THE PAST TWO DECADES have brought remarkable progress in our understanding of the molecular basis of hemophilia A. This disease, which has already been documented as a familial bleeding tendency in the fifth century,1 still persists as the most common hemorrhagic disorder, affecting 1 in approximately 5,000 males.2 Hemophilia has been associated with deficiency of a plasma component since 1937, when Patek and Taylor3 showed that the clotting defect of hemophilic plasma could be corrected by plasma of a normal individual. This component was called “antihemophilic factor,” or “factor VIII” according to the more recent nomenclature. Subsequent studies using preparations enriched in factor VIII activity have established factor VIII as being the cofactor of activated factor IX in the factor X–activating complex of the intrinsic coagulation pathway.4 However, the molecular entity of factor VIII has remained unidentified until the early 1980s, when the protein was purified to complete homogeneity, and its cDNA was cloned.5-8 This breakthrough has triggered numerous studies on the genetic and molecular basis of hemophilia A, and consequently our knowledge on the structure and function of the factor VIII protein has been rapidly expanding since then.
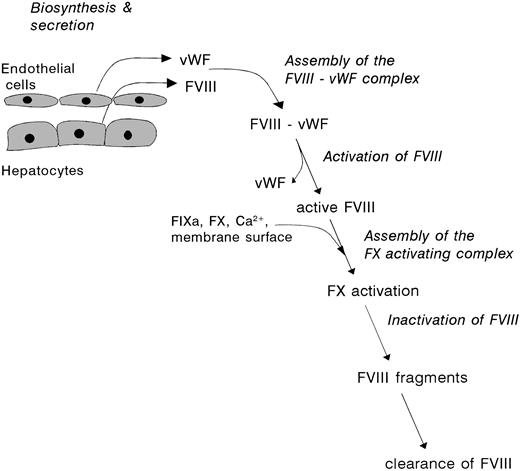
The present review focuses on the “life cycle” of factor VIII, which comprises the sequence of events between biosynthesis and clearance of the protein (see Fig 1). These processes are discussed in view of our current knowledge on factor VIII structure and function, with particular reference to the proteolytic modulation of factor VIII, and its assembly into the factor X activating complex.
The lifespan of factor VIII. Factor VIII is synthesized by various tissues, including liver, kidney, and spleen, as an inactive single-chain protein. After extensive posttranslational processing, factor VIII is released into the circulation as a set of heterodimeric proteins. This heterogenous population of factor VIII molecules readily interacts with vWF, which is produced and secreted by vascular endothelial cells. Upon triggering of the coagulation cascade and subsequent generation of serine proteases, factor VIII is subject to multiple proteolytic cleavages. These cleavages are associated with dramatic changes of the molecular properties of factor VIII, including dissociation of vWF and development of biological activity. After conversion into its active conformation, and participation in the factor X activating complex, activated factor VIII rapidly looses its activity. This process is governed by both enzymatic degradation and subunit dissociation.
The lifespan of factor VIII. Factor VIII is synthesized by various tissues, including liver, kidney, and spleen, as an inactive single-chain protein. After extensive posttranslational processing, factor VIII is released into the circulation as a set of heterodimeric proteins. This heterogenous population of factor VIII molecules readily interacts with vWF, which is produced and secreted by vascular endothelial cells. Upon triggering of the coagulation cascade and subsequent generation of serine proteases, factor VIII is subject to multiple proteolytic cleavages. These cleavages are associated with dramatic changes of the molecular properties of factor VIII, including dissociation of vWF and development of biological activity. After conversion into its active conformation, and participation in the factor X activating complex, activated factor VIII rapidly looses its activity. This process is governed by both enzymatic degradation and subunit dissociation.
BIOSYNTHESIS AND SECRETION OF FACTOR VIII
Factor VIII gene.
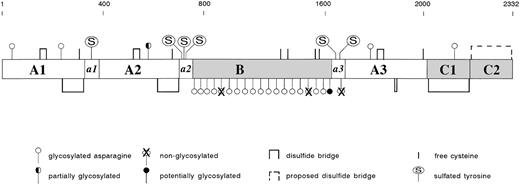
The gene of factor VIII is located at the tip of the long arm of the X chromosome.5 It spans over 180 kb, and as such is one of the largest genes known. Its transcription may require several hours assuming a transcription rate of 10 nucleotides per second, and yields a 9-kb mRNA product.5 The factor VIII gene comprises 26 exons, which encode a polypeptide chain of 2351 amino acids.6-8 This includes a signal peptide of 19 and a mature protein of 2332 amino acids. Analysis of the deduced primary structure determined from the cloned factor VIII cDNA showed the presence of a discrete domain structure: A1-a1-A2-a2-B-a3-A3-C1-C26-8 (Fig2). The A domains display approximately 30% homology to each other. These domains further display a similar extent of homology to the copper-binding protein ceruloplasmin and to factor V, the cofactor in the prothrombinase complex.9 The A domains are bordered by short spacers (a1, a2, and a3) that contain clusters of Asp and Glu residues, the so-called acidic regions. The C domains are structurally related to the C domains of factor V. In addition, the lipid-binding lectin discoidin I, human and murine milk fat globule proteins, and a putative neuronal cell adhesion molecule from Xenopus laevis share amino acid sequence similarity to the factor VIII C domains.10-12 The B domain is unique in that it exhibits no significant homology with any other known protein.
Biosynthesis of factor VIII.
Several tissues have the potential of expressing the factor VIII gene. Factor VIII mRNA has been demonstrated in a variety of tissues, including spleen, lymph nodes, liver, and kidney.13-15Transplantation studies in hemophilic animals showed that organs such as lung and spleen indeed contribute to the presence of circulating factor VIII.16,17 However, the liver most likely provides the primary source of factor VIII. This view is supported by liver perfusion18,19 and by liver transplantation studies in both animals and humans.20-22 A number of cases concerning hemophilic patients have been reported in which transplantation resulted in sustained, normalized levels of factor VIII.21 22
Several lines of evidence indicate that, within the liver, hepatocytes are the major factor VIII–producing cells. First, factor VIII mRNA is present in hepatocytes but not in sinusoidal cells.13Second, the promotor region of the factor VIII gene comprises responsive elements that are characteristic for hepatocyte-specific expression.23 Finally, in immuno-ultrastructural studies factor VIII protein was detected in the rough endoplasmatic reticulum and the Golgi apparatus of hepatocytes.24 It should be mentioned that other reports showed the presence of factor VIII in hepatic endothelium using histochemical techniques.25-27This is unexpected because these cells appear to lack factor VIII mRNA. It is possible that the latter observation reflects surface binding of factor VIII or internalization rather than factor VIII biosynthesis.
Secretion of factor VIII.
Studies on factor VIII biosynthesis and secretion have been limited by the lack of human cell lines that properly express significant amounts of factor VIII. Analysis of the factor VIII secretion process has therefore been restricted to autologous gene expression.28These studies showed that, in general, factor VIII is poorly expressed. Low expression is associated with a low level of steady-state mRNA29 and inefficient secretion.30 The initial stage of secretion involves the translocation of the mature 2332 amino acid polypeptide into the lumen of the endoplasmatic reticulum (ER), where N-linked glycosylation occurs. Within the ER, factor VIII appears to interact with a number of chaperone proteins, including calreticulin, calnexin, and the Ig-binding protein (BiP).31-34 Due to the interaction with these chaperone proteins, a significant proportion of the factor VIII molecules is retained within the ER, thereby limiting the transport of factor VIII to the Golgi apparatus. The mechanism reponsible for the transport from the ER to the Golgi apparatus is not elucidated yet. However, recent studies indicate that this step involves an intracellular membrane lectin: endoplasmatic reticulum-Golgi intermediate compartment-53 (ERGIC-53).35
Within the Golgi apparatus, factor VIII is subject to further processing, including modification of the N-linked oligosaccharides to complex-type structures, O-linked glycosylation, and sulfation of specific Tyr-residues (Fig 2). In addition, factor VIII is among the many proteins that undergoes intracelullar proteolysis.36-39 The middle part and the carboxyterminal region of the B domain comprise a motif (Arg-X-X-Arg), which is similar to the motif that is recognized by intracellular proteases of the subtilisin-like family.38 39 The responsible endoprotease mediating intracellular factor VIII proteolysis, however, has not been identified. The Arg-X-X-Arg motif allows proteolysis at Arg1313 and at Arg1648. The latter event disrupts the covalent linkage of the factor VIII heavy chain (A1-a1-A2-a2-B) and light chain (a3-A3-C1-C2), giving rise to the heterodimeric molecule that circulates in plasma.
The factor VIII heavy and light chain remain noncovalently associated through the A1 and A3 domain in a metal-ion–dependent manner.7,40-42 Considering the structural homology of factor VIII to the copper-binding protein ceruloplasmin, it is not surprising that copper ions have been found in factor VIII as well.43,44 One molecule of copper is present per molecule of factor VIII. Most likely, the copper-ion binding site is composed of residues His265, Cys310, His315, and Met320 within the A1 domain.44 Binding of copper ions to this site may allow the A1 domain to adopt an A3-domain binding conformation. Alternatively, copper ions may directly bridge the A1 and A3 domain by interacting with both domains simultaneously. Whether only copper ions are involved in the association between heavy and light chain is unclear. Whereas in the absence of other metal ions copper ions are ineffective in promoting reassembly of dissociated heavy and light chain, calcium or manganese ions are considerably more efficient in this respect.45-47a However, specific activity of factor VIII dimers that were reassociated in the presence of calcium ions, is markedly enhanced upon the addition of copper ions.47a Apparently, copper ions serve an auxiliary role to enhance cofactor function of factor VIII. These observations suggest that multiple sites may be involved in the association between heavy and light chain. Irrespective of the precise mechanism, it is clear that metal ions serve an important role in maintaining the heterodimeric structure of secreted factor VIII.
Factor VIII secretion and hemophilia A.
Aberrant biosynthesis or secretion may result from several defects. Obviously, gross deletions or rearrangements may result in impaired transcription, RNA processing, or translation. No data are reported on the secretion of such gene products or on the stability within the circulation provided that these gene products are actually secreted. Defective secretion may further be caused by apparently minor gene defects like single missense mutations. Frequently known missense mutations associated with low levels of factor VIII are located in codon 2307, resulting in replacement of Arg2307 by Gln or Leu.48,49 Both mutations have been analyzed using recombinant factor VIII mutants expressed in mouse fibroblasts50 and COS-1 monkey kidney cells.51Both proteins appear to be functionally normal, but are poorly secreted. The majority of the retained mutants are targeted into an ER-associated degradation pathway. The mechanism responsible for intracellular retention of these mutated factor VIII molecules is unknown.
Reduced levels of factor VIII protein may also result from defects located outside the factor VIII gene. One striking example concerns patients having combined factor V and VIII deficiency. The gene responsible for combined factors V and VIII deficiency has been mapped to the long arm of chromosome 18, between markers D18S849 and D18S1103,52,53 whereas the genes for factors V and VIII are located at chromosomes 1 and X, respectively. Recently, the unknown gene has been idenfied to encode the intracellular membrane lectin ERGIC-53, a resident protein of the ER-Golgi intermediate compartment.35 Indeed, affected individuals displayed mutations in this gene, in association with a complete lack of expression of ERGIC-53.35 Apparently, ERGIC-53 contributes to the secretion process of factors V and VIII, presumably by acting as a chaperone protein. Identification of the underlying mechanism should provide more insight in the intracellular routing and secretion of both cofactors.
ASSEMBLY OF THE FACTOR VIII-VON WILLEBRAND FACTOR (vWF) COMPLEX
Binding sites for vWF.
Immediately after its release into the circulation, the factor VIII heterodimer interacts with its carrier protein vWF to form a tight, noncovalent complex. Each monomer of the multimeric vWF protein is able to bind one factor VIII molecule with high affinity (kd < 0.5 nmol/L).54-57 In vivo, however, the stoichiometry is limited by the number of factor VIII molecules present, resulting in approximately a 1:50 ratio.
Two peptide regions of factor VIII are implicated to be involved in binding vWF: one at the aminoterminal end of intact factor VIII light chain,55-59 and one at the carboxyterminal end (residues 2303-2332).57,60-62 Using proteolytically derived fragments of factor VIII light chain, it was shown that both these individual regions are capable of binding vWF.57 However, the affinity of these fragments for vWF is markedly lower compared with the intact factor VIII heterodimer (two and three orders of magnitude for the aminoterminal and carboxyterminal end, respectively).57Apparently, both ends of factor VIII light chain act synergistically in the binding of vWF.
With respect to vWF binding to the aminoterminal region of factor VIII light chain, it appears that residues 1649 to 1671, thus including sulfated residue Tyr1664, are dispensable for vWF binding.57,63,64 In addition, the Arg1689-Ser1690 cleavage site has to be intact,57 suggesting that residues carboxyterminal of this thrombin cleavage site contribute to binding as well. Recombinant factor VIII synthesized in the presence of an inhibitor of sulfation displays reduced binding to vWF,63,65 suggesting a role for sulfated Tyr1680 in the interaction with the carier protein. Indeed, replacement of Tyr1680 by Phe results in loss of high-affinity binding to vWF, allowing only a low-affinity interaction.63,65 66 The presence of Tyr1680 in its sulfated form thus contributes to optimal binding of factor VIII to vWF.
Factor VIII interactions modulated by vWF.
One functional aspect of factor VIII-vWF complex formation may be to prevent premature binding of factor VIII to components of the factor X–activating complex. For instance, binding of factor VIII light chain to factor IXa is inhibited by vWF.67 Because the affinity of factor VIII for vWF exceeds that for factor IXa by approximately 100-fold,56 67 factor VIII highly favors vWF binding over factor IXa binding. The mechanism by which vWF inhibits factor IXa binding is not yet elucidated. Because binding of both proteins by factor VIII light chain seems to involve distinct parts of the molecule, it is unlikely that vWF competes with factor IXa for binding to the same site. Mechanisms that could contribute to inhibition include sterical hindrance and alteration of the factor VIII conformation so that factor IXa cannot bind.
Whereas factor IXa and vWF bind at different sites, vWF and phospholipids both bind to the C2-domain region 2303-2332.62,68 Using a human anti–factor VIII antibody, it has been shown that these sites, although in very close proximity, do not completely overlap.69 Nevertheless, the close proximity of these sites may explain the observations that binding of factor VIII to vWF is incompatible with factor VIII binding to membrane surfaces.70-73 It should be noted that, in comparison with noncomplexed factor VIII, factor VIII in complex with vWF is less susceptible to proteolytic attack by various lipid-binding proteases.74-77 These vitamin K–dependent serine proteases, which include activated protein C (APC) and factor Xa, require to assemble with factor VIII at a membrane surface for efficient proteolysis of factor VIII.76,78 In contrast, thrombin displays proteolytic activity independent of a membrane surface. Indeed, vWF does not protect factor VIII against cleavage by thrombin.79-81 Cleavage of factor VIII by thrombin results in loss of vWF binding and conversion of factor VIII into its active conformation.
vWF and hemophilia A.
The association with vWF serves an important role in factor VIII physiology, as vWF functions as a stabilizer of the heterodimeric structure of factor VIII.45,82-84 The physiological relevance of complex formation is particularly apparent in patients with von Willebrand disease (vWD) (type 3), who have no detectable vWF protein. Not only do these patients have a secondary deficiency of factor VIII, but they also have a considerably reduced half-life of intravenously administred factor VIII.83,85-88 This phenotype is also observed in patients with vWD with the so-called Normandy variant, which is defined as type 2N. Despite normal levels of circulating vWF, factor VIII levels are severely reduced.89 90 These patients harbor a mutation in the factor VIII–binding domain of vWF, which results in defective binding to vWF.
With regard to factor VIII, two distinct basepair substitutions have been reported that are associated with impaired complex assembly.49,91 92 Both mutations result in the replacement of one single amino acid, Tyr1680, and are associated with hemophilia A. So far, no mutations located within the C2 domain region have been reported that are associated with reduced affinity for vWF.
ACTIVATION OF FACTOR VIII
Cleavage sites associated with factor VIII activation.
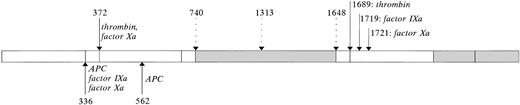
Within the factor X–activating complex, the proteolytic activity of factor IXa is markedly enhanced by factor VIII. It has been well established that proteolysis of factor VIII is required for the generation of its cofactor activity.79,93-97 The uncleaved factor VIII procofactor lacks the ability to enhance factor IXa activity.47,98 Enzymes that are able to endow factor VIII with its cofactor activity include thrombin and factor Xa.79,93,98-102 Thrombin cleaves factor VIII at one specific site within the light chain, Arg1689, and at two sites in the heavy chain: Arg372 and Arg740(Fig 3).79Proteolysis of factor VIII heavy chain by factor Xa involves three sites: Arg336, and the two thrombin-cleavage sites Arg372 and Arg740(Fig 3).79 With regard to factor VIII light chain, factor Xa is able to cleave at Arg1689, a site that is shared with thrombin, but also at Arg1721, a site that is specific for factor Xa.79 It should be noted that this site is cleaved in human factor VIII, but not in porcine factor VIII.103 It has been unclear whether cleavage at Arg1721 contributes to factor VIII activation or inactivation, because prolonged incubation with factor Xa results in loss of factor VIII activity in parallel with cleavages at positions 336 and 1721.79 This has been resolved by studying reassociated dimers of intact factor VIII heavy chain with either thrombin– or factor Xa–cleaved factor VIII light chain.47 The resulting factor VIII dimers were functionally indistinguishable, demonstrating that factor Xa–cleavage of the light chain is not associated with inactivation. It has been reported that factor Xa–activated factor VIII displays less activity than thrombin-activated factor VIII.79,98 102 Because cleavage of the light chain cannot be responsible for this phenomenon, this is most likely due to additional factor Xa–cleavage at Arg336in the factor VIII heavy chain.
The factor VIII protein. Mature factor VIII consists of 2332 amino acids, which are arranged in a discrete domain structure: A1 (residues 1-336), A2 (373-710), B (741-1648), A3 (1690-2019), C1 (2020-2172), and C2 (2173-2332). The A domains are bordered by acidic regions a1 (337-372), a2 (711-740), and a3(1649-1689). Disulfide Bridges: Using B-domainless factor VIII, seven disulfide bonds have been identified: residues 153 and 179, 248 and 329 (A1 domain), 528 and 554, 630 and 711 (A2 domain), 1832 and 1858, 1899 and 1903 (A3 domain), and 2021 and 2169 (C1 domain).196Within the C2 domain, residues 2174 and 2326 most likely also form a disulfide bridge. Free cysteine-residues have been identified at positions 310, 692, and 2000.196 Cys528 and Cys1858 may be present as free cysteines, because these residues are reactive toward a sulfhydryl-specific fluorphor.197 With regard to the Cys-residues in the B-domain it is unknown whether they are free or linked.N-Linked Glycosylation: Factor VIII contains 25 consensus sequences (Asn-Xxx-Thr/Ser) that allow N-linked glycosylation. Using either full-length or B-domainless factor VIII, the majority of these sites have been shown to be glycosylated: residues 42 and 239 (A1 domain), residues 757, 784, 828, 900, 963, 1001, 1005, 1055, 1066, 1185, 1255, 1259, 1282, 1300, 1412, and 1442 (B domain), residue 1810 (A3 domain), and residue 2118 (C2 domain).198-200Nonglycosylated residues are present at positions 943 and 1384 (B domain) and at position 1685 (a3 acidic region). Residue 582 (A2 domain) has been reported to be nonglycosylated in two studies,199,200 whereas one study reported this residue to be partially glycosylated.198 Finally, it remains to be investigated whether residue 1512 (B domain) is glycosylated. Tyrosine Sulfation: The acidic regions contain consensus sequences that allow sulfation of Tyr-residues at positions 346 (a1 region), 718, 719, 723 (a2 region), 1664, and 1680 (a3 region). Analysis using recombinant proteins established that all sites indeed can be sulfated.
The factor VIII protein. Mature factor VIII consists of 2332 amino acids, which are arranged in a discrete domain structure: A1 (residues 1-336), A2 (373-710), B (741-1648), A3 (1690-2019), C1 (2020-2172), and C2 (2173-2332). The A domains are bordered by acidic regions a1 (337-372), a2 (711-740), and a3(1649-1689). Disulfide Bridges: Using B-domainless factor VIII, seven disulfide bonds have been identified: residues 153 and 179, 248 and 329 (A1 domain), 528 and 554, 630 and 711 (A2 domain), 1832 and 1858, 1899 and 1903 (A3 domain), and 2021 and 2169 (C1 domain).196Within the C2 domain, residues 2174 and 2326 most likely also form a disulfide bridge. Free cysteine-residues have been identified at positions 310, 692, and 2000.196 Cys528 and Cys1858 may be present as free cysteines, because these residues are reactive toward a sulfhydryl-specific fluorphor.197 With regard to the Cys-residues in the B-domain it is unknown whether they are free or linked.N-Linked Glycosylation: Factor VIII contains 25 consensus sequences (Asn-Xxx-Thr/Ser) that allow N-linked glycosylation. Using either full-length or B-domainless factor VIII, the majority of these sites have been shown to be glycosylated: residues 42 and 239 (A1 domain), residues 757, 784, 828, 900, 963, 1001, 1005, 1055, 1066, 1185, 1255, 1259, 1282, 1300, 1412, and 1442 (B domain), residue 1810 (A3 domain), and residue 2118 (C2 domain).198-200Nonglycosylated residues are present at positions 943 and 1384 (B domain) and at position 1685 (a3 acidic region). Residue 582 (A2 domain) has been reported to be nonglycosylated in two studies,199,200 whereas one study reported this residue to be partially glycosylated.198 Finally, it remains to be investigated whether residue 1512 (B domain) is glycosylated. Tyrosine Sulfation: The acidic regions contain consensus sequences that allow sulfation of Tyr-residues at positions 346 (a1 region), 718, 719, 723 (a2 region), 1664, and 1680 (a3 region). Analysis using recombinant proteins established that all sites indeed can be sulfated.
Limited proteolysis of factor VIII. The major part of factor VIII circulates as a set of heterogenous dimers, consisting of a light (a3-A3-C1-C2) and heavy chain (A1-a1-A2-a2-B). The heavy chain is variably sized due to limited proteolysis within the B domain. Some of these cleavages may occur intracellularly at positions 1313 and 1648 (open downward arrows). Factor VIII can be converted into its active form by proteolysis in both the heavy and light chain by various serine proteases (closed downward arrows), including thrombin and factor Xa. Because proteolysis by factor Xa but not thrombin is inhibited by vWF, thrombin is probably the physiological activator of factor VIII. Proteolytic degradation of factor VIIIa proceeds through cleavages within the A1 and A2 domains by various serine proteases (upward arrows), and results in release of the a1 acidic region and bisecting of the A2 domain. In contrast to what has previously been assumed, cleavages within the light chain by factor IXa or factor Xa do not result in inactivation of factor VIII, but contribute to the development of factor VIII cofactor activity.
Limited proteolysis of factor VIII. The major part of factor VIII circulates as a set of heterogenous dimers, consisting of a light (a3-A3-C1-C2) and heavy chain (A1-a1-A2-a2-B). The heavy chain is variably sized due to limited proteolysis within the B domain. Some of these cleavages may occur intracellularly at positions 1313 and 1648 (open downward arrows). Factor VIII can be converted into its active form by proteolysis in both the heavy and light chain by various serine proteases (closed downward arrows), including thrombin and factor Xa. Because proteolysis by factor Xa but not thrombin is inhibited by vWF, thrombin is probably the physiological activator of factor VIII. Proteolytic degradation of factor VIIIa proceeds through cleavages within the A1 and A2 domains by various serine proteases (upward arrows), and results in release of the a1 acidic region and bisecting of the A2 domain. In contrast to what has previously been assumed, cleavages within the light chain by factor IXa or factor Xa do not result in inactivation of factor VIII, but contribute to the development of factor VIII cofactor activity.
Because activation of factor VIII involves proteolysis of both its heavy and light chain, it is of interest to compare the relative contribution of each cleavage to the development of factor VIII cofactor function. The contribution of cleavage at Arg740to factor VIII activation is limited, because mutations at this position do not interfere with factor VIII activation or function.97 Selective cleavage of factor VIII at Arg372 by a snake venom–derived protease generates a factor VIII molecule that displays 60% of the activity of fully activated factor VIII.80 Factor VIII dimers cleaved within the light chain only have 25% to 30% of the activity displayed by fully activated factor VIII.47,104 These findings allow the conclusion that both cleavage at Arg372 and Arg1689 are required to exert full cofactor activity. This view is supported by the observation that recombinant factor VIII mutants containing replacements at either Arg372 or Arg1689 are unable to correct the clotting time of factor VIII–deficient plasma.97
Acidic regions.
With regard to the cleavage sites at positions 372, 740, and 1689, it is noteworthy that these residues are located at the carboxyterminal end of an acidic sequence interconnecting adjacent domains. These acidic regions (a1, a2, and a3, see Fig 2) contain several Tyr residues in a sequence that meets the consensus features for tyrosine sulfation.105,106 Sulfated Tyr residues may serve a role in various processes, including protein-protein interactions.106 Because thrombin is known to interact with a variety of acidic, sulfated sequences, it has been proposed that acidic regions within factor VIII serve a role in thrombin activation of factor VIII.107 Replacement of Tyr346 (a1 region) or Tyr1664(a2 region) indeed results in factor VIII mutants that are activated by thrombin less efficiently.66 It should be mentioned that deletion of Tyr1664 leaves the activation kinetics of factor VIII by thrombin unaffected.108 109
Replacement of Tyr-residues by Phe in the a2 acidic region (residues 718, 719, and 723) did not affect thrombin activation, but rather resulted in mutants that displayed reduced ability to stimulate factor IXa enzymatic activity.66 In contrast, recombinant factor VIII variants (both full-length and B-domainless) containing nonsulfated Tyr at these positions are activated normally and display full cofactor activity.110,111 Although both approaches reveal apparently contrasting findings with regard to the effect of Tyr-sulfation in the a2 region on factor VIII cofactor function, they allow the conclusion that sulfation of these Tyr-residues does not contribute to activation of factor VIII. However, the acidic nature of this a2 sequence appears to be of importance for activation of factor VIII. First, a monoclonal antibody directed against a2 inhibits thrombin activation of factor VIII, but does not interfere with factor VIII cofactor activity.112,113 Secondly, deletion of a2 or part thereof in B-domainless factor VIII results in molecules that require higher thrombin concentrations than normal factor VIII for efficient activation, but display normal factor IXa cofactor activity.110,112 Interestingly, in both cases it was observed that reduced thrombin activation was caused by a reduced cleavage efficiency at Arg372 and Arg1689, suggesting that the a2 acidic region influences cleavage in remote regions in the factor VIII molecule. This hypothesis is supported by observations using a factor VIII chimera with a replacement in the a2 region. In this chimeric molecule, residues 716-736 of a2 have been replaced by a sequence that is known to have a high affinity for thrombin, ie, the amino acid sequence 51-80 of heparin cofactor II.114 This chimeric protein proved more potent than normal factor VIII in correcting the clotting time of factor VIII–deficient plasma. The increased intrinsic activity is caused by an increased rate of thrombin cleavage at Arg372 and Arg1689 compared with normal factor VIII. Thus, these findings suggest that the a2 region promotes proteolytic activation of factor VIII.
Apart from their role in thrombin activation, the acidic regionsa1 and a3 contribute to factor VIII function also in an additional manner. The a1 region has been described to be involved in maintaining the stability of the factor VIII heterotrimer115 and in binding of factor X116,117 (see Factor VIII Inactivation section). Thea3 region is important for high affinity binding of vWF (see Assembly of the Factor VIII-vWF Complex section). Further, factor VIII heterodimers consisting of uncleaved heavy and light chain do not posses any cofactor activity.47,48 In contrast, factor VIII exclusively cleaved at position 1689, thus lacking the a3region, displays significant cofactor activity (approximately 25% of fully activated factor VIII).47 104 Apparently, the acidic region a3 functions as an activation peptide that needs to be cleaved off for exposure of cofactor activity.
Defective activation and hemophilia A.
Mutations resulting in replacement of amino acids at the factor VIII activation sites should predispose to hemorraghic diathesis. Indeed, missense mutations at Arg372, Ser373, and Arg1689 are associated with hemophilia A.49,118-124 With regard to the Arg1689mutation, biochemical data predict residual activity to be at least half of that of normal factor VIII, as cleavage at Arg372accounts for approximately 60% of total activity.80However, residual activity appears to range between <1% and ≈12%, thus lower than expected.49 Several possibilities may be considered to explain this apparent discrepancy. First, due to substitution of Arg1689, the vWF binding site including sulfated Tyr1680 is not cleaved from the light chain. Therefore, the factor VIII–vWF complex may fail to dissociate upon thrombin treatment. As a consequence, factor VIII is unable to interact with factor IXa and phospholipids, and thus cannot assemble into the factor X–activating complex (see below). Secondly, in most of the documented patients with a mutation at Arg1689, this residue is replaced by a Cys.49 This Cys-residue has the potential to form an extra disulfide bridge within the factor VIII light chain.125 126 It seems conceivable that this results in misfolding within the factor VIII light chain, which precludes biological activity. Finally, residual activity may be affected by the type of amino acid substitution. This view is supported by the notion that a similar discrepancy exist for substitutions at Arg372. For instance, Arg372 to Pro substitutions exclusively result in severe hemophilia A, whereas Arg372 to His substitutions result in a mild to moderate phenotype.
ASSEMBLY OF THE FACTOR X–ACTIVATING COMPLEX
Regulation of complex assembly.
To activate factor X, factor IXa and factor VIIIa assemble into a membrane-bound complex. To maintain the hemostatic balance, this complex should only assemble after initiation of the coagulation cascade, implying that participation of factor VIII in this complex is subject to a delicate regulatory mechanism. In this regard, vWF plays a central role. Although factor IXa displays similar affinity for nonactivated and activated factor VIII,127 the factor IXa binding site within nonactivated factor VIII is unlikely to be accesible when factor VIII is in complex with vWF.67Furthermore, binding of factor VIII to the membrane surface is inhibited by vWF,70-73 suggesting that only activated factor VIII that is dissociated from vWF is able to bind to the membrane surface. However, it should be mentioned that the affinity of factor VIII for the membrane surface is dependent on the membrane composition, as affinities have been reported that differ 10- to 100-fold (10−9 to 10−11mol/L).72,128-131 Thus, under particular conditions the affinity of vWF and the membrane surface for factor VIII is similar. Apparently, a delicate balance may exist between factor VIII being in complex with vWF or the membrane surface. Because in direct binding studies vWF prevents binding of factor VIII to the membrane surface,70-73 a minor part of the factor VIII population probably is in complex with the membrane surface. This situation changes dramatically upon cleavage of factor VIII light chain, which results in a 1,600-fold decrease in affinity for vWF.57Because of this event, the balance will readily shift toward factor VIIIa binding to the membrane surface. This subsequently favors binding of factor IXa to factor VIIIa, which is no longer associated with vWF. Ultimately, this leads to the assembly of the membrane-bound factor VIIIa–factor IXa complex that activates factor X.
The role of the membrane surface in complex assembly.
The notion that in the absence of a membrane surface the generation of factor Xa by the factor VIIIa–factor IXa complex is negligible132 underscores the essential role of the membrane surface in the factor X–activating complex. The membrane surface may act in two distinct ways: first by positioning the enzyme-cofactor complex into an active conformation or, alternatively, by locating the enzyme and cofactor at the same site. At present, data have been reported that are in support of both mechanisms. On the one hand, it has been shown that the affinity of factor IXa for factor VIIIa is increased 2,000-fold in the presence of phospholipids,133 which suggests that the second mechanism is dominant. On the other hand, the affinity of (activated) factor VIII for factor IXa is reported to be similar in the presence (kd = 10−8 to 10−9mol/L)104,127,132,134,135 and absence (kd = 10−8 mol/L)67,132 of a phospholipid surface. Gilbert and Arena132 showed that in the presence of phospholipids the catalytic activity of the enzyme-cofactor complex is increased 1,500-fold. These data favor the view that the membrane surface positions the enzyme and cofactor in a conformation that allows efficient substrate cleavage.
Location of factor IXa interactive sites.
The interaction between factor VIIIa and factor IXa has been investigated in several elegant studies using factor IXa molecules that carry a fluorescent-label in the active site.127,134-139 It became evident that in the presence of phospholipids, factor VIIIa induces a conformational change in the factor IXa protease domain. In addition, maximal changes in the factor IXa protease domain require the presence of the factor VIIIa A2-domain,134 suggesting that the A2 domain contains a factor IXa interactive site. Indeed, studies using a series of synthetic peptides showed that factor IXa binding can be attributed to the A2-domain sequence 558 to 565.137Furthermore, a region within the carboxyterminal part of the A2 domain (residues 698 to 710) also has been proposed to comprise a factor IXa–binding site.140 141
Besides factor VIII heavy chain, the light chain also contributes to factor IXa binding. In equilibrium binding studies, isolated factor VIII light chain proved to bind factor IXa with high affinity.67 Moreover, factor VIII light chain and the intact factor VIII heterodimer are indistinguishable in terms of affinity for factor IXa, indicating that high-affinity binding to factor IXa is mediated by the factor VIII light chain. Binding of factor VIII light chain to factor IXa was found to be inhibited by the A3-domain directed monoclonal antibody CLB-CAg A, a strong inhibitor of factor VIII activity.67,113,142 By using synthetic peptides, the A3-domain sequence 1811 to 1818 has been identified as a site that binds factor IXa.142 Thus, interaction with factor IXa involves at least three sites on factor VIII: residues 558-565, 698-710, and 1811-1818.
Three-dimensional model of factor VIII and factor IXa.
Three-dimensional representations of the enzyme and cofactor have been published, based on factor IXa crystallography143 and factor VIII homology modeling144,145(Fig 4). So far, it is unknown which residues in the factor IXa molecule are involved in binding factor VIII. However, the location of these sites should fit with the location of their counterparts on the factor VIII molecule. Although the amino acid numbering suggests that the factor IXa–binding regions are located in completely different parts of the factor VIII protein, the three-dimensional model indicates that the factor IXa–binding sites are in close vicinity, and are exposed at the same side of the molecule (Fig 4). Matching of the factor VIII and factor IXa models suggests that factor IXa comprises distinct sites involved in factor VIIIa binding, interacting with the A2 or A3 domain. Factor VIIIa binding appears to be mediated by both the factor IXa light chain and protease domain. The involvement of the protease domain is in agreement with the observation by Bajaj et al146,147 that the stimulation of factor IXa proteolytic activity by factor VIIIa is inhibited by a monoclonal antibody directed against the protease domain residues 231-265. This suggests that this protease domain region comprises a factor VIII–binding site. However, recombinant factor IXa molecules in which residues in the antibody-binding epitope have been mutated combine a strongly reduced affinity for the antibody with normal biological activity.148 Because normal activity is associated with normal factor VIII binding, these findings leave the exact location of the factor VIII binding site in the factor IXa heavy chain unidentified. Irrespective of its precise location, this site on the factor IXa heavy chain presumably interacts with the factor VIII A2 domain, because this domain induces the largest change in the conformation of the factor IXa active site.134
Model of the factor VIII and factor IXa molecules. Shown are representiations of porcine factor IXa (Protein Data Bank accession code 1pfx) and the triplicated A-domains of human factor VIII (Hemophilia A web site, http://europium.mrc.rpms.ac.uk), which are derived from crystallography and homology modeling, respectively. Factor IXa binding region in the factor VIII A3 domain (residues 1811-1818) is shown in white, whereas the binding regions in the A2 domain (residues 558-565 and 698-710) are shown in dark and light blue, respectively (space-filling representations). These sites are in close vicinity, and are exposed at the same side of the molecule. The factor VIII A2 domain is required to induce significant changes within the factor IXa protease domain, indicating that it binds to the factor IXa protease domain. The A3 domain of factor VIII has been proposed to interact with the factor IXa light chain. Within the factor IXa light chain, residues 12, 64, 69, 78, 92, and 94 (see refs 150 to 155) are indicated (red, space-filling representation). These residues have been reported to be associated with an abnormal response to factor VIIIa in factor X activation.
Model of the factor VIII and factor IXa molecules. Shown are representiations of porcine factor IXa (Protein Data Bank accession code 1pfx) and the triplicated A-domains of human factor VIII (Hemophilia A web site, http://europium.mrc.rpms.ac.uk), which are derived from crystallography and homology modeling, respectively. Factor IXa binding region in the factor VIII A3 domain (residues 1811-1818) is shown in white, whereas the binding regions in the A2 domain (residues 558-565 and 698-710) are shown in dark and light blue, respectively (space-filling representations). These sites are in close vicinity, and are exposed at the same side of the molecule. The factor VIII A2 domain is required to induce significant changes within the factor IXa protease domain, indicating that it binds to the factor IXa protease domain. The A3 domain of factor VIII has been proposed to interact with the factor IXa light chain. Within the factor IXa light chain, residues 12, 64, 69, 78, 92, and 94 (see refs 150 to 155) are indicated (red, space-filling representation). These residues have been reported to be associated with an abnormal response to factor VIIIa in factor X activation.
Assuming an interaction between the factor IXa protease domain and the factor VIII A2 domain, it seems conceivable that the A3 domain interacts with a region within the factor IXa light chain. This is in line with recent observations that the light chain of factor VIII binds to the light chain of factor IXa.138,149 Furthermore, mutations within the factor IXa light chain have been described that are associated with an abnormal response to factor VIIIa in factor X activation.150-154 These mutations are dispersed over the factor IXa light chain, indicating that multiple sites may contribute to binding of the factor VIII A3 domain. Alternatively, some mutations may destabilize the factor VIII binding site by affecting the conformation of the factor IXa light chain. This latter possibility has been reported for two distinct factor IX mutations.154 155
Collectively, by combining biochemical data with the three-dimensional models of factor IXa and factor VIII, it appears that the factor VIII A2 domain binds to the factor IXa heavy chain, and the factor VIII A3 domain to the factor IXa light chain (Fig 4). It is of importance to realize that for factor VIII as well as for factor IXa the current models not fully represent the biologically active molecules. The factor IXa structure has been determined in the absence of calcium ions,143 which are obligatory for optimal exposure of the factor VIII light chain binding site and of the catalytic centre.133,155 The factor VIII model provided by Pemberton et al145 comprises the A domains only and lacks the B and C domains and the acidic domain spacers. It cannot be excluded that these domains affect the structure of the A domains. In addition, this factor VIII model does not distinguish between the inactive procofactor or activated factor VIII. It is obvious that both factor VIII species will have different structural properties, because only factor VIII, which is cleaved at specific positions, is able to stimulate factor IXa activity. Despite these restrictions, both the factor VIII and factor IXa model provide an important basis for proper selection of residues that may be investigated for their contribution in the assembly of the factor VIII–factor IX complex.
Factor IXa binding and hemophilia A.
Inspection of the hemophilia A database shows that several mutations have been reported that are in or close to the factor IXa binding sites: codons 558, 565, and 566, codons 698, 701, and 704 in the heavy chain, and codons 1789, 1796, 1823, and 1825 in the factor VIII light chain.49 It seems reasonable to assume that the bleeding tendency which is associated with these mutations is caused by suboptimal assembly of the factor IXa–factor VIIIa complex. With regard to the 558 and 566 mutation, this view has been supported by studies using recombinant factor VIII.156 Interestingly, a mutation outside the factor IXa–binding regions, ie, at codon Arg527, also has been reported to be associated with inefficient stimulation of factor IXa proteolytic activity.157,158 Examination of the three-dimensional model shows that Arg527 is located in the immediate vicinity of the factor IXa binding sequence 558 to 565.145 The exposure of this factor IXa binding site may be affected by substitution of Arg527. Alternatively, Arg527 may be part of an extensive factor IXa–binding interface, involving multiple sites of the A2 domain.
INACTIVATION OF FACTOR VIII
Mechanisms of inactivation.
Downregulation of the factor X–activating complex may involve inactivation or inhibition of either the enzyme factor IXa or the cofactor, factor VIIIa. Inactivation of the cofactor comprises two distinct pathways: proteolytic degradation and spontaneous dissociation. Once activated, factor VIII cofactor activity is rapidly lost.135,159-162 Compared with activated factor VIII, the procofactor is markedly more stabile, which is illustrated by its dissociation rate being 100-fold lower (kdiss ≈4 to 6 × 10−4 s−1 and 4 × 10−6 s−1 for factor VIIIa and factor VIII, respectively).47,163 The intrinsic instability of factor VIIIa can be attributed to the weak interaction between the A2 domain and the metal ion-linked A1/A3-C1-C2 dimer.164-166 The kd for this interaction is approximately 0.2 μmol/L.42 Because this value exceeds the factor VIII concentration in plasma 100- to 1,000-fold, equilibrium is in favor of the inactive, dissociated state of factor VIIIa.
Proteolytic degradation of factor VIIIa involves cleavages in the heavy chain at positions 336 and 562 by various enzymes, such as factor IXa, factor Xa, and APC.74-79,167-170 Cleavage at position 336 in factor VIIIa releases a1, the acidic sequence that interconnects the A1 and A2 domain. Because of this release, the A2 domain dissociates more rapidly from the factor VIIIa heterotrimer.115 This acidic spacer has been proposed to comprise a binding site for the substrate factor X,116,117indicating that release of this site results in impaired substrate binding. Thus, cleavage at Arg336 affects both intramolecular (A2 domain dissociation) and intermolecular (factor VIII–factor X) interactions. Arg562, which is part of the A2 domain sequence that comprises a factor IXa interactive site, is exclusively cleaved by APC.168 It seems conceivable that loss of cofactor activity due to cleavage at this site reflects the loss of the ability to interact with factor IXa.
One intriguing question is whether proteolytic degradation or spontaneous dissociation dominates the inactivation of factor VIII in vivo. At present the relative contribution of each mechanism to factor VIII inactivation is not fully understood, although some reports indicate that spontaneous dissociation is dominant.162,171,172 This view is underscored by the observation that dissociation of the factor VIIIa heterotrimer may be accelerated by binding of the A2 domain to the low-density lipoprotein receptor-related protein.173 To describe the process of factor VIIIa inactivation, it should further be considered that factor IXa plays a dual role. It stabilizes factor VIIIa by linking the A2 domain to the A3 domain,137,142,159 and protects factor VIII against inactivation by APC.136,167,174 On the other hand, under certain conditions factor IXa may inactivate factor VIIIa by cleavage at position 336,169 170 a site that is shared with factor Xa and APC. The fact that factor IXa is involved both in stabilization and in inactivation of factor VIII complicates a final assessment of the regulatory role of factor IXa in intrinsic factor Xa formation.
Defects in factor VIII inactivation.
Impaired inactivation of factor VIIIa or its homologue factor Va may be associated with a disturbed balance between procoagulant and anticoagulant systems. With respect to factor Va this view is supported by the notion that mutation at Arg506, a site that is cleaved by APC, predisposes to venous thromboembolism.175-178 It has been investigated whether patients displaying venous thromboembolism carry analogous mutations at the APC cleavage sites in factor VIII (ie, Arg336 and Arg562).179,180 However, this association has not been observed, which suggests that mutations at these positions are rare. Alternatively, such mutations may not predispose to thrombotic disorders, indicating that proteolytic inactivation of factor VIIIa is less important than inactivation of factor Va with regard to the hemostatic balance. This would be in agreement with the fact that murine factor VIII lacks the inactivation site at position 336.15 In addition, in vitro data using genetically engineered factor VIII with mutations in the APC cleavage sites showed that these mutations were not associated with reduced clotting times in APC-resistance assays.181
Although APC-resistant factor VIII molecules have not been identified in patients, the possibility remains open that APC-resistance may modulate factor VIII inactivation in an indirect manner. Inactivation of factor VIII by APC is enhanced in the presence of the APC-cofactor protein S.74,167 Several investigators have reported that the factor V procofactor enhances the cofactor effect of protein S in factor VIII inactivation.162,182,183 This link between factor V and factor VIII inactivation becomes even more apparent by the finding of Váradi et al,183 who reported that factor V, which carries the Arg506 to Gln mutation, has impaired cofactor activity in APC– and protein S–dependent factor VIII inactivation. The physiological significance of this observation remains unclear. However, the possibility that APC-resistance also affects factor VIII inactivation is challenging and deserves further study.
Another intriguing finding is that APC-resistant factor V has the potential to bypass the absence of factor VIII activity to some extent in in vitro thrombin generation studies.184 Therefore, it cannot be excluded that hemophilic patients which carry the factor V Arg506 to Gln mutation display a less severe phenotype than expected. This indeed has been shown for some hemophilic patients as described by Nichols et al.185 However, Arbini et al186 did not find an association between the presence of APC-resistant factor V and the severity of the bleeding tendency in a population of 295 hemophilic patients. The ability of APC-resistant factor V to bypass factor VIII deficiency may be restricted to specific, thus far unrecognized conditions.
CLEARANCE OF FACTOR VIII
At present, little is known about the mechanism by which factor VIII is cleared from the circulation. Obviously, vWF serves an important role, because in patients with severe vWD the factor VIII half-life is considerably decreased.83,86-88 As vWF protects factor VIII against proteolytic degradation in vitro (see Assembly of the Factor VIII–vWF Complex section), it cannot be excluded that the decreased half-life in the absence of vWF factor is associated with proteolytic degradation of factor VIII. However, experimental data in support of this possibility are lacking thus far. Another explanation for the rapid clearance could be binding of noncomplexed factor VIII to the surface of cells. In this respect it is of interest to mention that hepatic endothelial cells have been reported to contain factor VIII protein, but not its mRNA (see Biosynthesis and Secretion of Factor VIII section). It may be noteworthy that the copper-binding protein ceruloplasmin, which is structurally related to the factor VIII A domains, is internalized in the hepatic endothelial cells through a receptor-mediated process.187 It seems possible that factor VIII is taken up by hepatic endothelium by a similar mechanism.
Because the effect of factor V proteolysis on its survival has been investigated in a nonhuman primate model,188 it is of interest to compare factor VIII survival with that of factor V. Whereas the half-life of the factor V procofactor is approximately 14 hours, the half-life of its thrombin-activated derivative is dramatically different. The heavy and light chain are cleared very rapidly (t1/2 < 20 minutes). The half-life for factor V light chain is remarkably close to the half-life of 10 minutes reported for the isolated light chain of factor VIII after infusion into hemophilic dogs.189 Perhaps, clearance of both proteins involves a similar mechanism. Clearance of the factor V activation peptide, ie, the B domain, is considerably slower (t1/2 > 30 hours),188 suggesting a previously unrecognized role of this domain in preventing premature clearance of factor V. Whether this is also true for the factor VIII B domain is not clear. The half-lifes of normal factor VIII and B-domain deleted factor VIII are similar in hemophilic patients190 as well in hemophilic dogs,87,191 192 indicating that the B domain does not contribute to factor VIII survival. However, it is important to note that these data reflect factor VIII survival in the presence of vWF. Therefore, it would be of interest to investigate survival of B-domainless factor VIII in patients with vWD type 3.
FUTURE DIRECTIONS
Structure-function studies have contributed significantly to our current understanding of factor VIII biology and the molecular background of hemophilia A. As such this has provided the basis for the development of second generation recombinant factor VIII molecules that may find a therapeutical application. Pertinent to this point are the B-domain–deleted factor VIII variants, which are subject to clinical (factor VIII-SQ)190 or preclinical108,191,192testing. Furthermore, various factor VIII variants have been designed which in the future may be particular useful in the treatment of hemophilia A. Variants have been described which are less prone to inhibitor neutralization.193,194 Other examples include variants with enhanced hemostatic potency114 or stability.195
Despite the rapid accumulation of information regarding the structure and function of factor VIII, a number of questions remain to be answered. For instance, it is still unclear why factor VIII becomes a potent cofactor of factor IXa once it is cleaved within the heavy and light chain. What is the structural basis for such a dramatic increase in biological activity? How does factor VIIIa, together with the membrane surface, push factor IXa into an extremely potent configuration? It seems obvious that studies on three-dimensional modeling techniques based on crystal structures of factor VIII or factor VIII fragments complexed with their ligands (eg, factor IX) should provide a solid basis for a better understanding of the molecular aspects of factor VIII function and dysfunction. Because both factor VIII and factor IX are now potentially available in substantial quantities, determination of the crystal structure of complexes comprising wild-type or mutant proteins should be feasible and undoubtedly will lead to significant advances in this field. This approach will also facilitate the design of antagonistic inhibitors of the factor VIII system, which could provide novel anticoagulants for the treatment of thrombotic disorders.
The question of why exposure to factor VIII concentrates is associated with allo-immune reponses in some of the hemophilia A patients remains unanswered. What is the mechanism that causes these adverse reactions? Why and how is the immune system challenged under these conditions? Why do certain specific polypeptide regions of the factor VIII molecule play a prominent role in these immune reactions? Finally, little is known about the origin and expression of factor VIII at the cellular level. Which mechanisms trigger the specific cellular responses that govern elevations of the factor VIII plasma level under a variety of clinical conditions? Similarly, the mechanism of factor VIII clearance is an unexplored subject. Finding the answers to these questions is not only of fundamental, merely scientific appeal, but also has the potential of further improving our current strategies for the treatment of hemophilia A.
ACKNOWLEDGMENT
We express our gratitude to Drs W.G. van Aken, J. Voorberg, O.D. Christophe, and K. Fijnvandraat for helpful discussions and critical reading of the manuscript. We also thank Dr G. Kemball-Cook for providing the coordinates of the factor VIII.
REFERENCES
Author notes
Address reprint requests to Peter J. Lenting, PhD, Department of Plasma Protein Technology, CLB, Sanquin Blood Supply Foundation, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail: P_Lenting@clb.nl.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal