TPH mRNA+ Cells . | HDC mRNA+ Cells . | MC-CPA mRNA+ Cells . | |
| +/+ CMCs | 88 ± 4 | 70 ± 4 | 72 ± 2 | 91 ± 2 |
| mi/mi CMCs | 6 ± 3-151 | 13 ± 1-151 | 79 ± 6 | 93 ± 2 |
| mi/mi CMCs overexpressing +-MITF | 73 ± 8 | 68 ± 3 | 79 ± 2 | 93 ± 2 |
| mi/mi CMCs overexpressing mi-MITF | 7 ± 3-151 | 11 ± 3-151 | 82 ± 1 | 90 ± 2 |
| Cells . | Proportion to Alcian Blue+ Cells (%)* . | |||
|---|---|---|---|---|
| Gr B mRNA+ Cells . | TPH mRNA+ Cells . | HDC mRNA+ Cells . | MC-CPA mRNA+ Cells . | |
| +/+ CMCs | 88 ± 4 | 70 ± 4 | 72 ± 2 | 91 ± 2 |
| mi/mi CMCs | 6 ± 3-151 | 13 ± 1-151 | 79 ± 6 | 93 ± 2 |
| mi/mi CMCs overexpressing +-MITF | 73 ± 8 | 68 ± 3 | 79 ± 2 | 93 ± 2 |
| mi/mi CMCs overexpressing mi-MITF | 7 ± 3-151 | 11 ± 3-151 | 82 ± 1 | 90 ± 2 |
*Mean ± SE of three experiments.
P < .01 by t-test when compared with the value of +/+ CMCs.
Recovery of Gr B and TPH expression after the introduction of +-MITF cDNA into mi/mi CMCs. Five micrograms of total RNA from +/+ CMCs, mi/mi CMCs, and mi/miCMCs overexpressing +-MITF were fractionated on 1% agarose gels and subjected to hybridization with the MITF, Gr B, and TPH probes. The +-MITF cDNA introduced into mi/mi CMCs appeared to be transcribed abundantly. Reprobing with the β-actin and MC-CPA probes showed that equal loading had been achieved.
Recovery of Gr B and TPH expression after the introduction of +-MITF cDNA into mi/mi CMCs. Five micrograms of total RNA from +/+ CMCs, mi/mi CMCs, and mi/miCMCs overexpressing +-MITF were fractionated on 1% agarose gels and subjected to hybridization with the MITF, Gr B, and TPH probes. The +-MITF cDNA introduced into mi/mi CMCs appeared to be transcribed abundantly. Reprobing with the β-actin and MC-CPA probes showed that equal loading had been achieved.
Gr B shows a distinct expression profile among multiple granzymes.
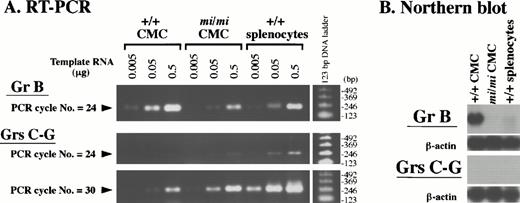
The cDNA sequences of granzymes (Grs) B-G are highly similar to one another. Because the Gr B probe used in the Northern blot analysis (Figs 3 and 4) was a full-length Gr B cDNA (clone no. 11), it will crossreact with transcripts from all of the Gr genes. To discriminate Gr B transcripts from those of other Grs, we performed RT-PCR analysis (Fig 5A). We designed two sets of oligonucleotide primers: one set was Gr B–specific and was expected to amplify the 235-bp cDNA fragment derived from the Gr B 3′ untranslated region (UTR); the other set was common to Grs C-G and was expected to amplify the 264-bp (Grs C, E, F, and G) and/or 276-bp (Gr D) cDNA fragment derived from the Grs C-G open reading frame. A series of diluted total RNAs (0.5, 0.05, and 0.005 μg) from +/+ CMCs,mi/mi CMCs, and +/+ PWM-stimulated splenocytes were reverse-transcribed, and the resulting cDNAs were PCR-amplified with the Gr B– or Gr C-G–specific primers. Half of the respective PCR products were analyzed on agarose gels. The specificity of the reaction was demonstrated by the presence of a single band of the expected size among the PCR products and by DNA sequencing. The intensity of the bands corresponding to Gr B or Grs C-G increased proportionally with the amount of template RNA. This showed that PCR amplification was semiquantitative throughout the entire PCR reaction. After only 24 PCR cycles, amplification of the Gr B–specific cDNA was easily detectable on agarose gels, and Gr B expression in mi/mi CMCs appeared to be less than one tenth of that seen in +/+ CMCs (Fig 5A). This observation was compatible with the result of Northern blot analysis shown in Fig 3. On the other hand, amplification of the bands corresponding to Grs C-G was hardly detectable in either +/+ ormi/mi CMCs after 24 cycles using a set of the primers common to Grs C-G. At an increased PCR cycle number of 30, bands of about 270 bp in size became visible for both types of cells (Fig 5A). Judging from the band intensity, Gr C-G expression was slightly higher inmi/mi CMCs than in +/+ CMCs. Because the Gr B 3′UTR sequence, which was PCR-amplified with Gr B–specific primers, lacks homology to the cDNA sequences of other Grs, a fragment containing this sequence is a useful probe for the specific detection of the Gr B transcript. When we reassessed Gr B expression in CMCs by Northern blot analysis using this Gr B–specific probe, we obtained a result (Fig 5B) similar to that shown in Fig 3, where full-length Gr B cDNA was used as a probe. In contrast, the probe prepared from a PCR product common to the other Grs did not detect any transcript after the same exposure time (5 hours). These results showed that the expression profile of Gr B is distinct from those of other Grs, and that Gr B gene expression inmi/mi CMCs is drastically reduced.
Expression profile of the Gr B gene and the other Grs of the murine Gr B locus in +/+ and mi/mi CMCs. (A) Serially diluted total RNA (0.5, 0.05, and 0.005 μg) from +/+ andmi/mi CMCs, and splenocytes of +/+ mice were reverse-transcribed and PCR-amplified with the Gr B–specific (uppermost panel) or Gr C-G–specific (lower two panels) primers. The PCR was stopped after the indicated number of cycles, loaded on 2% agarose gels, and stained with ethidium bromide. As a positive control, +/+ splenocytes were used after PWM-stimulation. Molecular size standards are shown on the right. (B) Five micrograms of total RNA from +/+ and mi/mi CMCs, and +/+ splenocytes stimulated with PWM were fractionated on 1% agarose gels and fixed onto nylon membranes. The PCR-amplified DNA fragment seen in (A) was used as a probe to detect the Gr B–specific (upper panel) and the Gr C-G–specific (lower panel) transcripts. Reprobing with the β-actin probe showed that equal loading had been achieved.
Expression profile of the Gr B gene and the other Grs of the murine Gr B locus in +/+ and mi/mi CMCs. (A) Serially diluted total RNA (0.5, 0.05, and 0.005 μg) from +/+ andmi/mi CMCs, and splenocytes of +/+ mice were reverse-transcribed and PCR-amplified with the Gr B–specific (uppermost panel) or Gr C-G–specific (lower two panels) primers. The PCR was stopped after the indicated number of cycles, loaded on 2% agarose gels, and stained with ethidium bromide. As a positive control, +/+ splenocytes were used after PWM-stimulation. Molecular size standards are shown on the right. (B) Five micrograms of total RNA from +/+ and mi/mi CMCs, and +/+ splenocytes stimulated with PWM were fractionated on 1% agarose gels and fixed onto nylon membranes. The PCR-amplified DNA fragment seen in (A) was used as a probe to detect the Gr B–specific (upper panel) and the Gr C-G–specific (lower panel) transcripts. Reprobing with the β-actin probe showed that equal loading had been achieved.
Impaired expression of TPH but not of HDC in mi/mi CMCs.
TPH and HDC are the rate-limiting enzyme of the serotonin and histamine synthesis, respectively29,51; both chemical mediators were stored in the basophilic granules of mast cells.34 Although the TPH gene was transcriptionally downregulated in mi/mi CMCs, the HDC gene was equally transcribed between +/+ and mi/mi CMCs (Fig 3). The proportion of the HDC mRNA+ CMCs was comparable among +/+ CMCs, mi/mi CMCs, mi/mi CMCs overexpressing +-MITF, and mi/mi CMCs overexpressingmi-MITF (Table 1). The serotonin content was significantly smaller in mi/mi CMCs than in +/+ CMCs (Table2). In contrast, the histamine content was comparable between +/+ and mi/mi CMCs (Table 2). The introduction of +-MITF cDNA to mi/mi CMCs increased the serotonin content to nearly normal levels, but the introduction ofmi-MITF cDNA to mi/mi CMCs did not. The introduction of +- or mi-MITF cDNA to mi/mi CMCs did not affect the histamine content (Table 2). Since mast cells take up serotonin from media,52 53 we next evaluated the serotonin uptake by +/+ and mi/mi CMCs. The amount of serotonin that was taken up from the media was comparable between +/+ and mi/mi CMCs (Table3).
Concentration of Serotonin and Histamine in +/+ CMCs, mi/mi CMCs, and mi/mi CMCs Overexpressing +- ormi-MITF
| Cells . | Concentration (nmol/106 cells)* . | |
|---|---|---|
| Serotonin . | Histamine . | |
| +/+ CMCs | 14.6 ± 0.3 | 2.8 ± 0.5 |
| mi/mi CMCs | 2.8 ± 0.1† | 3.8 ± 1.2 |
| mi/mi CMCs overexpressing +-MITF | 15.6 ± 1.3 | 4.1 ± 0.2 |
| mi/mi CMCs overexpressing mi-MITF | 4.8 ± 0.5† | 6.6 ± 0.6 |
| Cells . | Concentration (nmol/106 cells)* . | |
|---|---|---|
| Serotonin . | Histamine . | |
| +/+ CMCs | 14.6 ± 0.3 | 2.8 ± 0.5 |
| mi/mi CMCs | 2.8 ± 0.1† | 3.8 ± 1.2 |
| mi/mi CMCs overexpressing +-MITF | 15.6 ± 1.3 | 4.1 ± 0.2 |
| mi/mi CMCs overexpressing mi-MITF | 4.8 ± 0.5† | 6.6 ± 0.6 |
*Means ± SE of three experiments.
P < .01 by t-test when compared with the value of +/+ CMCs.
Binding of +-MITF to CANNTG motifs in the 5′-flanking sequences of the Gr B and TPH genes.
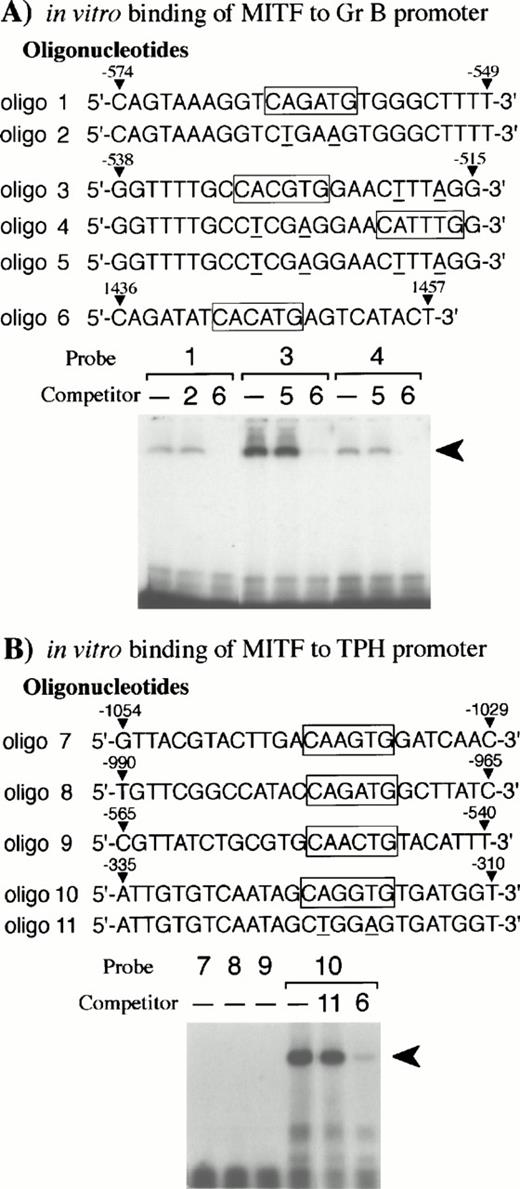
Bleackley et al54,55 reported that two positive regulatory elements (nt −682 to −427 and nt −243 to −112)48were present in the Gr B 5′-flanking region. Since the basic domain of the bHLH-Zip family recognizes the CANNTG motif,56,57 we searched for this motif in the two positive elements. We found three CANNTG motifs in the 5′ element (nt −682 to −427)48and examined whether the +-MITF protein bound each CANNTG motif by EGMSA. As shown in Fig 6A,the +-MITF protein bound all of the three CANNTG motifs. The binding was inhibited by the excess amount of oligo 6 containing a CACATG motif, but not of oligonucleotides containing mutated motifs (CAGATG between nt −563 and −558 to CTGAAG, CACGTG between nt −530 and −525 to CTCGAG, and CATTTG between nt −521 and −516 to CTTTAG)48 (Fig 6A).
EGMSA using GST-+-MITF fusion protein. (A) Six kinds of oligonucleotides (oligo 1 to 6) were synthesized: oligo 1 to 5 were derived from the Gr B gene promoter and oligo 6 from the gp49B gene promoter.50 In vitro binding of oligo 6 to +-MITF was confirmed in the other experiment (data not shown). CANNTG motifs are boxed and the mutations introduced into the motifs are shown by underlines. The labeled oligonucleotide containing the CAGATG motif (oligo 1), the CACGTG motif (oligo 3), or the CATTTG motif (oligo 4) was used as a probe. The probes were incubated with purified GST-+-MITF fusion protein in the presence or absence of unlabeled oligonucleotide competitors. To compete the in vitro binding between the three CANNTG motifs (oligo 1, 3, and 4) and GST-MITF fusion protein, excess amount of unlabeled oligo 2, oligo 5, or oligo 6 was added. The arrowhead indicates the complex obtained with the labeled oligonucleotides and GST-MITF fusion protein. (B) Five kinds of oligonucleotides (oligo 7 to 11) were synthesized based on the promoter sequence of the TPH gene. The labeled oligonucleotide containing the CAAGTG motif (oligo 7), the CAGATG motif (oligo 8), the CAACTG motif (oligo 9), or the CAGGTG motif (oligo 10) was used as a probe. Competition for the binding of GST-+-MITF to the labeled oligo 10 was also examined. The excess amount of a nonlabeled oligo 6 or an oligonucleotide mutated at the CAGGTG motif (to CTGGAG, oligo 11) was added.
EGMSA using GST-+-MITF fusion protein. (A) Six kinds of oligonucleotides (oligo 1 to 6) were synthesized: oligo 1 to 5 were derived from the Gr B gene promoter and oligo 6 from the gp49B gene promoter.50 In vitro binding of oligo 6 to +-MITF was confirmed in the other experiment (data not shown). CANNTG motifs are boxed and the mutations introduced into the motifs are shown by underlines. The labeled oligonucleotide containing the CAGATG motif (oligo 1), the CACGTG motif (oligo 3), or the CATTTG motif (oligo 4) was used as a probe. The probes were incubated with purified GST-+-MITF fusion protein in the presence or absence of unlabeled oligonucleotide competitors. To compete the in vitro binding between the three CANNTG motifs (oligo 1, 3, and 4) and GST-MITF fusion protein, excess amount of unlabeled oligo 2, oligo 5, or oligo 6 was added. The arrowhead indicates the complex obtained with the labeled oligonucleotides and GST-MITF fusion protein. (B) Five kinds of oligonucleotides (oligo 7 to 11) were synthesized based on the promoter sequence of the TPH gene. The labeled oligonucleotide containing the CAAGTG motif (oligo 7), the CAGATG motif (oligo 8), the CAACTG motif (oligo 9), or the CAGGTG motif (oligo 10) was used as a probe. Competition for the binding of GST-+-MITF to the labeled oligo 10 was also examined. The excess amount of a nonlabeled oligo 6 or an oligonucleotide mutated at the CAGGTG motif (to CTGGAG, oligo 11) was added.
The 5′-flanking sequence of the mouse TPH gene was reported by Stoll and Goldman.31 We isolated the fragment between nt −1054 and +86 (+1 shows the transcription initiation site),31which contained four CANNTG motifs. The +-MITF bound one of four CANNTG motifs (CAGGTG between nt −322 and nt −317),31 whereas the +-MITF did not bind the remaining three CANNTG motifs (CAAGTG between nt −1041 and −1036, CAGATG between nt −977 and −972, CAACTG between nt −552 and −547)31 (Fig 6B). The excess amount of oligo 6 containing a CACATG motif inhibited the binding of +-MITF, whereas the excess amount of the oligonucleotide that had the mutation at the CAGGTG motif (CAGGTG to CTGGAG) did not inhibit the binding of +-MITF (Fig 6B). The +-MITF did bind to oligo 1 but did not bind to oligo 8 (Fig 6A and B); nevertheless, both possessed the identical core sequence, CAGATG. Presumably flanking sequences affect the binding.
+-MITF directly transactivated the Gr B and TPH promoters.
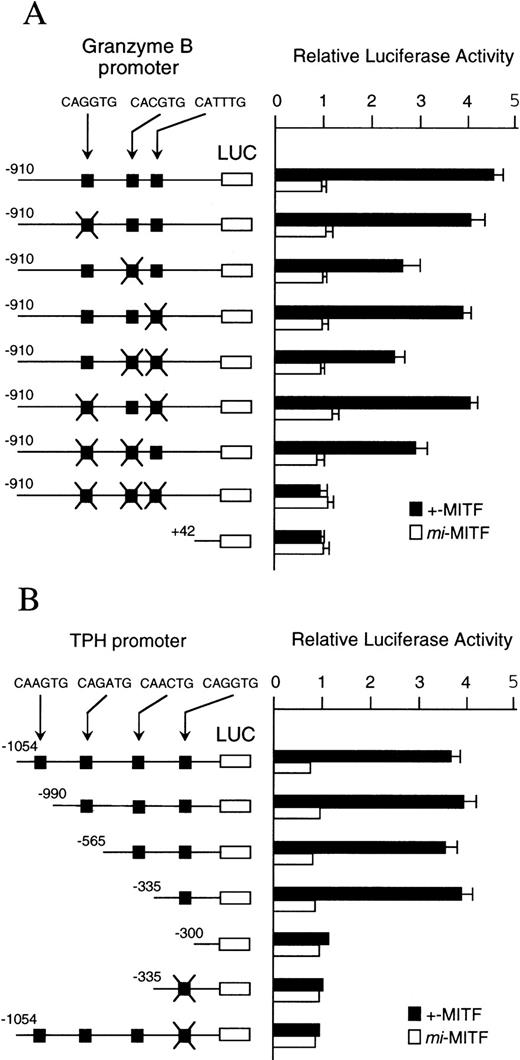
The binding specificity of +-MITF protein for three CANNTG motifs of the Gr B promoter and for one of four motifs of the TPH promoter suggested that these motifs may mediate the transactivation effect of +-MITF. We elucidated this possibility by the transient cotransfection assay. The 5′-flanking sequences of the Gr B (nt −910 to +42, +1 is the transcription start site)48 were cloned upstream of the luciferase gene. This luciferase construct was cotransfected into NIH/3T3 fibroblasts along with the expression plasmid containing +-MITF or mi-MITF cDNA. The coexpression of +-MITF increased the luciferase activity approximately fivefold (Fig7A). In contrast, coexpression ofmi-MITF did not increase luciferase activity at all. Then, we introduced mutations into the three CANNTG motifs as is the case of oligonucleotides used in EGMSA. Of the seven kinds of mutated reporter plasmids, three kinds of plasmids bearing mutations at either CACGTG motif, at both CACGTG and CATTTG motifs, or at both CAGATG and CATTTG motifs, showed significant reductions in luciferase activity. When mutations were introduced into all of the three motifs, the transactivation effect of +-MITF was abolished. When mi-MITF was expressed, no transactivation effect was detected using any mutated reporter, either. These results indicate that +-MITF transcriptionally activates the Gr B gene by recognizing three CANNTG motifs, of which the CACGTG motif appears to be particularly important.
(A) The effect of coexpression of +-MITF ormi-MITF cDNA on the luciferase activity under the control of the normal or mutated Gr B promoter. Three square black boxes represent CANNTG motifs between nt −910 and +42. The boxes with X have mutated motifs: CAGATG (nt −563 to −558) motif is mutated to CTGAAG, CACGTG (nt −530 to −525) to CTCGAG, CATTTG (nt −521 to −516) to CTTTTG. The open and filled bars represent the mean ± SE of the relative luciferase activities obtained by three independent experiments. (B) The effect of coexpression of +-MITF ormi-MITF cDNA on the luciferase activity under the control of the normal, deleted, or mutated TPH promoter. Four square black boxes represent CANNTG motifs between nt −1054 and +86. The boxes with X have a mutated motif: CAGGTG (nt −322 to −317) to CTGGAG. The data represent the mean ± SE of three experiments. In some cases, the SE was too small to be shown by the bars.
(A) The effect of coexpression of +-MITF ormi-MITF cDNA on the luciferase activity under the control of the normal or mutated Gr B promoter. Three square black boxes represent CANNTG motifs between nt −910 and +42. The boxes with X have mutated motifs: CAGATG (nt −563 to −558) motif is mutated to CTGAAG, CACGTG (nt −530 to −525) to CTCGAG, CATTTG (nt −521 to −516) to CTTTTG. The open and filled bars represent the mean ± SE of the relative luciferase activities obtained by three independent experiments. (B) The effect of coexpression of +-MITF ormi-MITF cDNA on the luciferase activity under the control of the normal, deleted, or mutated TPH promoter. Four square black boxes represent CANNTG motifs between nt −1054 and +86. The boxes with X have a mutated motif: CAGGTG (nt −322 to −317) to CTGGAG. The data represent the mean ± SE of three experiments. In some cases, the SE was too small to be shown by the bars.
To examine the transactivation effect of +-MITF for the TPH gene, the promoter region and a part of the first exon of the TPH gene (nt −1054 to +86)31 was cloned upstream of the luciferase gene. We also constructed the deleted reporter plasmids containing the TPH promoter starting from nt −990, −565, −335, or −330. The coexpression of +-MITF with the reporter plasmid containing the CAGGTG motif increased the luciferase activity (Fig 7B). In contrast, the coexpression of +-MITF and the reporter plasmid without the CAGGTG motif did not increase the luciferase activity. The mutation of the CAGGTG motif to CTGGAG completely abolished the transactivation activity of +-MITF. Themi-MITF did not transactivate any reporter plasmids (Fig 7B).
DISCUSSION
In the present study we attempted to isolate as many target genes as possible that are transcriptionally upregulated by MITF. For this purpose we prepared a subtracted cDNA library of high quality using +/+ and mi/mi CMCs. From the +/+ CMC cDNA library, the clones carrying inserts complementary to mi/mi CMC mRNAs were removed by hybridization. When we characterized the resulting subtracted cDNA library by application of Southern and Northern analyses, we found that it might contain +/+ CMC-specific clones at high frequencies. When 400 clones were randomly selected from the subtracted cDNA library and analyzed, 22 genes were found to be transcribed preferentially in +/+ CMCs: 6 corresponded to Gr B, TPH, ST2L, gp49B, MMCP-5, and -6, and 16 were unknown genes. MMCP-5 and -6 genes have already been described as +/+-specific genes and targets of MITF.5,8 9 This result enabled us to judge that the subtraction method described here was successful enough to answer our purpose.
By +-MITF introduction, EGMSA and transient cotransfection assay, we showed that MITF directly involved in transcriptional activation of the Gr B and TPH genes through recognition of the CANNTG motifs. The CAGGTG motif located between nt −322 and −31731 was essential for the activation of the TPH promoter. Reed et al58analyzed the mouse TPH promoter in the TPH gene-expressing tumor mast cell line P815-HTR. Because P-815-HTR cells expressed +-MITF (Morii E, unpublished data, May 1997), the result that the deletion between nt −343 and nt −25531 containing the CAGGTG motif decreased the promoter activity in P815-HTR cells is consistent with the present result. Pham et al59 determined the genomic structure of the murine Gr B locus on chromosome 14, where Gr B is located at the 5′ end and followed by the genes for Grs C, F, G, D, and E, cathepsin G, and MMCP-2. They also showed that all of the Grs in this locus seem to share a common regulatory pattern in natural killer (NK) cells. From these observations they proposed a model to explain the regulatory mechanism of multiple Gr gene expression. The model comprised a regulatory element, located upstream of the Gr gene cluster, and a responsive sequence for this element within each of the Gr promoters. Interaction between the two would guarantee balanced expression of the genes making up the Gr locus. They presumed this common regulatory element to be another example of a locus control region of a previous report.60 We have shown here that mutation of the binding sites for MITF reduced the expression of the Gr B promoter-reporter construct by 80%. However, this reduction is inferior to that seen in Northern blot analyses (Figs 3 and 4), where the reduction of expression appeared to be at least 100-fold or more. This inconsistency would be explained if the promoter of the Gr B gene contained, in addition to the MITF binding sites, the additional and responsive element proposed in the above model. If this was the case, then full activation of the Gr B gene would depend not only on MITF interaction with the three CANNTG motifs in the promoter region, but also on an interaction of another responsive sequence with a putative common regulatory element. Whether MITF also regulates the Gr B gene in CTLs and NK cells remains to be addressed. Stechschulte et al14 reported that mi/mi spleen cells were significantly less effective than +/+ ones when assessed for cytotoxic activity against YAC-1 cells. They attributed this impaired cytotoxity to a decreased number of large granular lymphocytes. Here we could present another reason that Gr B expression also reduced in CTLs and NK cells of mi/mi mice as is the case in mast cells.
In addition to the production of serotonin by mast cells, the uptake of exogenous serotonin by mast cells has been reported.52,53Because CMCs were cultured with FCS which contained substantial amount of serotonin,53 the different serotonin content between +/+ CMCs and mi/mi CMCs may be ascribed to their different potential of serotonin incorporation. Since the amount of serotonin uptake by +/+ CMCs was comparable to that of mi/mi CMCs (Table3), the decreased serotonin content in mi/mi CMCs was due to the impaired synthesis, which may be caused by the deficient transcription of the TPH gene. However, there is a possibility that it was caused by deficient expression of c-kit, since Ziegler et al61 reported that recombinant SCF (100 ng/mL) increased the serotonin content in CMCs and since we previously reported that the c-kit expression and the reactivity to SCF were impaired inmi/mi CMCs.7 16 Although the SCF concentration in our experimental condition appeared to be too low to increase serotonin synthesis, we could not completely rule out the possibility.
Histamine is also a chemical mediator that is preformed and stored in the basophilic granules of mast cells.34 Histamine content and the amount of HDC mRNA were comparable between +/+ andmi/mi CMCs. The +-MITF did not appear to be involved in the HDC expression and in the histamine synthesis. We previously reported that the content of histamine per skin mast cell decreased in mi/mimice.17 The skin mast cells contain heparin proteoglycan to which histamine is considered to be bound, and the content of heparin per mi/mi skin mast cell also decreased.17 The decreased histamine content of mi/mi skin mast cells may be due to the decrease of heparin proteoglycan. In contrast to skin mast cells, CMCs did not contain heparin.20,62This may explain that histamine content decreased inmi/mi skin mast cells but not in mi/mi CMCs. However, it must be pointed out that the histamine content of +/+ CMCs was only 1% that of +/+ skin mast cells.22 Heparin proteoglycan may be necessary to store histamine in high concentration, as in the case of +/+ skin mast cells.
We and others showed that six genes encoding tyrosinase,9,63 tyrosinase-related protein-1,64MMCP-6,5,8 MMCP-5,9 c-kit,7and p75 NGF receptor10 are targets of MITF. Here we have identified the Gr B and TPH genes as a novel target of MITF. According to our estimates, the subtracted cDNA library described in this study contains 240 independent clones whose expression is downregulated inmi/mi CMCs. Thus, the four genes, Gr B, TPH, ST2L, and gp49B, may represent only a small proportion of the total number of possible genes under the control of MITF. Further investigation of the subtracted cDNA library will allow the identification of a greater number of MITF target genes, and permit a critical assessment of the biological significance of the transcriptional cascades mediated by MITF.
ACKNOWLEDGMENT
The authors thank M. Kobori (Yamanouchi Pharmaceutical Company Ltd, Tsukuba, Japan) for technical advice on the preparation of subtracted cDNA library, Dr T. Nakaji (Kobe University, Kobe, Japan) for screening of the plasmids and DNA sequencing, Dr S. Nagata (Osaka University, Osaka, Japan) for pEF-BOS, Dr K. Nakajima (Osaka University) for pSPLuc, and Drs S. Nomura (Osaka University) and M. Yamamoto (Tsukuba University, Tsukuba, Japan) for valuable discussions.
A.I. and E.M. contributed equally to this work.
Supported by grants from the Ministry of Education, Science and Culture, Japan and grants from the Osaka Cancer Society, the Naito Foundation, Ryoichi Naito Foundation, and Kenko-kagaku Foundation.
Address reprint requests to Hiroshi Nojima, PhD, Department of Molecular Genetics, Research Institute for Microbial Diseases, Osaka University, Yamada-oka 3-1, Suita, Osaka 565, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal