Hemophilia B is a leading target for gene therapy because current therapy is not optimal. Hence, a murine model of factor IX (F. IX) deficiency was generated to develop gene therapy strategies for hemophilia B. A targeting vector was created by replacing a 3.2-kb segment of the gene encompassing the catalytic domain with a phosphoglycerokinase promoter-driven neomycin resistant (neor) gene cassette. The transfected embryonic stem cell clones generated chimeric male mice, and germ line transmission of the inactivated F. IX gene was observed in their offsprings. Southern analysis confirmed the mutant genotype in hemizygous male and carrier female mice. F. IX transcripts were not detected in liver RNA isolated from hemizygous mice, and lower levels of F. IX mRNA were noted in carrier female mice when compared with those of normal litter mates. As expected, the mean F. IX coagulant titer of affected male mice was 2.8 U/dL (n = 10), while the mean F. IX titer of carrier female mice was 35 U/dL (n = 14), compared with 69 U/dL (n = 9) for the normal female mice and 92 U/dL (n = 22) for normal male and female litter mates. Further, the tail bleeding time of hemizygous mice was markedly prolonged (>3 hours) compared with those of normal and carrier female litter mates (15 to 20 minutes). Seven of 19 affected male mice died of exsanguination after tail snipping, and two affected mice died of umbilical cord bleeding. Currently, there are 10 affected mice surviving at 4 months of age. Aside from the factor IX defect, the carrier female and hemizygous male mice had no liver pathology by histologic examination, were fertile, and transmitted the F. IX gene mutation in the expected Mendelian frequency. Taken together, we have generated a F. IX knockout mouse for evaluation of novel gene therapy strategies for hemophilia B.
HEMOPHILIA B, AN X-LINKED clotting disorder that affects one in 30,000 males, is caused by a deficiency of functional clotting factor IX (F. IX) protein.1-3 The human F. IX gene is a single copy gene residing on X-chromosome q 27.1.4 The complete sequence has been determined5 and is at least 34 kb in size, consisting of eight exons (a to h) and seven introns. Exon a encodes the signal peptide, exons b and c encode the propeptide and gamma-carboxy glutamyl domain, exons d and e encode epidermal growth factor (EGF)-like domains, exon f encodes the activation domain where proteolytic processing of the mature F. IX molecule occurs, and exons g and h encode the catalytic domain of the F. IX protein. The human F. IX cDNA is about 2.8 kb in size, and has a long 3′ untranslated tail, the function of which is still not known. The mouse F. IX cDNA sequence is known6 and there exists a 68% sequence homology between mouse and human F.IX protein at the amino acid level. Most of the human mutations are found within the 2.2-kb coding region and are due to either transitions (from purine to purine or pyrimidine to pyrimidine) or transversion (purine to pyrimidine) at the CpG dinucleotide region and other sites, while deletions and insertions account for only 15% of all factor IX mutations.7-10 The catalytic region comprises the largest domain and accounts for the largest number of F. IX mutations responsible for the moderately severe to severe forms of hemophilia B in humans. Because hemophilia B is a prospective target for gene therapy, we generated a F.IX knock-out mouse model for testing of gene therapy vectors and for further study of the F.IX gene function.
MATERIALS AND METHODS
Mouse F. IX gene isolation and construction of the targeting vector.
An 18.6-kb mouse F. IX genomic clone was isolated from a 129/Sv lambda Fix II phage library (Stratagene, La Jolla, CA) by screening with a mouse factor IX cDNA11 and it contained three exons corresponding to the last three exons (activation domain and catalytic domain) of the human F. IX gene. From the genomic clone, 7.0-kb Xba I, 8.0-kb Xba I, and 11.6-kb XhoI-Not I fragments were respectively subcloned into the pBluescript II SK vector and mapped using standard techniques. The catalytic domain (exon g and h) was disrupted by its replacement with a neomycin resistance gene cassette driven by a phosphoglycerol kinase promoter (PGK-neor).12 The targeting vector was made from the 11.6-kb lambda clone by replacement of a 3.2-kbBamHI fragment, which contained exon g and exon h with the 1.6-kb Xho I fragment of PGK-neor. A negative selection marker13 was created by subcloning a 2.1-kbXho I fragment containing the HSV-tk gene from pXhoIMC1tk (gift from Dr Paul Hasty, M.D. Anderson Cancer Center, Houston, TX) into the Xho I site of the 11.6-kb lambda clone. Hence, the resultant vector had a 5.7-kb region of homology with the F.IX gene at the 5′ end and a 2.6-kb homology at the 3′ end, a PGK-neor cassette replacing exons g and h, with a HSV-tk cassette at its Xho I site.
Generation of the F. IX-deficient mice.
The targeting vector was linearized by Not I digestion, electroporated into the CCE line of embryonic stem (ES) cells (from K. Lyons, UCLA, Los Angeles, CA) (1 × 107cells), and selected with G418 (0.4 mg/mL) and ganciclovir (2 μmol/L).14-16 The G418 resistant colonies were selected, expanded, and screened for homologous recombination by Southern blot technique.17 Genomic DNA from the ES cell clones was digested with Xba I, electrophoresed on a 0.6% agarose gel, and transferred to a nylon membrane. The membrane was hybridized with a 0.5-kb 5′ external probe (Fig 1, Probe A) isolated by XhoI digestion of the pBluescript II SK lambda 8 vector. The sizes of theXbaI fragments of the wild-type (WT) gene and the mutant (MT) allele were 8.0-kb and 6.4-kb, respectively (Fig 2, Probe A). AnHindIII-NotI 1.5-kb 3′ probe (Fig 1, Probe B) also confirmed the sizes of 3.9 kb for WT and 4.6 kb for the MT after digestion of the DNA with Xba I (Fig 2, Probe B). The presence of the single integration event was confirmed by hybridization with aPst I-Xba I 0.637-kb neo fragment without the endogenous phosphoglycerol kinase promoter (data not shown). The targeted ES cell clones were then injected into blastocysts derived from C57BL6/J mice, and transferred into the uteri of pseudopregnant CD-1 female (USC Transgenic Core Facility). The chimeric males were mated with C57BL6/J females, and germ-line transmission of the ES cell-derived phenotype was determined by the presence of an agouti coat color and further confirmed by Southern analysis of DNA. The carrier female mice of the first (F1) generation were again mated with C57BL6/J males to obtain males with F. IX null alleles, with the genotype again confirmed by Southern analysis. The genotypes of the carrier female and hemizygous male mice were determined by digestion of the tail DNA with XbaI and hybridized with the same 5′ external (Probe A) and 3′ probe (Probe B) as described earlier.
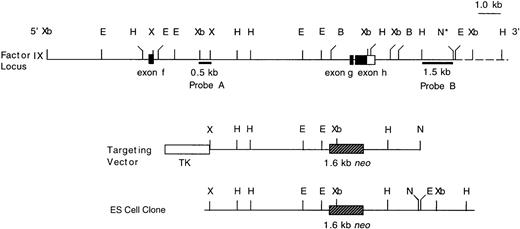
Targeted disruption of the murine F. IX gene by homologous recombination. An 18.6-kb genomic map of the mouse F. IX gene showing exons f, g, and h. The targeting vector was made from the 11.6-kb Xho I-Not I fragment of the lambda clone. The targeted allele contained a neo gene inserted into exon g and exon h. The 3.2-kb BamHI fragment covering exons g and h and the introns was replaced by a 1.6-kb neor cassette. The 2.1-kb thymidine kinase cassette was cloned into the Xho I site. A 0.5-kb fragment (Probe A), external to the Xho I site of the targeting vector, and an HindIII-Not I 1.5-kb fragment (Probe B) were used to screen the ES cell clones and mouse tail DNA. A 637-bp neo gene probe was also used to screen the same clones (data not shown). B, BamHI; E, EcoRI; H,HindIII; N, Not I; X, Xho I; Xb, Xba I; TK, thymidine kinase gene; neo, neomycin gene. The dotted line represents a portion of the genomic map, which was not included in the λ clone, but was identified by subsequent mapping after Southern blot hybridization of ES cell and mouse DNA. The * (asterisk) on theNot I site denotes that it is not part of the clone λ.
Targeted disruption of the murine F. IX gene by homologous recombination. An 18.6-kb genomic map of the mouse F. IX gene showing exons f, g, and h. The targeting vector was made from the 11.6-kb Xho I-Not I fragment of the lambda clone. The targeted allele contained a neo gene inserted into exon g and exon h. The 3.2-kb BamHI fragment covering exons g and h and the introns was replaced by a 1.6-kb neor cassette. The 2.1-kb thymidine kinase cassette was cloned into the Xho I site. A 0.5-kb fragment (Probe A), external to the Xho I site of the targeting vector, and an HindIII-Not I 1.5-kb fragment (Probe B) were used to screen the ES cell clones and mouse tail DNA. A 637-bp neo gene probe was also used to screen the same clones (data not shown). B, BamHI; E, EcoRI; H,HindIII; N, Not I; X, Xho I; Xb, Xba I; TK, thymidine kinase gene; neo, neomycin gene. The dotted line represents a portion of the genomic map, which was not included in the λ clone, but was identified by subsequent mapping after Southern blot hybridization of ES cell and mouse DNA. The * (asterisk) on theNot I site denotes that it is not part of the clone λ.
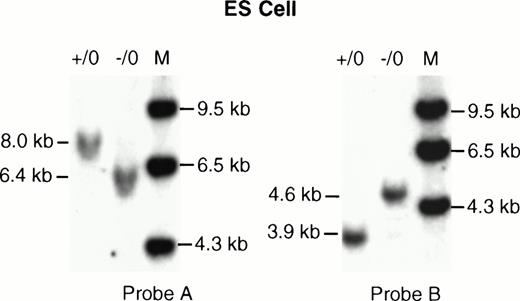
Southern analysis of the ES cell clones. (A) The ES cell clone (D11) and 129/Sv mouse DNA were digested with Xba I and hybridized with probe A. The wild-type clone (+/0) showed an 8.0-kb fragment, the recombinant disrupted clone (−/0) showed a 6.4-kb band. (B) The same DNA samples were digested with XbaI and hybridized with probe B. The wild-type clone (+/0) showed a 3.9-kb band, while the mutant allele (−/0) showed a 4.6-kb band.
Southern analysis of the ES cell clones. (A) The ES cell clone (D11) and 129/Sv mouse DNA were digested with Xba I and hybridized with probe A. The wild-type clone (+/0) showed an 8.0-kb fragment, the recombinant disrupted clone (−/0) showed a 6.4-kb band. (B) The same DNA samples were digested with XbaI and hybridized with probe B. The wild-type clone (+/0) showed a 3.9-kb band, while the mutant allele (−/0) showed a 4.6-kb band.
F. IX coagulant assay.
Mouse plasma coagulant F.IX titer was measured using a modification of the kaolin partial thromboplastin time technique,18-20 with human F.IX-deficient plasma as substrate (GK 927327P1; George King Biomedical Co, Overland Park, KS). Blood samples were collected by tail snipping from 4-week-old and 6-month-old mice and mixed with 9:1 vol/vol whole blood: 3.8% sodium citrate. Reference standard plasma (F.IX coagulant titer: 97 U/dL; Pacific Hemostasis, Huntersville, NC) was used as the standard for determination of the F.IX coagulant titer.
Histopathologic examination of hematoxylin-esoin stained sections from formalin-fixed liver tissue was conducted.
Northern analysis.
Total RNA was prepared from liver tissue of 4-week-old and 6-month-old mice, using the one-step method with RNAzol reagent (Telstar Inc, Friendswood, TX) according to the manufacturer's instructions. Twenty-five micrograms of total RNA was electrophoresed on 0.8% agarose gel containing 6% formaldehyde, transferred to nylon membrane (Amersham, Arlington Heights, IL) and cross-linked to the membrane by ultraviolet (UV) light (Stratalinker, Stratagene). The membrane was prehybridized at 65°C with Rapid-Hyb buffer (Amersham) for 15 minutes and hybridized with a 644-bp radiolabeled probe, which was isolated from mouse F. IX cDNA by digestion with EcoRV and Xho I. The fragment was similar to the 537-bp probe, which has been shown to give a better signal than the whole cDNA.21 The mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) probe was used as internal control.
Reverse transcription-coupled polymerase chain reaction (RT-PCR) analysis.
Total RNA was extracted from liver tissue of 4-week-old and 6-month-old mice using RNAzol reagent. Briefly, 5 μg of total RNA was treated with Superscript II Rnase H-Reverse Transcriptase (GIBCO-BRL, Gaithersburg, MD) in a 20-μL reaction volume with random hexamers. PCR amplifications were done with Gene Amp PCR system 9600 (Perkin-Elmer, Norwalk, CT) and Taq DNA polymerase (Qiagen Inc, Santa Clarita, CA), using 2.0 μL of cDNA solution in an incubation volume of 50 μL. PCR amplifications were performed at 94°C for 2 minutes followed by 30 cycles at 94°C for 1 minute, 55°C for 45 seconds, 72°C for 45 seconds, and final extension of the PCR product at 72°C for 7 minutes. Mouse factor IX primers were chosen from exon f (5′-GTCACTGAAAGTAGTGAA-3′) and exon h antisense (5′-GACGTACCGGGAAACCTTAGT-3′). The mouse 18S ribosomal primer (an internal control) was purchased from Ambion, Austin, TX. The PCR products were analyzed by loading 4 μL for F. IX and 2 μL for 18S from 50 μL reaction volume on a 1.6% agarose gel and visualized by ethidium bromide staining.
RESULTS
Targeted inactivation of the mouse F. IX gene.
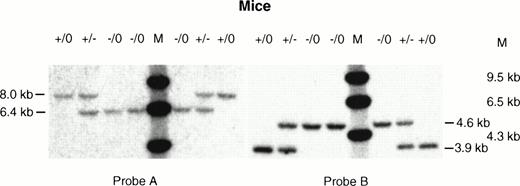
A targeting vector was designed using the genomic clone of the mouse F. IX gene isolated from the 129/Sv mouse genomic library. This vector carries a 3.2-kb deletion spanning exon g and exon h. Both the exons and part of the introns were replaced by a 1.6-kb neomycin resistance gene as a positive selection marker (Fig1). Negative selection against random integration was conferred by a herpes simplex virus thymidine kinase (HSV-tk) gene.13 ES cells were electroporated with the targeting vector and 200 double-resistant colonies were picked and screened by Southern blot analysis. Nine positive targeted clones were identified based on the predicted size of the targeted allele (Fig2, Probe A and Probe B). Hybridization with the neor probe eliminated the possibility of an additional integration event of the targeting vector (data not shown). One of the selected clones (D11) was then injected into C57Bl6/J blastocysts, which generated one chimeric female and two chimeric male mice. Germline carrier females were obtained by mating the chimeric males with C57Bl6/J females. Because the F.IX gene is located on the X-chromosome, only the heterozygous F. IX carrier female mice with mutant alleles were obtained in the first generation. The carrier females, which were mated with normal C57Bl6/J male mice, transmitted the mutant F. IX allele to the male progeny generating affected hemizygous male mice (Fig 3, Probe A and Probe B) (Table 1). The F. IX-deficient male mice were again mated with normal mice and the mutant allele was transmitted to 50% of their progenies from four different matings (Table 2). Further, histologic examination of the liver samples from two different litters generated from this mating showed normal liver architecture with no liver pathology (data not shown).
Southern analysis of tail DNA from hemizygous male and carrier female mice. Tail DNA from normal male (+/0), affected male (−/0), and carrier female (+/−) mice were digested withXba I and hybridized with (A) probe A and (B) probe B. The expected sizes of the bands were the same as those detected in the ES cell clones (see Fig 2).
Southern analysis of tail DNA from hemizygous male and carrier female mice. Tail DNA from normal male (+/0), affected male (−/0), and carrier female (+/−) mice were digested withXba I and hybridized with (A) probe A and (B) probe B. The expected sizes of the bands were the same as those detected in the ES cell clones (see Fig 2).
Generation of Mutant F. IX Hemizygous Male Mice
| Carrier Female (F1) D11 . | Normal Male . | Litter Size . | F2 . | |||
|---|---|---|---|---|---|---|
| Hemophilic Male . | Carrier Female . | Wild-Type . | ||||
| Male . | Female . | |||||
| #39 | C57BI6 | 7 | 2 | 1 | 2 | 2 |
| #40 | C57BI6 | 9 | — | 5 | 2 | 2 |
| #46 | C57BI6 | 8 | 3 | — | 3 | 2 |
| #58 | C57BI6 | 8 | 3 | 1 | 3 | 1 |
| #62 | C57BI6 | 7 | 3 | 1 | 1 | 2 |
| #66 | C57BI6 | 6 | 3 | 1 | 1 | 1 |
| Total | 45 | 14 | 9 | 12 | 10 | |
| Carrier Female (F1) D11 . | Normal Male . | Litter Size . | F2 . | |||
|---|---|---|---|---|---|---|
| Hemophilic Male . | Carrier Female . | Wild-Type . | ||||
| Male . | Female . | |||||
| #39 | C57BI6 | 7 | 2 | 1 | 2 | 2 |
| #40 | C57BI6 | 9 | — | 5 | 2 | 2 |
| #46 | C57BI6 | 8 | 3 | — | 3 | 2 |
| #58 | C57BI6 | 8 | 3 | 1 | 3 | 1 |
| #62 | C57BI6 | 7 | 3 | 1 | 1 | 2 |
| #66 | C57BI6 | 6 | 3 | 1 | 1 | 1 |
| Total | 45 | 14 | 9 | 12 | 10 | |
Generation of Carrier Females From Hemophilic Male Mice
| Hemophilic Male (F2) . | Normal Female . | Litter Size . | F3 . | |
|---|---|---|---|---|
| Carrier Female . | Wild-Type Male . | |||
| #108 | C57BI6/J | 7 | 5 | 2 |
| #107 | C57BI6/J | 7 | 2 | 5 |
| #118 | C57BI6/J | 8 | 5 | 3 |
| #46-12 | C57BI6/J | 7 | 2 | 5 |
| Total | 29 | 14 | 15 | |
| Hemophilic Male (F2) . | Normal Female . | Litter Size . | F3 . | |
|---|---|---|---|---|
| Carrier Female . | Wild-Type Male . | |||
| #108 | C57BI6/J | 7 | 5 | 2 |
| #107 | C57BI6/J | 7 | 2 | 5 |
| #118 | C57BI6/J | 8 | 5 | 3 |
| #46-12 | C57BI6/J | 7 | 2 | 5 |
| Total | 29 | 14 | 15 | |
F. IX transcript levels in normal, carrier, and affected mice.
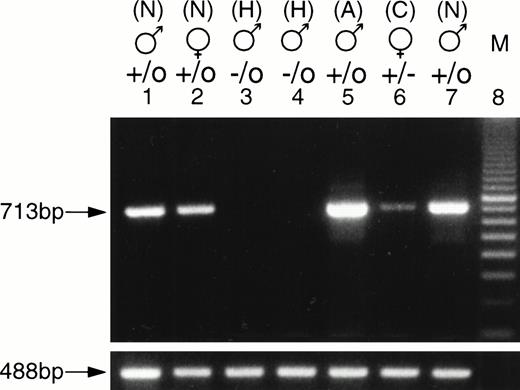
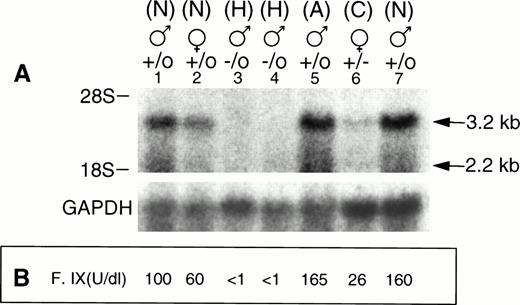
To evaluate the expression of the F. IX gene in the F. IX knockout mice, RT-PCR was done using total RNA extracted from 4-week-old carrier female, hemizygous male, littermate control and 6-month-old normal mice. The PCR amplified product of 713 bp from normal 4-week-old male and female mice was comparable to the adult level F. IX PCR product (Fig 4). The carrier female littermate showed a reduced level of F. IX PCR product, while the affected hemizygous male showed no detectable PCR product. Northern blot analysis also showed the major 3.2 kb and minor 2.2 kb transcripts of F. IX. As expected, affected male mice had no detectable F. IX mRNA, while the carrier female had reduced F. IX mRNA levels (Fig 5A). The results were normalized relative to GAPDH levels (Table 3).
RT-PCR analysis of the F. IX transcripts. Total RNA was isolated from the livers of 4-week-old carrier female, affected male, normal litter mate, and normal adult male mice. PCR amplifications were performed for 30 cycles under the following conditions: 94°C for 1 minute, 55°C for 45 seconds, and 72°C for 45 seconds using an F. IX exon f sense primer and an exon h antisense primer. The length of the PCR product corresponding to the exon f and exon h portion of the F. IX transcripts is 713-bp (lanes 1 to 7). Lanes 1 and 7, nos. 90 and 95, normal (N) males; lane 2, no. 92, normal (N) female; lanes 3 and 4, nos. 89 and 91, hemophilic (H) males; lane 5, normal 6-month-old adult (A) male, lane 6, no. 93, carrier (C) female; lane 8, 100-bp ladder; mouse 18S ribosomal primers were used as loading control. The PCR fragment obtained by amplification of the 18S rRNA transcript for all samples is 488 bp and was shown below the corresponding lanes.
RT-PCR analysis of the F. IX transcripts. Total RNA was isolated from the livers of 4-week-old carrier female, affected male, normal litter mate, and normal adult male mice. PCR amplifications were performed for 30 cycles under the following conditions: 94°C for 1 minute, 55°C for 45 seconds, and 72°C for 45 seconds using an F. IX exon f sense primer and an exon h antisense primer. The length of the PCR product corresponding to the exon f and exon h portion of the F. IX transcripts is 713-bp (lanes 1 to 7). Lanes 1 and 7, nos. 90 and 95, normal (N) males; lane 2, no. 92, normal (N) female; lanes 3 and 4, nos. 89 and 91, hemophilic (H) males; lane 5, normal 6-month-old adult (A) male, lane 6, no. 93, carrier (C) female; lane 8, 100-bp ladder; mouse 18S ribosomal primers were used as loading control. The PCR fragment obtained by amplification of the 18S rRNA transcript for all samples is 488 bp and was shown below the corresponding lanes.
(A) Northern analysis of the F. IX transcripts. Total liver RNA (25 μg) samples from 4-week-old carrier female, hemizygous male, normal litter mate, and normal adult male mice were loaded into 0.8% agarose-formaldehyde gel and transferred to a nylon membrane. The membrane was hybridized with a 0.644-kb factor IX cDNA as described in Materials and Methods. The normal sizes of the F. IX transcripts are 3.2 kb and 2.2 kb. Lanes 1 and 7, nos. 90 and 95, normal (N) males; lane 2, no. 92, normal (N) female; lanes 3 and 4, nos. 89 and 91, hemophilic (H) males; lane 5, control 6-month-old adult (A) male; lane 6, no. 93, carrier (C) female; below, hybridization of the filter with a GAPDH probe as internal standard. (B) Plasma F. IX coagulant titer, expressed as U/dL, of carrier females, hemizygous male, and normal mice are shown in corresponding lanes.
(A) Northern analysis of the F. IX transcripts. Total liver RNA (25 μg) samples from 4-week-old carrier female, hemizygous male, normal litter mate, and normal adult male mice were loaded into 0.8% agarose-formaldehyde gel and transferred to a nylon membrane. The membrane was hybridized with a 0.644-kb factor IX cDNA as described in Materials and Methods. The normal sizes of the F. IX transcripts are 3.2 kb and 2.2 kb. Lanes 1 and 7, nos. 90 and 95, normal (N) males; lane 2, no. 92, normal (N) female; lanes 3 and 4, nos. 89 and 91, hemophilic (H) males; lane 5, control 6-month-old adult (A) male; lane 6, no. 93, carrier (C) female; below, hybridization of the filter with a GAPDH probe as internal standard. (B) Plasma F. IX coagulant titer, expressed as U/dL, of carrier females, hemizygous male, and normal mice are shown in corresponding lanes.
Quantitation of F. IX Message by Laser Densitometric Scanning
| Lane . | Area (AU × mm) . | Normalized Message (1):(2) . | |
|---|---|---|---|
| F. IX Transcript (1) . | GAPDH Transcript (2) . | ||
| 1 | 2.25 | 0.58 | 3.87 |
| 2 | 1.22 | 0.59 | 2.06 |
| 3 | 0.23 | 0.93 | 0.24 |
| 4 | 0.23 | 0.73 | 0.31 |
| 5 | 2.48 | 0.69 | 3.59 |
| 6 | 0.83 | 1.62 | 0.51 |
| 7 | 2.95 | 1.54 | 1.91 |
| Lane . | Area (AU × mm) . | Normalized Message (1):(2) . | |
|---|---|---|---|
| F. IX Transcript (1) . | GAPDH Transcript (2) . | ||
| 1 | 2.25 | 0.58 | 3.87 |
| 2 | 1.22 | 0.59 | 2.06 |
| 3 | 0.23 | 0.93 | 0.24 |
| 4 | 0.23 | 0.73 | 0.31 |
| 5 | 2.48 | 0.69 | 3.59 |
| 6 | 0.83 | 1.62 | 0.51 |
| 7 | 2.95 | 1.54 | 1.91 |
All of the lanes from 1 to 7 of Fig 5A were scanned by Laser Densitometer (Ultroscan XL; LKB, Bromma, Sweden). The F. IX major transcript band was considered in each case. The area for the background level was 0.20. AU, absorbance unit, height is expressed in AU; position is expressed in mm.
Phenotypic analysis of F. IX-deficient mice.
In most cases, the number of pups born from each litter was normal (six to eight pups), and the litter mates had no structural abnormalities. Two affected male mice died 1 day after birth of umbilical cord hemorrhage. To confirm the occurrence of F. IX gene inactivation, tail bleeding time and plasma F. IX coagulant titers were measured in hemizygous males, carrier females, and normal litter mates at 1 month of age (Fig 5B). The normal littermates, normal female mice, and carrier female mice had clotting times of 15 to 20 minutes after tail snipping and mean plasma F. IX titers of 92, 69, and 35 U/dL, respectively. The mean F. IX titer of carrier females was significantly lower than those of normal female mice (P < .003). In contrast, the affected male mice had tail clotting times of more than 3 hours and a mean plasma F. IX titer of 2.8 U/dL (Table 4). Seven of 19 affected male mice died of excessive blood loss after tail snipping, and two affected mice died at 2 days of age of umbilical cord bleeding. Currently, 10 affected mice are surviving at 4 months of age.
F. IX Coagulant Assays in Normal, Carrier Female, and Affected Male Mice
| Sample . | No. . | Factor IX Coagulant Titer, U/dL . | |
|---|---|---|---|
| Mean ± SD . | Range . | ||
| Normal (males and females) | 22 | 92.0 ± 30.0 | 60-165 |
| Normal females | 9 | 69.0 ± 15.9 | 60-97 |
| Carrier females | 14 | 35.0 ± 7.9 | 26-49 |
| Affected males | 10 | 2.8 ± 1.5 | <1-63-150 |
| Sample . | No. . | Factor IX Coagulant Titer, U/dL . | |
|---|---|---|---|
| Mean ± SD . | Range . | ||
| Normal (males and females) | 22 | 92.0 ± 30.0 | 60-165 |
| Normal females | 9 | 69.0 ± 15.9 | 60-97 |
| Carrier females | 14 | 35.0 ± 7.9 | 26-49 |
| Affected males | 10 | 2.8 ± 1.5 | <1-63-150 |
Abbreviation: SD, standard deviation.
Although appropriate care was taken to obtain freely flowing blood, the coagulant activity detected may reflect the activation of the coagulation cascade during blood collection from contamination with tissue fluid.
DISCUSSION
Hemophilia B is a leading target for somatic gene therapy because current therapy is suboptimal, and this clotting disorder would be an excellent model for gene transfer strategies requiring systemic delivery of gene products. The clinical manifestations of hemophilia B may be mild, moderate, or severe. Persons with severe F. IX deficiency have plasma levels of <1 U F. IX/dL and develop frequent spontaneous hemarthroses, which can be crippling, and are susceptible to life-threatening hemorrhage, which would be fatal if untreated. Patients with F. IX levels of 2 to 5 U/dL have moderately severe hemophilia B, while patients with 5 to 30 U/dL have mild hemophilia B and have prolonged bleeding only after surgery or severe trauma.22 F. IX replacement therapy is the mainstay of treatment, requiring repeated transfusions of plasma-derived and recently recombinant F. IX preparations. Transfusion of blood-derived F. IX products is associated with the risk of viral transmission including human immunodeficiency virus (HIV)-1 and hepatitis viruses. Plasma-derived, as well as recombinant F. IX preparations, are costly and are not affordable in 80% of the world. Hence, therapy is frequently reactive, and quality of life is impaired without sufficient replacement therapy. In recent years, successful albeit transient gene therapy approaches have been reported using adenoviral, retroviral vectors, or adeno-associated viral vectors.18-20,23-31There has been limited success in delivering the human F. IX gene with retroviral vectors due to inefficient gene transfer,26 and failure of repeated adenoviral vector infusions due to immune responses against viral gene products have been reported.32 Initial promising progress has been recently reported with recombinant adeno-associated viral (rAAV) vectors.33
The canine hemophilia B model has long been used to test the safety and efficacy of F. IX concentrates, and recently, of adenoviral F. IX vectors.3,26 29 These animals are expensive to breed and are used for the testing of F. IX therapies before a clinical trial. A mouse model of hemophilia B would be an alternative animal model for testing of various gene therapy strategies, as mice are inexpensive, easy to breed, and have a much shorter gestational period than the dog.
In this study, we report the generation of a mouse model of hemophilia B by targeted inactivation of the mouse F. IX gene. The catalytic domain, including both exons g and h of the mouse F. IX gene, was selected for targeted disruption because mutations in this domain account for the largest number of cases of hemophilia B. Two other groups have reported successful generation of F. IX knock-out mice. Wang et al34 generated an F. IX-deficient mouse using a similar approach by targeted inactivation of exon h of the F. IX gene. In contrast, Lin et al35 used the plug-socket gene targeting method to generate the hemophilia B mouse, wherein a functional neomycin gene and a partially deleted hypoxanthine phosphoribosyl transferase minigene replaced the promoter through exon 3 of the F. IX gene. In the latter mouse model, the frequency of the hemizygous phenotype was only 41%, which is less than the expected frequency for affected males. In our study (Table 1), the frequency of male offsprings with the F. IX mutation was 50%. This finding confirms that a F. IX mutation within the catalytic domain is not embryologically lethal.34 Moreover, we provide further characterization of the hemophilic phenotype and additional information relating to the fertility and postnatal survival from two generations of hemophilic mice (Table 2), which was not described previously. The affected male mice were not distinguishable from the carrier or WT litter mates on the basis of size, activity, or fertility. Histologic examination of liver sections from affected male mice showed absence of liver pathology. Southern analysis confirmed the genotypes of the hemizygous male and carrier female mice. F. IX transcripts were not detected in liver RNA isolated from the hemizygous mice, while lower levels of F. IX mRNA were noted in liver RNA from carrier female mice compared with those of normal litter mates. Targeted disruption of the catalytic domain of the murine F. IX gene resulted in the creation of a murine model of hemophilia B with the affected male mice having a phenotype of severe to moderately severe hemophilia B. Further, carrier female mice had lower F. IX titers than normal litter mates. Seven of 19 affected mice died of exsanguination after tail snipping, two affected mice died of umbilical cord bleeding, and 10 affected mice are alive at 4 months of age. Taken together, we confirm that targeted disruption of the catalytic domain of the F. IX gene results in the generation of a mouse model for severe to moderately severe hemophilia B, which provides a valuable tool for studying the function of the F. IX gene and for developing novel gene therapy strategies for hemophilia B.
Our studies are limited to the genotypic and phenotypic analysis of F. IX gene expression in newborn, 4-week-old, and 4-month-old knock-out mice. Future studies will analyze F. IX gene expression with age in the affected and carrier female mice, compared with normal litter mates. Further, transgenic animals expressing F. IX constructs driven by tissue-specific promoters would be mated with the F. IX-deficient mice to test the efficiency of the gene delivery system in rescuing the bleeding phenotype in a transgenic setting. Studies are also in progress to test the efficacy of various retro-, adeno-, and adeno-associated vectors bearing human F. IX constructs driven by tissue-specific or virus-based promoter/enhancers both in vivo and ex vivo. Finally, the murine model of severe hemophilia B could be used for testing the safety and efficacy of new F. IX concentrates and in studies of immunologic tolerance.
ACKNOWLEDGMENT
The authors are grateful to D.H. Zhu for technical assistance in the ES cell cultures and to Dr F.L. Hall for helpful suggestions in writing this manuscript.
Supported in parts by the USC Center for Liver Diseases, Grant No. DK-93-024 from the National Institute of Digestive, Diabetes, and Kidney Diseases (awarded to E.M.G.) (Pilot/Feasibility Project #4), Grant No. HL53713 from the National Heart Lung and Blood Institute (awarded to K.K.), Grant No. HD22416 from the Child Health and Human Development Institute (awarded to R.E.M.), the National Institutes of Health, and in part by a grant from Genetic Therapy Inc/Novartis (awarded to W.F.A.).
Address reprint requests to Erlinda M. Gordon, MD, 1441 Eastlake Ave, MS# 73, Norris Cancer Hospital, Rm 609, Los Angeles, CA 90033.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal