CD8+ T lymphocytes play a pivotal role in controlling human immunodeficiency virus (HIV)-1 replication in vivo. We have performed four-color flow cytometric analysis of CD8+peripheral blood lymphocytes (PBL) from 21 HIV-1 seronegative and 103 seropositive individuals to explore the phenotypic heterogeneity of CD8β-chain expression on CD8+ T lymphocytes and to clarify how its expression on CD8+ T lymphocytes may relate to acquired immunodeficiency syndrome (AIDS) clinical progression. We showed that the single monoclonal antibody (MoAb) 2ST8-5H7, directed against the CD8αβ-heterodimer, identifies CD8+ T lymphocytes as effectively as the conventional combination of anti-CD3 and anti-CD8α antibodies. However, we detected a significantly lower mean fluorescence (MF) of anti-CD8αβ staining on PBL from HIV-1 seropositive donors as compared with seronegative donors. In fact, CD8+ T lymphocytes from HIV-1–infected individuals with the lowest CD4 counts showed the lowest levels of CD8αβ MF. To explore further this change in CD8αβ expression, we assessed the expression of 14 different cell surface molecules on CD8αβ+ T lymphocytes of PBL from 11 HIV-1 seronegative and 22 HIV-1 seropositive individuals. The MF of anti-CD8αβ staining was significantly reduced on CD8+T lymphocyte subsets that showed immunophenotypic evidence of activation. The subset of lymphocytes expressing low levels of CD8αβ expressed higher levels of activation, adhesion, and cytotoxic-associated molecules and was predominantly CD45RO+ and CD28−. Finally, we monitored the expression of the CD8αβ-heterodimer on PBL of eight HIV-1–infected individuals over a 16-week period after the initiation of highly active antiretroviral therapy (HAART), including zidovudine (ZDV), lamivudine (3TC), and indinavir (IDV), and found a significant increase in the expression of the CD8αβ-heterodimer. These results suggest that antibodies recognizing the CD8αβ-heterodimer are useful tools to specifically identify CD8+ T lymphocytes. Moreover, the quantitative monitoring of CD8αβ expression allows the detection of discrete CD8+ T lymphocyte subsets and may be useful for assessing the immune status of individuals infected with HIV-1.
STUDIES OF ACQUIRED immunodeficiency syndrome (AIDS) immunopathogenesis suggest that CD8+ T lymphocytes play a major role in controlling the replication of human immunodeficiency virus (HIV)-1. CD8+ T lymphocytes can inhibit HIV replication in autologous CD4+ T lymphocytes, both by soluble factor/chemokine release and through lysis of infected cells.1-4 Virus-specific cytotoxic lymphocytes (CTL) are readily demonstrated at a high frequency in HIV-infected individuals in a variety of anatomic compartments, including the peripheral blood, lymph node, spleen, cerebral spinal fluid, skin, and in mucosal tissues.5-9 The best immunologic correlate of the early containment of AIDS virus replication in man and monkeys is the emergence of a potent virus-specific CTL response.10-12Finally, the investigations of Rinaldo et al13 suggested that a long-term clinical nonprogressor status is correlated with a high frequency HIV-specific CTL response. Consistent with these functional studies, the characterization of CD8+ T lymphocytes of HIV-1–infected individuals has demonstrated a significant increase in the expression by these cells of molecules associated with chronic activation.14 15
The human CD8 molecule is composed of two distinct polypeptide chains that pair on the cell surface either as a CD8αα-homodimer or as a CD8αβ-heterodimer.16-19 These forms of the CD8 molecule are differentially expressed on functionally distinct CD8+lymphocyte subsets. Four distinct subpopulations of CD8+lymphocytes have been described based on the form of the CD8 dimer expressed by cells: (1) T-cell receptor (TCR)αβ+CD8αβ+CD3+ T lymphocytes, (2) TCRαβ+CD8αα+CD3+ T lymphocytes, (3) TCRγδ+CD8αα+ CD3+ T lymphocytes, and (4) CD8αα+CD3- natural killer (NK) cells. TCRαβ+ T lymphocytes recognize antigen in a major histocompatibility complex (MHC) class I–restricted fashion and function to eliminate replicating intracellular pathogens and tumors.20,21 TCRγδ+ T lymphocytes and NK cells function for the most part in an MHC class I–unrestricted fashion.22 23
The majority of monoclonal antibodies used to define CD8+ T cells recognize epitopes on the CD8α-chain.24 Thus, specific identification of CD8+ T cells by flow cytometry requires the use of both anti-CD3 and anti-CD8 monoclonal antibodies (MoAbs) to exclude CD8+ NK cells.25-27 However, this two- antibody combination limits further subset analysis of lymphocytes to one or two additional reagents when three- or four-color flow cytometric subset studies are performed. In the present study, we show that a MoAb, 2ST8-5H7, which is directed against a conformational epitope of the CD8αβ-heterodimer comprised of domains of both the CD8α and CD8β-chain,16,17 24 can be used as a single gating reagent to define specifically CD8+ T lymphocytes in peripheral blood of HIV-1 seronegative and HIV-1 seropositive donors. Furthermore, we have assessed peripheral blood lymphocytes (PBL) of normal and HIV-1–infected individuals for CD8+ T lymphocyte expression of the CD8β-chain, as well as molecules associated with activation, adhesion, maturation, and cytotoxic effector function.
MATERIALS AND METHODS
Patient and control blood specimens.
EDTA-anticoagulated peripheral blood specimens were obtained from 21 healthy volunteers and 103 HIV-1–infected individuals. HIV-1 seropositive individuals provided written informed consent before donating blood or blood was evaluated from specimens available in the course of routine clinical testing. HIV-1 seronegative blood donors provided verbal consent. Blood samples from eight HIV-1–infected individuals participating in the prospective open-label AIDS Clinical Trials Group (ACTG) study 343 were obtained at baseline and at weeks 4, 8, and 16 after starting treatment. These patients received highly active antiretroviral therapy (HAART), including zidovudine (ZDV; Glaxo Wellcome, Inc, Research Triangle Park, NC) 300 mg twice daily, lamivudine (3TC; Glaxo Wellcome, Inc) 150 mg twice daily, and indinavir (IDV; Merck & Co, Inc, Whitehouse Station, NJ) 800 mg three times a day.
MoAbs and sample preparation.
The MoAbs used for this study (all from Coulter Corp, Miami, FL) were directly conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), phycoerythrin-Texas red (ECD), or allophycocyanin (APC) (Table 1). The following four-color combinations were used: (1) CD8α-FITC, CD4-PE, CD8αβ-ECD, and CD3-APC; (2) CD8α-FITC, TCRγδ-PE, CD8αβ-ECD, and CD3-APC; (3) CD8α-FITC, CD56-PE, CD8αβ-ECD, and CD3-APC; (4) CD8α-FITC, C1.7-PE, CD8αβ-ECD, and CD3-APC; (5) CD8α-FITC, HLA-DR-PE, CD8αβ-ECD, and CD3-APC; (6) CD8α-FITC, CD11a-PE, CD8αβ-ECD, and CD3-APC; (7) CD8α-FITC, CD28-PE, CD8αβ-ECD, and CD3-APC; (8) CD8α-FITC, CD38-PE, CD8αβ-ECD, and CD3-APC; (9) CD8α-FITC, CD45RA-PE, CD8αβ-ECD, and CD3-APC; (10) CD8α-FITC, CD45RO-PE, CD8αβ-ECD, and CD3-APC; (11) CD8α-FITC, CD49d-PE, CD8αβ-ECD, and CD3-APC; (12) CD8α-FITC, CD57-PE, CD8αβ-ECD, and CD3-APC; (13) CD8α-FITC, CD58-PE, CD8αβ-ECD, and CD3-APC; (14) CD8α-FITC, CD62L-PE, CD8αβ-ECD, and CD3. In addition, a two-color reagent (CD45-FITC and CD14-PE) was used for verifying the recovery and purity of the lymphocytes within the light scatter gates according to the Centers for Disease Control (CDC) guidelines for performing CD4+ T-cell determinations in persons infected with HIV-1.25 Aliquots of 100 μL of EDTA-anticoagulated blood from each donor were incubated with each of these reagent combinations for 15 minutes at room temperature before lysis and fixation using Coulter Immunoprep Reagent System and Q-prep Workstation (Coulter, Miami, FL). To reduce the background level of staining, the Q-Prep procedure was modified, and lysed samples were washed with 1.0 mL phosphate-buffered saline (PBS) and centrifuged for 3 minutes at 300g. The supernatants were decanted, cells were resuspended in 0.5 mL PBS containing 1% paraformaldehyde and maintained for 24 hours at 4°C before flow cytometric analysis.
MoAbs Used
| Reactivity . | Function/Association . | Clone Name . | μg/mL . |
|---|---|---|---|
| Cytotoxic function | C1.7 | 5.0 | |
| HLA-DR, -DP, -DQ* | Antigen presentation (MHC class II) | 9-49 | 2.5 |
| CD3 | Peptide/MHC class antigen binding, Signal transduction | UCHT1 | 10.0 |
| CD4 | Coreceptor (MHC class II) | SFCI12T4D11 | 0.7 |
| CD8α | α-chain of MHC class I coreceptor | SFC121Thy2D3 | 2.5 |
| CD8αβ | αβ-chains of MHC class I coreceptor | 2ST8-5H7 | 5.0 |
| CD11a | Adhesion | S6F1LDB11LDH10 | 5.0 |
| CD14 | Monocytes/macrophages | 116 | 1.4 |
| CD28 | Costimulatory molecule | 4B10 | 10.0 |
| CD38 | Unknown function, activation | LS198-4-3 | 2.5 |
| CD45 | Signal transduction | KC56 | 10.0 |
| CD45RA | Naive cells | 2H4LDH11LDB9 | 3.0 |
| CD45RO | Memory cells | UCHL1 | 3.0 |
| CD49d | Adhesion | 8F2 | 20.0 |
| CD56 | Adhesion | N901 | 2.5 |
| CD57 | Unknown function | NC1 | 10.0 |
| CD58 | Adhesion | AICD58 | 3.3 |
| CD62L | Adhesion | LAM1-3 | 10.0 |
| TCRγδ | Gamma/delta T-cell receptor | Immu 510 | 1.6 |
| Reactivity . | Function/Association . | Clone Name . | μg/mL . |
|---|---|---|---|
| Cytotoxic function | C1.7 | 5.0 | |
| HLA-DR, -DP, -DQ* | Antigen presentation (MHC class II) | 9-49 | 2.5 |
| CD3 | Peptide/MHC class antigen binding, Signal transduction | UCHT1 | 10.0 |
| CD4 | Coreceptor (MHC class II) | SFCI12T4D11 | 0.7 |
| CD8α | α-chain of MHC class I coreceptor | SFC121Thy2D3 | 2.5 |
| CD8αβ | αβ-chains of MHC class I coreceptor | 2ST8-5H7 | 5.0 |
| CD11a | Adhesion | S6F1LDB11LDH10 | 5.0 |
| CD14 | Monocytes/macrophages | 116 | 1.4 |
| CD28 | Costimulatory molecule | 4B10 | 10.0 |
| CD38 | Unknown function, activation | LS198-4-3 | 2.5 |
| CD45 | Signal transduction | KC56 | 10.0 |
| CD45RA | Naive cells | 2H4LDH11LDB9 | 3.0 |
| CD45RO | Memory cells | UCHL1 | 3.0 |
| CD49d | Adhesion | 8F2 | 20.0 |
| CD56 | Adhesion | N901 | 2.5 |
| CD57 | Unknown function | NC1 | 10.0 |
| CD58 | Adhesion | AICD58 | 3.3 |
| CD62L | Adhesion | LAM1-3 | 10.0 |
| TCRγδ | Gamma/delta T-cell receptor | Immu 510 | 1.6 |
*Referred in Table 4 as HLA-DR.
Flow cytometry.
Samples were analyzed on a Coulter EPICS Elite ESP equipped with argon and helium neon lasers, a gated amplifier, and a 120 μm flow cell tip. The instrument was run at high bandwidth and alignment was controlled on a daily basis using DNA-CHECK EPICS Alignment Fluorospheres (Coulter Corp) to maintain the same sensitivity levels during the entire study. Linear performance in each channel was controlled using the EPICS Immuno-Brite Standards Kit (Coulter Corp). The sensitivity of the photo multiplier tube for detection of the ECD-characteristic fluorescence was controlled during the entire study with a single lot of anti-CD8αβ–ECD stained Coulter CYTO-TROL control cells (Coulter Corp). Voltage and compensation levels were established using both unstained cells for adjusting the negative/background levels of fluorescence to the first log step and single-color stained cells for adjusting spectral overlap. A total of 5,000 lymphocytes were analyzed in a manually set acquisition gate and positive cutoffs for fluorescence were set to the first log step to include less than 1% of nonstaining cells. Data analysis was performed using the EPICS Elite software version 4.02 (Coulter Corp). Absolute numbers of lymphocyte subsets were calculated using routine diagnostic lymphocyte counts obtained from the same blood specimens analyzed on a Coulter Hematology Analyzer T-540 (Coulter Corp).
Statistical analysis and data presentation.
All results showing percent or absolute count data in Tables 2-4 are expressed as median (25th percentile, 75th percentile). Comparisons of the three data values per row in Tables 2and 3 were first done using the Kruskal-Wallis test.28 If the results of the Kruskal-Wallis test were statistically significant (P < .05), the three data pairs per row were compared by the Dunn test.29 The crude significance level used for the Dunn tests within each row of Tables 2 and 3 was P < .01 to account for the three tests being done in each row. This method was also used to obtain theP values in Fig 1C (P < .01).
Percentage of CD8α+ or CD8αβ+ Cells Within the CD8α+ T Cell, CD8αbright T Cell, NK Cell, TCRγδ+ T Cell, and CD4+ T-Cell Subsets
| Gated Cells . | Assessed for Expression of: . | HIV-1 Seronegative (n = 21) . | HIV-1 Seropositive >200 CD4+cells/μL (n = 58) . | HIV-1 Seropositive <200 CD4+ cells/μL (n = 45) . |
|---|---|---|---|---|
| Median % of CD8α+ or CD8αβ+ cell subset within gated cells (25%, 75%) . | ||||
| CD8α+ T cells | CD8αβ | 90.8 (81.1, 92.6) | 96.9 (94.8, 98.0)* | 97.6 (95.3, 98.3)* |
| CD8αbright T cells | CD8αβ | 97.1 (94.7, 98.3) | 99.4 (98.6, 99.6)* | 99.2 (98.6, 99.6)* |
| NK cells | CD8α | 46.7 (41.1, 60.7) | 28.2 (23.2, 35.3)* | 25.2 (19.2, 36.4)* |
| NK cells | CD8αβ | 1.7 (0.9, 2.5)*,† | 1.7 (0.5, 3.5)* | 0.9 (0.6, 1.3)† |
| TCRγδ+ T cells | CD8α | 26.0 (23.0, 34.5) | 43.0 (34.7, 52.0)* | 41.9 (24.7, 55.0)* |
| TCRγδ+ T cells | CD8αβ | 4.2 (1.8, 6.4) | 17.6 (12.2, 27.5)* | 13.2 (9.7, 24.5)* |
| CD4+ T cells | CD8α | 1.5 (0.9, 2.0) | 3.8 (2.4, 6.8)* | 4.9 (3.0, 8.0)* |
| CD4+ T cells | CD8αβ | 0.6 (0.4, 0.8) | 1.8 (1.2, 3.0) | 3.5 (2.2, 5.3) |
| Gated Cells . | Assessed for Expression of: . | HIV-1 Seronegative (n = 21) . | HIV-1 Seropositive >200 CD4+cells/μL (n = 58) . | HIV-1 Seropositive <200 CD4+ cells/μL (n = 45) . |
|---|---|---|---|---|
| Median % of CD8α+ or CD8αβ+ cell subset within gated cells (25%, 75%) . | ||||
| CD8α+ T cells | CD8αβ | 90.8 (81.1, 92.6) | 96.9 (94.8, 98.0)* | 97.6 (95.3, 98.3)* |
| CD8αbright T cells | CD8αβ | 97.1 (94.7, 98.3) | 99.4 (98.6, 99.6)* | 99.2 (98.6, 99.6)* |
| NK cells | CD8α | 46.7 (41.1, 60.7) | 28.2 (23.2, 35.3)* | 25.2 (19.2, 36.4)* |
| NK cells | CD8αβ | 1.7 (0.9, 2.5)*,† | 1.7 (0.5, 3.5)* | 0.9 (0.6, 1.3)† |
| TCRγδ+ T cells | CD8α | 26.0 (23.0, 34.5) | 43.0 (34.7, 52.0)* | 41.9 (24.7, 55.0)* |
| TCRγδ+ T cells | CD8αβ | 4.2 (1.8, 6.4) | 17.6 (12.2, 27.5)* | 13.2 (9.7, 24.5)* |
| CD4+ T cells | CD8α | 1.5 (0.9, 2.0) | 3.8 (2.4, 6.8)* | 4.9 (3.0, 8.0)* |
| CD4+ T cells | CD8αβ | 0.6 (0.4, 0.8) | 1.8 (1.2, 3.0) | 3.5 (2.2, 5.3) |
All results are expressed as median % (25th percentile, 75th percentile). Statistical differences (P < .05) among the three blood donor groups per row were first analyzed by the Kruskal-Wallis test. If the results were found to be significant, each of the three data groups per row were analyzed by the Dunn test. CD8α or CD8αβ expression was assessed on gated cells defined as: CD8α+ T cells (CD8α+CD3+), NK cells (CD56+CD3−), TCRγδ+ T cells (TCRγδ+CD3+) and CD4+ T cells (CD4+CD3+). CD8αbright T cells were defined using a subjective tight gate around the main subpopulation of CD8α+ T cells, which express CD8α in high density as described.32 33
The differences between the two groups were NOT significant (P < .01) using the Dunn test.
The differences between the two groups were NOT significant (P < .01) using the Dunn test. All other data groups per row were significantly different.
Absolute Cell Counts of Total CD8α+ or CD8αβ+ Cells of CD8α+ T Cells, CD8αbright T Cells, NK Cells, TCRγδ+ T Cells, and CD4+ T Cells
| Assessed Cell Subset . | HIV-1 Seronegative (n = 21) . | HIV-1 Seropositive >200 CD4+ cells/μL (n = 58) . | HIV-1 Seropositive <200 CD4+ cells/μL (n = 45) . |
|---|---|---|---|
| Absolute Cell Counts (25%, 75%) . | |||
| CD8α+ T cells | 544 (371, 624)* | 1028 (838, 1389) | 762 (422, 932)* |
| CD8αβ+CD8α+ T cells | 463 (345, 560)* | 981 (784, 1300) | 733 (415, 903)* |
| CD8αbright T cells | 461 (331, 552)* | 942 (768, 1289) | 712 (408, 899)* |
| CD8αβ+CD8αbright T cells | 444 (313, 528) | 934 (752, 1276) | 700 (404, 891) |
| NK cells | 260 (155, 429) | 72 (36, 138)* | 82 (53, 163)* |
| CD8α+ NK cells | 154 (72, 190) | 18 (10, 33)* | 30 (11, 41)* |
| CD8αβ+ NK cells | 4 (3, 8) | 1 (0, 3)* | 1 (0, 2)* |
| TCRγδ+ T cells | 74 (37, 128)† | 90 (53, 141)† | 64 (33, 94)† |
| TCRγδ+CD8α+ T cells | 20 (12, 31)* | 40 (22, 58) | 25 (15, 39)* |
| TCRγδ+CD8αβ+ T cells | 3 (1, 5) | 16 (10, 27) | 10 (5, 13) |
| CD4+ T cells | 857 (779, 965) | 408 (330, 525) | 102 (58, 150) |
| CD4+CD8α+ T cells | 12 (8, 14)* | 14 (8, 29)* | 4 (3, 8) |
| CD4+CD8αβ+ T cells | 5 (3, 7)* | 7 (4, 12)* | 3 (2, 5) |
| Assessed Cell Subset . | HIV-1 Seronegative (n = 21) . | HIV-1 Seropositive >200 CD4+ cells/μL (n = 58) . | HIV-1 Seropositive <200 CD4+ cells/μL (n = 45) . |
|---|---|---|---|
| Absolute Cell Counts (25%, 75%) . | |||
| CD8α+ T cells | 544 (371, 624)* | 1028 (838, 1389) | 762 (422, 932)* |
| CD8αβ+CD8α+ T cells | 463 (345, 560)* | 981 (784, 1300) | 733 (415, 903)* |
| CD8αbright T cells | 461 (331, 552)* | 942 (768, 1289) | 712 (408, 899)* |
| CD8αβ+CD8αbright T cells | 444 (313, 528) | 934 (752, 1276) | 700 (404, 891) |
| NK cells | 260 (155, 429) | 72 (36, 138)* | 82 (53, 163)* |
| CD8α+ NK cells | 154 (72, 190) | 18 (10, 33)* | 30 (11, 41)* |
| CD8αβ+ NK cells | 4 (3, 8) | 1 (0, 3)* | 1 (0, 2)* |
| TCRγδ+ T cells | 74 (37, 128)† | 90 (53, 141)† | 64 (33, 94)† |
| TCRγδ+CD8α+ T cells | 20 (12, 31)* | 40 (22, 58) | 25 (15, 39)* |
| TCRγδ+CD8αβ+ T cells | 3 (1, 5) | 16 (10, 27) | 10 (5, 13) |
| CD4+ T cells | 857 (779, 965) | 408 (330, 525) | 102 (58, 150) |
| CD4+CD8α+ T cells | 12 (8, 14)* | 14 (8, 29)* | 4 (3, 8) |
| CD4+CD8αβ+ T cells | 5 (3, 7)* | 7 (4, 12)* | 3 (2, 5) |
All results are expressed as median absolute cell counts/μL whole blood (25th percentile, 75th percentile). Statistical differences among the three blood donor groups per row were first analyzed by the Kruskal-Wallis test. If the results were found to be significant (P < .05), each of the three data groups per row were analyzed by the Dunn test. Gated cell subsets were defined as: CD8α+ T cells (CD8α+CD3+), CD8αβ+CD8α+ T cells (CD8αβ+CD8α+CD3+), NK cells (CD56+CD3−), TCRγδ+ T cells (TCRγδ+CD3+), and T-helper cells (CD4+CD3+). CD8αbright T cells were defined using a subjective tight gate around the main subpopulation of CD8α+ T cells, which express CD8α in high density as described.32 33
The differences between the two groups were not significant (P < .01) using the Dunn test. All other data groups per row were significantly different.
These data values were not statistically different using the Kruskal-Wallis test.
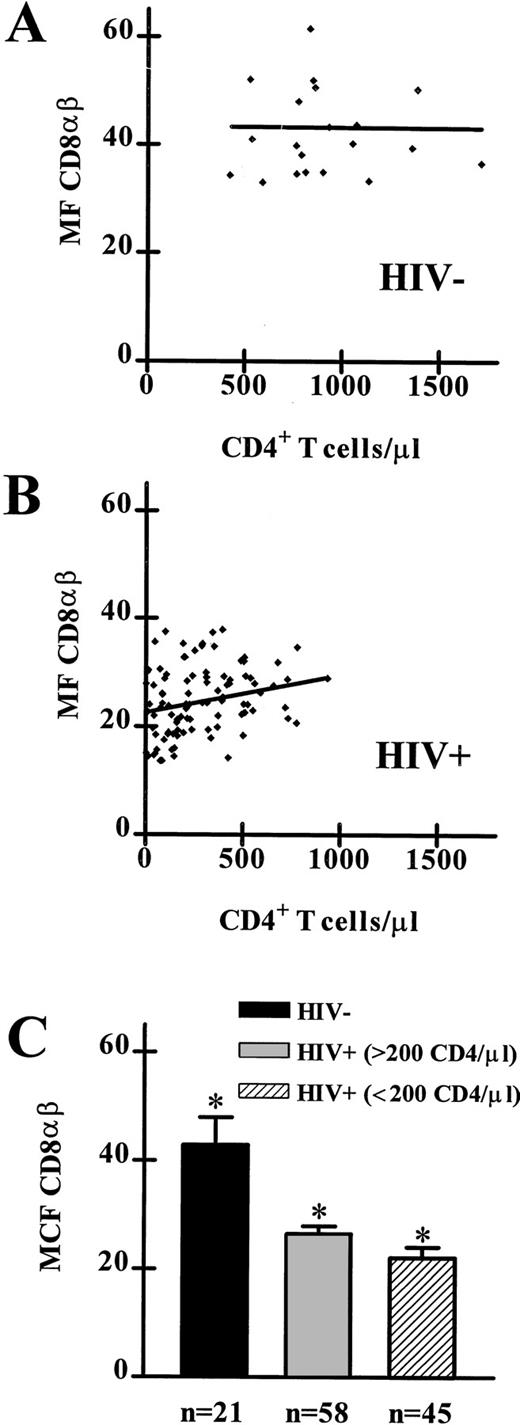
CD8αβ expression on PBL from HIV-1 seronegative and HIV-1 seropositive individuals. (A) The linear regression analysis of CD8αβ MF and absolute CD4+T-lymphocyte counts from data obtained from 21 HIV-1 seronegative individuals showed no significant deviation from a slope of 0. (B) The linear regression analysis of CD8αβ MF and absolute CD4+ T-lymphocyte counts from data obtained from 103 HIV-1 seropositive individuals showed a significant deviation from a slope of 0 (P < .01). (C) CD8αβ MF on PBL from 21 HIV-1 seronegative individuals, 58 HIV-1 seropositive individuals (> 200 CD4+ T lymphocytes/μL), and 45 HIV-1 seropositive individuals (< 200 CD4+ T lymphocytes/μL). Significant differences were observed between all three groups (HIV-1 seronegative v either HIV-1 seropositive patient group:P < .0001; between HIV-1 seropositive patient groups:P < .01).
CD8αβ expression on PBL from HIV-1 seronegative and HIV-1 seropositive individuals. (A) The linear regression analysis of CD8αβ MF and absolute CD4+T-lymphocyte counts from data obtained from 21 HIV-1 seronegative individuals showed no significant deviation from a slope of 0. (B) The linear regression analysis of CD8αβ MF and absolute CD4+ T-lymphocyte counts from data obtained from 103 HIV-1 seropositive individuals showed a significant deviation from a slope of 0 (P < .01). (C) CD8αβ MF on PBL from 21 HIV-1 seronegative individuals, 58 HIV-1 seropositive individuals (> 200 CD4+ T lymphocytes/μL), and 45 HIV-1 seropositive individuals (< 200 CD4+ T lymphocytes/μL). Significant differences were observed between all three groups (HIV-1 seronegative v either HIV-1 seropositive patient group:P < .0001; between HIV-1 seropositive patient groups:P < .01).
The comparison of the mean fluorescence (MF) of anti-CD8αβ staining of PBL from all three blood donor groups is shown in Fig 1. Linear regression analyses were performed relating CD8αβ MF to absolute counts of CD4+ T lymphocytes/μL blood (Fig 1A and B). The F test was used to test whether the slope was significantly different from 0.30 The Wilcoxon matched pairs31 test was used for comparison of the CD8αβ MF of PBL from the eight HIV-1 seropositive study subjects receiving HAART.
Within each of the three blood donor groups in Table 4 the anti-CD8αβ staining of the two different CD8αβ+ T lymphocyte subsets was investigated using the Wilcoxon matched pairs test (paired within each patient). For example, the test at the top of the leftmost column in Table 4 compared the MF of anti-CD8αβ staining for HLA-DR+ versus HLA-DR− CD8+ T lymphocytes from HIV-1 seronegative individuals. Statistical tests in Table 4 were considered significant if the P values were < .05. These P values were not adjusted for multiple comparisons.
CD8αβ+ Lymphocyte Subsets: MF (25%, 75%)
| Assessed Subset . | HIV-1 Seronegative n = 11 . | HIV-1 Seropositive (>200 CD4) n = 11 . | HIV-1 Seropositive (<200 CD4) n = 11 . |
|---|---|---|---|
| HLA-DR | |||
| + | 29 (29, 35) 3-150 | 18 (15, 22) | 15 (13, 17) |
| − | 45 (40, 53) | 21 (17, 25) | 16 (13, 18) |
| C1.7 | |||
| + | 31 (26, 37) 3-150 | 19 (19, 22) 3-150 | 15 (13, 15) |
| − | 48 (38, 55) | 25 (20, 27) | 18 (16, 23) |
| CD11a | |||
| + | 29 (24, 32) 3-150 | 18 (17, 20) 3-150 | 15 (13, 18) 3-150 |
| − | 52 (50, 71) | 31 (25, 35) | 24 (22, 32) |
| CD28 | |||
| + | 42 (42, 49) 3-150 | 28 (21, 30) 3-150 | 21 (19, 25) 3-150 |
| − | 29 (25, 33) | 20 (18, 21) | 16 (13, 17) |
| CD38 | |||
| + | 50 (42, 52) | 21 (19, 22) | 17 (14, 20) |
| − | 42 (36, 47) | 23 (21, 26) | 21 (17, 23) |
| CD45RA | |||
| + | 52 (46, 62) 3-150 | 22 (18, 25) | 20 (18, 22) |
| − | 28 (24, 32) | 23 (20, 24) | 16 (14, 20) |
| CD45RO | |||
| + | 27 (24, 32) 3-150 | 21 (18, 22) | 17 (13, 18) |
| − | 56 (42, 64) | 21 (18, 24) | 18 (17, 22) |
| CD49d | |||
| + | 35 (33, 42) 3-150 | 22 (19, 23) 3-150 | 18 (15, 19) |
| − | 63 (53, 77) | 31 (23, 34) | 19 (16, 33) |
| CD56 | |||
| + | 19 (17, 23) 3-150 | 14 (13, 18) 3-150 | 16 (14, 19) |
| − | 45 (40, 52) | 22 (21, 24) | 18 (15, 21) |
| CD57 | |||
| + | 22 (19, 29) 3-150 | 17 (16, 18) 3-150 | 13 (12, 14) 3-150 |
| − | 46 (43, 52) | 26 (23, 28) | 20 (18, 24) |
| CD58 | |||
| + | 31 (29, 39) 3-150 | 22 (21, 24) | 18 (15, 20) |
| − | 61 (53, 63) | 24 (20, 26) | 19 (17, 23) |
| CD62L | |||
| + | 47 (37, 56) 3-150 | 25 (21, 29) 3-150 | 20 (16, 22) |
| − | 25 (21, 30) | 20 (19, 21) | 16 (13, 17) |
| Assessed Subset . | HIV-1 Seronegative n = 11 . | HIV-1 Seropositive (>200 CD4) n = 11 . | HIV-1 Seropositive (<200 CD4) n = 11 . |
|---|---|---|---|
| HLA-DR | |||
| + | 29 (29, 35) 3-150 | 18 (15, 22) | 15 (13, 17) |
| − | 45 (40, 53) | 21 (17, 25) | 16 (13, 18) |
| C1.7 | |||
| + | 31 (26, 37) 3-150 | 19 (19, 22) 3-150 | 15 (13, 15) |
| − | 48 (38, 55) | 25 (20, 27) | 18 (16, 23) |
| CD11a | |||
| + | 29 (24, 32) 3-150 | 18 (17, 20) 3-150 | 15 (13, 18) 3-150 |
| − | 52 (50, 71) | 31 (25, 35) | 24 (22, 32) |
| CD28 | |||
| + | 42 (42, 49) 3-150 | 28 (21, 30) 3-150 | 21 (19, 25) 3-150 |
| − | 29 (25, 33) | 20 (18, 21) | 16 (13, 17) |
| CD38 | |||
| + | 50 (42, 52) | 21 (19, 22) | 17 (14, 20) |
| − | 42 (36, 47) | 23 (21, 26) | 21 (17, 23) |
| CD45RA | |||
| + | 52 (46, 62) 3-150 | 22 (18, 25) | 20 (18, 22) |
| − | 28 (24, 32) | 23 (20, 24) | 16 (14, 20) |
| CD45RO | |||
| + | 27 (24, 32) 3-150 | 21 (18, 22) | 17 (13, 18) |
| − | 56 (42, 64) | 21 (18, 24) | 18 (17, 22) |
| CD49d | |||
| + | 35 (33, 42) 3-150 | 22 (19, 23) 3-150 | 18 (15, 19) |
| − | 63 (53, 77) | 31 (23, 34) | 19 (16, 33) |
| CD56 | |||
| + | 19 (17, 23) 3-150 | 14 (13, 18) 3-150 | 16 (14, 19) |
| − | 45 (40, 52) | 22 (21, 24) | 18 (15, 21) |
| CD57 | |||
| + | 22 (19, 29) 3-150 | 17 (16, 18) 3-150 | 13 (12, 14) 3-150 |
| − | 46 (43, 52) | 26 (23, 28) | 20 (18, 24) |
| CD58 | |||
| + | 31 (29, 39) 3-150 | 22 (21, 24) | 18 (15, 20) |
| − | 61 (53, 63) | 24 (20, 26) | 19 (17, 23) |
| CD62L | |||
| + | 47 (37, 56) 3-150 | 25 (21, 29) 3-150 | 20 (16, 22) |
| − | 25 (21, 30) | 20 (19, 21) | 16 (13, 17) |
All results are expressed as median channel of MF (25th percentile, 75th percentile) of CD8αβ+ lymphocyte subset. CD8αβ+ lymphocytes were gated according to their lymphocyte light scatter characteristics and expression of CD8αβ-heterodimer.
Indicated data groups of median MF of anti-CD8αβ staining of CD8+ lymphocyte subsets showed significant differences (P < .05) using the Wilcoxon matched pairs test.
All statistical analyses were performed using Microsoft Excel software version 5.0 (Microsoft Corp, Redmond, WA) and GraphPad PRISM software version 2.01 (GraphPad Software, San Diego, CA). Data presentation was performed using WinMDI software version 2.3 (Joseph Trotter, La Jolla, CA) and Microsoft PowerPoint software version 4.0c (Microsoft Corp).
RESULTS
The absolute CD4+ T lymphocyte counts of the HIV-1 seropositive blood donors ranged from 1 to 1,359 cells/μL. Of the 103 HIV-1 seropositive donors analyzed, the 58 with > 200 CD4+ T cells/μL were designated as early stage of disease and the 45 donors with < 200 CD4+ T cells/μL were designated as late stage of disease. The CD4 counts of the 21 healthy, HIV-1 seronegative donors all fell within the expected normal range for our laboratory. For the purpose of those studies, lymphocyte subsets were defined as follows: CD8+ T lymphocytes (CD8α+CD3+), NK cells (CD56+CD3−), TCRγδ+ T lymphocytes (TCRγδ+CD3+), and T-helper cells (CD4+CD3+).
Expression of CD8α and CD8αβ on PBL of HIV-1 seronegative and seropositive individuals.
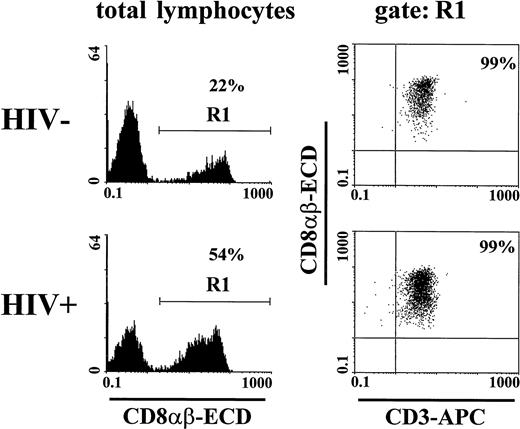
We found that CD8αβ-heterodimer positive lymphocytes were almost all CD3+ (99%, Fig 2). We sought to clarify which subset of CD8+ lymphocytes express the CD8αβ-heterodimer in HIV-1 seronegative and seropositive individuals. A median of 91% of CD8α+CD3+ T lymphocytes of HIV-1 seronegative blood donors and 97% (<200 CD4+ T cells/μL) or 98% (<200 CD4+ T cells/μL) of HIV-1 seropositive blood donors expressed the CD8αβ-heterodimer (Table 2). It has previously been suggested that the subset of CD8bright T lymphocytes predominantly contains true MHC class I–restricted CD8+ T lymphocytes with potential cytotoxic function.32 33 We found that almost all CD8αbright cells were CD8αβ+ (median, 97% of the HIV-1 seronegative and 99% of the HIV-1 seropositive blood donors) (Table 2).
Restricted expression of CD8αβ on CD3+T cells. Lymphocytes from an HIV-1 seronegative and a seropositive individual were gated according to light scatter characteristics and expression of CD8αβ (gate R1). Only a minor fraction (<1%) of CD8αβ+ cells did not express the CD3 molecule.
Restricted expression of CD8αβ on CD3+T cells. Lymphocytes from an HIV-1 seronegative and a seropositive individual were gated according to light scatter characteristics and expression of CD8αβ (gate R1). Only a minor fraction (<1%) of CD8αβ+ cells did not express the CD3 molecule.
The median differences in the absolute cell counts between all CD8α+CD3+ T lymphocytes and the CD8αβ+ T lymphocytes were less than 57 cells/μL (HIV-1 seronegative donors, 56; HIV-1 seropositive donors > 200 CD4+ T cells/μL, 38; HIV-1 seropositive donors < 200 CD4+ T cells/μL, 21). The median differences between the absolute cell counts of bright CD8α+ CD3+ T lymphocytes and the CD8αβ+ T lymphocytes within this subset were less than 10 cells/μL (HIV-1 seronegative donors, 9; HIV-1 seropositive donors donors > 200 CD4+ T cells/μL, 7; HIV-1 seropositive donors < 200 CD4+ T cells/μL, 5).
The CD8αβ molecule was only rarely expressed on NK cells (Tables 2and 3). Less than 1.7% of NK cells in PBL of HIV-1 seronegative and seropositive donors expressed the CD8αβ-heterodimer (Table 2). In the small subset of peripheral blood T lymphocytes expressing TCRγδ fewer cells expressed the CD8αβ-heterodimer than the CD8αα-homodimer (Tables 2 and 3). We found no significant difference in the absolute counts of TCRγδ+ T cells between these groups of subjects. However, more (percent and absolute counts) CD8αβ+TCRγδ+ T lymphocytes were observed in HIV-1–infected than in normal donors; higher numbers of this cell subset were also seen in patients with > 200 CD4+ T cells/μL than in individuals with < 200 CD4+ T cells/μL (Table 3). The CD8αβ-heterodimer was only expressed on a subpopulation of the small subset of CD4+ T-helper cells, which expressed the CD8α-chain. We observed a biologically negligible increase in the percentage of CD8αβ+CD4+ T lymphocytes in the blood of HIV-1–infected individuals and a decrease in absolute counts of CD8αβ+CD4+ T lymphocytes in donors with < 200 CD4+ T cells/μL (Tables 2 and 3).
These observations indicate that the MoAb 2ST8-5H7, directed against the CD8αβ-heterodimer, identifies most CD8+ T lymphocytes in HIV-1 seronegative donors, and virtually all CD8+ T lymphocytes in HIV-1 seropositive individuals. Therefore, we used this anti-CD8αβ MoAb in place of the conventional antibody combination anti-CD3 and anti-CD8α to detect CD8+ T lymphocytes in the four-color analyses in these studies. By using this MoAb, an additional fluorescence channel was available in multiparameter flow cytometric analysis for characterization of discrete CD8+ T lymphocyte subsets.
MF of CD8αβ-heterodimer expression on PBL of HIV-1 seronegative and seropositive individuals.
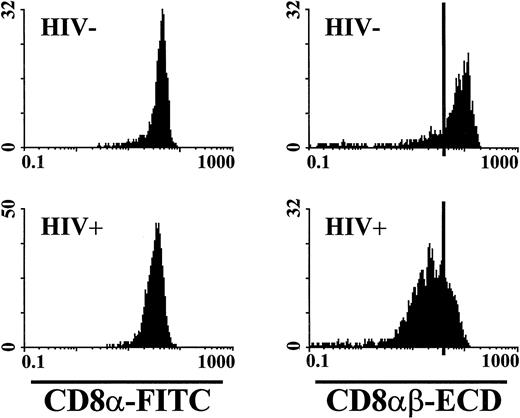
Interestingly, we observed a decrease in the MF of the binding of this CD8αβ-heterodimer–specific antibody on CD8+ T lymphocytes of 101 HIV-1 seropositive individuals as compared with 21 seronegative individuals. When analyzed for expression of the CD8α-chain using an anti-CD8α–specific MoAb, CD8+ T lymphocytes of HIV-1 seronegative and seropositive blood donors showed a similar MF. Data from representative blood donors are shown in Fig 3.
Relative fluorescence intensity of anti-CD8α and anti-CD8αβ staining. The fluorescence intensity of anti-CD8α and CD8αβ staining was assessed on CD8α+CD3+ T lymphocytes from an HIV-1 seronegative and a seropositive individual. The anti-CD8α staining showed a similar fluorescence intensity on T lymphocytes from investigated subjects. The CD8αβ expression on T lymphocytes from the HIV-1 seropositive individual was significantly reduced compared with the HIV-1 seronegative individual. The black bars in the CD8αβ-ECD histograms were set at channel 40 for use as a fluorescent reference point.
Relative fluorescence intensity of anti-CD8α and anti-CD8αβ staining. The fluorescence intensity of anti-CD8α and CD8αβ staining was assessed on CD8α+CD3+ T lymphocytes from an HIV-1 seronegative and a seropositive individual. The anti-CD8α staining showed a similar fluorescence intensity on T lymphocytes from investigated subjects. The CD8αβ expression on T lymphocytes from the HIV-1 seropositive individual was significantly reduced compared with the HIV-1 seronegative individual. The black bars in the CD8αβ-ECD histograms were set at channel 40 for use as a fluorescent reference point.
We sought to determine whether the extent of decrease in CD8αβ expression by PBL correlated with clinical disease status in the study subjects. Linear regression analysis of CD8αβ MF of PBL and absolute CD4 cell counts from the HIV-1 seronegative subjects showed no significant deviation from a slope of 0 (Fig 1A). However, we detected a significant positive correlation between CD8αβ MF and absolute CD4 cell counts (slope, 0.007) when we investigated the data values of the HIV-1 seropositive subjects (Fig 1B). In fact, we found significant differences (HIV-1 seronegative v either HIV-1 seropositive patient group: P < .0001; between HIV-1 seropositive patient groups: P < .01) in the CD8αβ MF between PBL of each pair of the three donor groups (median CD8αβ MF, 41 channel [HIV-1 seronegative]; 27 channel [HIV-1 seropositive with > 200 CD4+ T cells/μL]; and 21 channel [HIV-1 seropositive with < 200 CD4+ T cells/μL]) (Fig 1C).
Percentage and MF of CD8αβ+ T-cell subsets of PBL of HIV-1 seronegative and seropositive individuals.
The association of the intensity of the CD8αβ staining and cell expression of activation, adhesion, and maturation-associated molecules was then explored. Eleven blood specimens from each of the three groups of subjects were studied: HIV-1 seronegative donors, HIV-1 seropositive donors with > 200 CD4+ T lymphocytes/μL, and HIV-1 seropositive donors with < 200 CD4+ T lymphocytes/μL. Four-color analyses were performed on these samples using 14 different antibody combinations. We observed a heterogeneous expression of CD8αβ in PBL from subjects in all three donor populations. Based on this heterogeneity, CD8+ T lymphocytes could be divided into two groups: cells with a relative high MF of anti-CD8αβ staining and cells that showed only a low to intermediate MF of anti-CD8αβ staining.
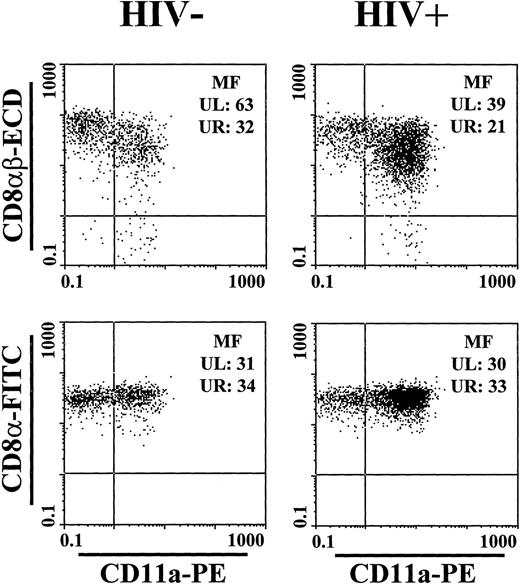
In PBL of HIV-1 seronegative individuals, we could demonstrate a pattern of expression of activation-, adhesion-, and maturation-antigens that was associated with the intensity of CD8αβ-specific staining. CD8αβ+ lymphocytes that expressed HLA-DR, C1.7, CD11a, CD45RO, CD49d, CD56, CD57, or CD58 showed a significantly lower CD8αβ MF than did lymphocytes not expressing these molecules (Table 4, Fig4). Furthermore, CD8αβ+ lymphocytes that expressed CD28, CD45RA, or CD62L showed a significantly higher CD8αβ MF than lymphocytes not expressing one of these molecules. These observations suggest that CD8+ T lymphocytes from normal donors can be divided into a fraction containing nonactivated cells with higher levels of CD8αβ expression and a fraction of activated cells with a lower level of CD8αβ expression.
CD8α, CD8αβ, and CD11a expression of CD8+ T lymphocytes from a representative HIV-1 seronegative and a seropositive individual. Lymphocytes were gated according to light scatter characteristics and expression of CD8α and CD3. CD8αβ MF from both individuals showed significantly more heterogeneity than CD8α MF. CD8αβ MF was lower on the CD11a+ subset.
CD8α, CD8αβ, and CD11a expression of CD8+ T lymphocytes from a representative HIV-1 seronegative and a seropositive individual. Lymphocytes were gated according to light scatter characteristics and expression of CD8α and CD3. CD8αβ MF from both individuals showed significantly more heterogeneity than CD8α MF. CD8αβ MF was lower on the CD11a+ subset.
In PBL of HIV-1–infected individuals, we similarly detected significant differences in the CD8αβ MF of specific T-cell subsets, although the differences were not as large as in HIV-1 seronegative individuals (Table 4, Fig 4). Lower levels of CD8αβ MF were seen on cells expressing C1.7, CD11a, CD49d, CD56, or CD57. Higher levels of anti-CD8αβ MF were seen on cells expressing CD28 or CD62L. Interestingly, in PBL of most HIV-1–infected blood donors, we did not find significant differences in CD8αβ MF on CD8+lymphocyte subsets which expressed either CD45RA or CD45RO (Table 4).
MF of CD8αβ+ PBL of HIV-1 seropositive individuals after initiation of HAART.
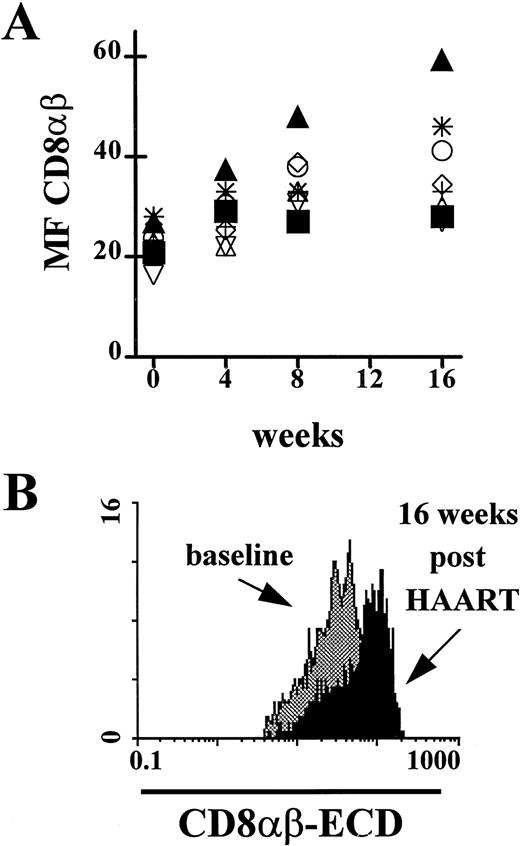
Blood specimens from eight HIV-1–infected study subjects receiving HAART were investigated for expression of CD8αβ MF. We found a significant increase in the MF of CD8αβ staining of PBL during the 16 weeks after initiation of therapy (median increase, 11 channel;P < .008 [two-sided Wilcoxon test]). One patient had an increase of only 5 channels, the other seven patients had an increase of at least 10 channels. The median absolute count of CD4+T lymphocytes increased from 500 to 647 cells/μL, and the median absolute count of CD8+ T lymphocytes from 910 to 960 cells/μL. Interestingly, the subject with the highest increase in CD4+ T lymphocytes (before treatment: 400 cells/μL and after treatment: 780 cells/μL) showed the greatest increase of CD8αβ MF (Fig 5B). In three individuals, the CD8αβ MF after 16 weeks of HAART was higher than the median CD8αβ MF in the HIV-1 seronegative individuals.
CD8αβ expression on PBL from HIV-1 seropositive individuals at baseline and after initiation of highly active antiretroviral therapy (HAART). (A) The expression of the CD8αβ-heterodimer over a 16-week treatment period showed a significant increase (P < .008). (▴) Subject with the largest increase in CD8αβ expression; (▪) subject with the smallest change in CD8αβ expression. (B) Change of CD8αβ expression on PBL from the HIV-1 seropositive individual in the cohort of treated patients with the largest increase in CD4+ T lymphocytes/μL blood and CD8αβ MF. The CD8αβ MF showed an increase of 32 channels (baseline, 27; 16 weeks post-HAART, 59).
CD8αβ expression on PBL from HIV-1 seropositive individuals at baseline and after initiation of highly active antiretroviral therapy (HAART). (A) The expression of the CD8αβ-heterodimer over a 16-week treatment period showed a significant increase (P < .008). (▴) Subject with the largest increase in CD8αβ expression; (▪) subject with the smallest change in CD8αβ expression. (B) Change of CD8αβ expression on PBL from the HIV-1 seropositive individual in the cohort of treated patients with the largest increase in CD4+ T lymphocytes/μL blood and CD8αβ MF. The CD8αβ MF showed an increase of 32 channels (baseline, 27; 16 weeks post-HAART, 59).
DISCUSSION
In most investigative studies and in routine clinical immunophenotyping, the expression of the CD8 molecule on human lymphocytes is determined with antibodies that recognize the α-chain of the CD8αα-homodimer and the CD8αβ-heterodimer. Some heterogeneity in the intensity of staining of cells using anti-CD8α antibodies has been observed in those studies.23,26,27,32,34 The difference in the intensity of anti-CD8α staining has been used to distinguish dim CD8α+ NK cells from bright CD8α+ T cells.35,36 However, these two CD8α+ cell fractions overlap. Therefore, the use of a MoAb specific for CD3 has been recommended in addition to an anti-CD8α MoAb to differentiate CD8+ T cells from non-T cells in routine immunophenotyping.25-27
In the present study, we have shown that a MoAb directed against the CD8αβ-heterodimer, 2ST8-5H7, binds almost exclusively to CD3+ T cells and recognizes nearly all CD8α+T cells from HIV-1 seronegative and seropositive individuals. A small subset of CD8+ T lymphocytes expresses only the CD8αα-homodimer and, presumably, matures through extrathymic pathways.37 Interestingly, this cell subset represents a significantly lower percentage of CD8+ T lymphocytes after HIV-1 infection. The MoAb, 2ST8-5H7, binds to virtually no NK cells, to substantially fewer TCRγδ+ T cells and CD4/CD8 coexpressing T cells than does a CD8α-chain–specific MoAb. These observations suggest that this CD8αβ-specific MoAb is an acceptable substitute for the anti-CD3 and anti-CD8α MoAb combination to detect CD8+ T lymphocytes.
Roederer et al38 have shown a small decrease in the density of CD8α expression on CD8+ T lymphocytes in PBL of HIV-1–infected individuals. We observed little difference in the CD8α MF between T lymphocytes from HIV-1 seronegative and seropositive individuals. However, the expression of the CD8β-chain was much more heterogeneous in PBL of those donors. The CD8αβ MF was greater in PBL of HIV-1 seronegative individuals than in PBL of HIV-1 seropositive individuals. Furthermore, PBL of HIV-1 seropositive individuals with more than 200 CD4+ T lymphocytes/μL blood showed a higher CD8αβ staining than PBL of HIV-1 seropositive individuals with fewer than 200 CD4+ T lymphocytes/μL blood. Moreover, using a panel of MoAbs directed against molecules associated with activation, adhesion, maturation, or cytotoxic function, we found that nonactivated CD8+ T lymphocytes from HIV-1 seronegative and seropositive individuals had significantly higher levels of anti-CD8αβ staining than did CD8+ T lymphocytes with phenotypic evidence of activation. The finding of a decreased cell surface expression of the CD8β-chain on activated CD8+ T lymphocytes is consistent with the previous observation that the expression of the CD8β-chain on CD8+T lymphocytes decreases following in vitro culture.19 39
Interestingly, the MF of CD8αβ staining of peripheral blood mononuclear cell (PBMC) from HIV-1 seropositive individuals did not correlate in all instances with the activation or maturation status of CD8α+ T cells, as determined by the expression of molecules associated with activation (HLA-DR and CD38) or maturation (CD45RA and CD45RO). This may be due to the fact that the cell surface expression of molecules associated with activation on PBMC of HIV-1–infected individuals does not fully reflect the functional activity of the cells. In addition, it is well known that the CD45RA/CD45RO naive/memory paradigm has not proven particularly useful in analyzing CD8+ T cells.40 A substantial fraction of CD45RA+ CD8+ T cells are probably not true naive cells.
Treatment of HIV-1 infections with HAART results in a substantial impact on viral load and peripheral blood CD4+ T lymphocyte counts.41-43 Significant changes in the CD8+ T lymphocyte subset in the peripheral blood of treated individuals have also been seen. An early increase and eventual fall in the number of circulating CD8+ T lymphocytes has been reported after initiation of HAART.41 Because persistent virus replication is responsible for driving the chronic activation of the immune system and HAART decreases this virus replication, treatment is associated with a decrease in the expression of activation-associated molecules, including HLA-DR and CD38, on CD8+ T lymphocytes.41 43 In the present study, we found that the CD8αβ MF in PBL of HIV-1–infected individuals significantly increases during the first 16 weeks of HAART. As we have shown that a relative high MF of staining of the CD8αβ-heterodimer is predominantly associated with nonactivated cells, our observation of treatment-associated increases in CD8αβ MF in PBL of HIV-infected individuals is consistent with a reduction in CD8+T-lymphocyte activation.
It is difficult to speculate as to the biologic ramifications of the decreased expression of CD8αβ on CD8α+ T cells of HIV-1–infected individuals because it is not clear whether CD8αα and CD8αβ have different cellular functions. It has been suggested that the β-chain of the CD8αβ-heterodimer increases the functional activity of the CD8 molecule.44 Others were unable to substantiate this observation.45 However, the observation made in the present study does indicate that CD8αα and CD8αβ expression are differentially regulated.
ACKNOWLEDGMENT
For excellent technical support in the antibody-fluorochrome conjugations and optimization of the reagent combinations used, we are indebted to Kirt Toussaint, Ed O'Connell, Lisa Edwards, and MaryLyn Monson. For donation of blood samples, we are grateful to all participating HIV-1 seronegative and seropositive blood donors. For collection of HIV-1 seropositive blood samples, we thank Beryl Chapman, Karen A. McLaughlin, and Dr Jennifer Adelson-Mitty.
Supported by the AACTG Developmental Immunology Award (NIH) AI 38858, the Coulter Corp, Miami, FL, and the German Bundesministerium für Forschung und Technologie AIDS program, Bonn, Germany.
Address reprint requests to Dr Med Jörn E. Schmitz, Division of Viral Pathogenesis, Department of Medicine, Beth Israel Deaconess Medical Center, RE113, 330 Brookline Ave, Boston, MA 02215.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal