Abstract
The t(3;21)(q26;q22) chromosomal translocation associated with blastic crisis of chronic myelogenous leukemia results in the formation of the AML1/Evi-1 chimeric protein, which is thought to play a causative role in leukemic transformation of hematopoietic cells. Here we show that AML1/Evi-1 represses growth-inhibitory signaling by transforming growth factor-β (TGF-β) in 32Dcl3 myeloid cells. The activity of AML1/Evi-1 to repress TGF-β signaling depends on the two separate regions of the Evi-1 portion, one of which is the first zinc finger domain. AML1/Evi-1 interacts with Smad3, an intracellular mediator of TGF-β signaling, through the first zinc finger domain, and represses the Smad3 activity, as Evi-1 does. We also show that suppression of endogenous Evi-1 in leukemic cells carrying inv(3) restores TGF-β responsiveness. Taken together, AML1/Evi-1 acts as an inhibitor of TGF-β signaling by interfering with Smad3 through the Evi-1 portion, and both AML1/Evi-1 and Evi-1 repress TGF-β–mediated growth suppression in hematopoietic cells. Thus, AML1/Evi-1 may contribute to leukemogenesis by specifically blocking growth-inhibitory signaling of TGF-β in the t(3;21) leukemia.
RECENT PROGRESS in elucidating the details of growth control networks has uncovered diverse functions of oncogenic proteins that act in the nucleus, including a variety of transcription regulators. Many of them have been recognized through their involvement in chromosomal translocations. The t(3;21)(q26;q22) translocation, seen in the blastic crisis of chronic myelogenous leukemia and myelodysplastic syndrome–derived leukemia,1,2generates a fusion product of the distinct transcription regulators, AML1 and Evi-1.3 In this translocation, Evi-1becomes juxtaposed to the AML1 gene and is transcriptionally activated as the AML1/Evi-1 chimera under control of theAML1 promoter. A resultant fusion protein, AML1/Evi-1, is envisioned to cause leukemic transformation of hematopoietic cells.
The AML1 gene is located on chromosome 21q22 and is recognized as the most frequent target of chromosomal translocations associated with human leukemias.4 AML1 belongs to a family of transcription factors which share high homology with a DNA-binding region designated as the Runt-domain present in the Drosophilapair-rule gene runt product.5 Through the Runt-domain, AML1 forms a heterodimeric complex with PEBP2β (also known as CBFβ), which does not bind to DNA by itself but enhances DNA binding of AML1.6,7 AML1 binds to and activates transcription from the enhancer core motifs (TGT/cGGT), which are present in numerous myeloid promoters and lymphoid enhancers.8-13 A dominant inhibitory form of AML1, which competitively interferes with DNA binding of intact AML1, can abrogate myeloid cell differentiation induced by granulocyte colony-stimulating factor (G-CSF),14 suggesting that AML1 performs a key role in hematopoietic differentiation. In the AML1/Evi-1 chimera, AML1 is disrupted at the end of the Runt-domain and fused with the entire Evi-1 protein.3
Evi-1 was first identified as a gene existing in a common locus of retroviral integration in myeloid tumors in AKXD mice.15This gene encodes a 145-kD nuclear-localized protein.16,17 Although Evi-1 expression is barely detectable in normal hematopoietic cells, its frequent transcriptional activation is documented in a subset of myeloid malignancies. The humanEvi-1 gene is located on chromosome 3q26, and rearrangements involving this region, including t(3;21), t(3;12), t(3;3), and inv(3), often activate Evi-1 expression in myeloid leukemias and myelodysplasias.18-21 Elevated expression of Evi-1also occurs without cytogenetically evident translocations in some myeloid malignancies.22 23 These facts suggest a critical role of Evi-1 in malignant transformation of hematopoietic cells as a dominant oncogene.
Structurally, Evi-1 possesses seven and three repeats of Cys2His2-type zinc finger motifs separated into two domains (ZF1-7 and ZF8-10)16 (Fig1). Some evidence suggests that Evi-1 works as a negative regulator of gene expression,24,25 while characteristics of Evi-1 as a transcriptional activator have also been described. We have reported that Evi-1 elevates intracellular AP-1 activity and stimulates the c-fos promoter through the second zinc finger domain,26 although the identity of authentic target genes that Evi-1 may directly regulate has not been determined yet. Thus far, several biological effects of Evi-1 have been described. As we have reported, Evi-1 causes cellular transformation when overexpressed in the Rat1 fibroblast cells.27 Overexpressed Evi-1 blocks granulocytic differentiation of a murine myeloid cell line induced by G-CSF.28 Forced expression of Evi-1 in normal hematopoietic progenitors renders a decrease in colony formation in response to erythropoietin.29 From these findings, Evi-1 is thought to possess the ability of growth promotion and differentiation block in some types of cells.
Structures of the AML1/Evi-1 protein and its derivatives are compared with Evi-1. ▵ZF1-7 is a deletion mutant of AML1/Evi-1 that lacks the first zinc finger domain of Evi-1, while ▵Rep is a mutant lacks the repression domain of Evi-1. Distinct functional domains of AML1/Evi-1 are presented. Zinc finger motifs are numbered 1-10.
Structures of the AML1/Evi-1 protein and its derivatives are compared with Evi-1. ▵ZF1-7 is a deletion mutant of AML1/Evi-1 that lacks the first zinc finger domain of Evi-1, while ▵Rep is a mutant lacks the repression domain of Evi-1. Distinct functional domains of AML1/Evi-1 are presented. Zinc finger motifs are numbered 1-10.
Proliferation and differentiation of cells are tightly regulated by a delicate balance of growth factors and growth inhibitory factors. Transforming growth factor-β (TGF-β) is one of the best characterized members of growth inhibitory factors. TGF-β can inhibit proliferation of a wide range of cell types including epithelial, endothelial, and hematopoietic cells.29-32 The intracellular components that transduce TGF-β signals into the nucleus have been unveiled in recent years. Genetic screens inDrosophila isolated a protein called MAD by its involvement in the signaling pathway of dpp.33 MAD-related proteins in vertebrates are designated as Smad and define a novel family that acts downstream of the receptors to mediate TGF-β signaling.34-36 Among Smad proteins, Smad2 and Smad3 are directly phosphorylated by the receptor kinases in response to TGF-β, form heteromeric complexes with Smad4, another member of the Smad family, and then are translocated into the nucleus.37-42Once in the nucleus, Smad complexes are thought to act as transcriptional activators.43-45 Recently we reported that Evi-1 antagonizes growth-inhibitory effects of TGF-β in the epithelial cells that are highly sensitive to TGF-β.46Two separate regions of Evi-1 are responsible for this repression, one of which is the first zinc finger domain. Through this domain, Evi-1 physically associates with Smad3, thereby suppressing the transcriptional activity of Smad3. Thus, the interaction between Evi-1 and Smad3 is critical for repression of TGF-β signaling.
Multiple mechanisms have been proposed for leukemogenesis in the t(3;21) leukemia. Our previous study showed that AML1/Evi-1 dominantly suppresses the transactivation by intact AML1, thereby leading to a block of myeloid cell differentiation.47 AML1/Evi-1 shows a higher affinity for PEBP2β/CBFβ than that of wild-type AML1, which may account for one of the dominant effects of AML1/Evi-1.48 We have also found that AML1/Evi-1 can increase AP-1 activity and transform Rat-1 cells with dependence on the second zinc finger domain of the Evi-1 portion.27 47 In this study, we show that AML1/Evi-1 blocks TGF-β–induced transactivation of the responsive promoters, as Evi-1 does. AML1/Evi-1 abrogates responses to growth-suppressive signaling of TGF-β in hematopoietic cells. Two regions of the Evi-1 portion are required for the AML1/Evi-1 repressor activity, one of which is the first zinc finger domain. Through this domain, AML1/Evi-1 physically interacts with Smad3, and this ability is necessary for its function in efficient inhibition of TGF-β signaling. Thus, AML1/Evi-1 may contribute to leukemogenesis by interfering with TGF-β–mediated growth inhibition.
MATERIALS AND METHODS
Plasmid constructions.
The cDNA of AML1/Evi-1 was identified and obtained from the SKH-1 cell line.3 The Evi-1 cDNA was obtained from the AML1/Evi-1 fusion cDNA.47AML1/Evi-1and Evi-1 were subcloned into the expression vectors, pME18S49 (pME-AML1/Evi-1 and pME-Evi-1) and pMV750 (pMV-AML1/Evi-1 and pMV-Evi-1), as described elsewhere.26,47 For construction of AML1/Evi-1 deletion mutant ΔZF1-7 (AML1/Evi-1ΔZF1-7), the fragment from theEcoRV to Nsp V sites of AML1/Evi-1 was replaced with that of Evi-1ΔZF1-7.46 AML1/Evi-1ΔRep was generated by replacing the fragment from the EcoRV to Nsp V sites of AML1/Evi-1 with that of Evi-1Δ(608-732).46 These mutants were inserted into the EcoRI site of pME18S. Construction of pMV-AML1/Evi-1ΔZF8-10 were described previously.47 For construction of expression vectors for Smad3-Flag and Smad4-Flag, theBamHI-HindIII fragment of Smad3-Flag and theEcoRI-HindIII fragment of Smad4-Flag were excised from those placed in pRK541 (kind gifts from R. Derynck, University of California at San Francisco). The resultantHindIII end of each fragment was changed into the Xho I end and subcloned into pCMV5 (a kind gift from J.L. Wrana, Hospital for Sick Children, Toronto, Ontario, Canada). p3TP-Lux was kindly provided by K. Miyazono (Department of Biochemistry, The Cancer Institute, Tokyo, Japanese Foundation for Cancer Research).42
Cell lines, transfections, and oligonucleotide treatments.
HepG2 and COS7 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal calf serum (FCS). MOLM-1 cells were maintained in PRMI1640 supplemented with 10% FCS. The 32Dcl3 cells were maintained in RPMI1640 with 10% FCS and 0.25 ng of murine interleukin-3 (IL-3) per mL. The 32Dcl3 cells were cultured in 5 ng of human G-CSF per mL instead of IL-3 when indicated. The 32Dcl3 cells were transfected with pMV7 or pMV-Evi-1, and stable transfectants were isolated as described previously.47 Establishment of the 32Dcl3 clones that stably express AML1/Evi-1 (A51 and A53) or AML1/Evi-1ΔZF8-10 (B13 and B18) was described elsewhere.47 Cell morphology of 32Dcl3 clones was determined by staining cytospin preparations by May-Grünwald-Giemsa solution. Phosphothioate oligonucleotides were transfected into MOLM-1 cells by using SuperFect Transfection Reagents (QIAGEN Inc, Valencia, CA) according to the manufacturer’s recommendation. The oligonucleotide sequences were as follows: sense, TATCGCTGCGAAGACTGTGA; antisense, TCACAGTCTTCGCAGCGATA. Transient transfection into COS7 cells was performed by the diethylaminoethyl (DEAE)-dextran method51 as described previously.27
Growth inhibition assays.
Cells were seeded in 96-well culture plates at a density of 1 × 104 per well in the complete medium supplemented with 10% FCS. TGF-β at the different concentrations was added to the cells 12 hours later and cells were incubated for 24 hours. During the last 2 hours cells were labeled with 1 μCi/mL [3H]thymidine (Amersham, Arlington Heights, IL). Thereafter, cells were obtained and 3H radioactivity was measured in a liquid scintillation β-counter (Aloka, Mitaka, Tokyo, Japan).
Western blot analysis, immunoprecipitation, and metabolic labeling.
Polyclonal antisera to Evi-1 were raised in rabbit against maltose-binding protein fusion of the protein as described previously.26 For detection of Evi-1 and Flag-tagged Smad3, cells indicated were lysed in the TNE buffer (10 mmol/L Tris-HCl pH 7.4, 150 mmol/L NaCl, 1% NP40, 1 mmol/L ethylenediamide-tetraacetic acid) containing phosphatase inhibitors (12.5 mmol/L β-glycerophosphate, 1 mmol/L sodium orthovanadate) and a cocktail of proteinase inhibitors (10 U/mL aprotinin, 1 mmol/L phenylmethylsulfonyl fluoride, 5 μg/mL leupeptin, 1 μg/mL pepstatin A, 2 mmol/L benzamidine, 1 μg/mL antipain, 1 μg/mL chymostatin, and 2 μg/mL soybean trypsin inhibitor). Protein concentrations of cell extracts were quantified using Protein Assay Dye (Bio-Rad, Hercules, CA). Whole-cell extracts containing 100 μg of protein were subjected to either 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for Evi-1 or 10% SDS-PAGE for Flag-tagged Smad3, and transferred to polyvinylidene difluoride membranes (Immobilon; Millipore, Bedford, MA). The membranes were blocked with 10% skim milk, treated with the anti–Evi-1 antibody or the anti-Flag M2 monoclonal antibody (Sigma, St Louis, MO), washed, and reacted with the donkey anti-IgG antibody coupled to horseradish peroxydase. The blots were visualized with the enhanced chemiluminescence method (Amersham). For the anti-Flag immunoprecipitation, cells were lysed in the TNE buffer and subjected to immunoprecipitation with anti-Flag followed by absorption to Protein G-Sepharose (Pharmacia, Uppsala, Sweden). Immunoprecipitates were then washed and separated by a 7.5% SDS-polyacrylamide gel. Subsequent detection of Evi-1 in the precipitates was carried out as described above.
Transcriptional response assays.
For TGF-β–inducible luciferase reporter assays, HepG2 cells were seeded at a density of 2 × 105 per 6-cm plate. Cells were transfected 18 hours after seeding with 5 μg of the reporter plasmids (p3TP-Lux) along with the effector plasmids (2.4 μg for pME-AML1/Evi-1 or the equivalent molar for their derivatives, and 2 μg for Flag-tagged Smads in pCMV5), using LipofectAMINE (GIBCO-BRL, Gaithersburg, MD) according to the manufacturer’s recommendation. For analysis of the luciferase activity derived from cotransfection with several expression plasmids, the equivalent-molar plasmids were transfected and the total amount of DNA in terms of weight was adjusted to be equal by adding the plasmid pUC13. As an internal control of transfection efficiency, a plasmid expressing β-galactosidase driven by SRα promoter (1 μg) was cotransfected. Cells were added 15 hours later with equal volume of DMEM containing 4% FCS, and incubated for additional 9 hours. Thereafter, cells were washed with phosphate-buffered saline twice and incubated for 24 hours in the absence or presence of TGF-β in DMEM containing 0.2% FCS. Cells were then obtained and assayed for the luciferase activity using the luciferase assay system (Promega, Madison, WI) and a luminometer (Lumat; Berthold, Badwildbod, Germany). To control transfection efficiencies, the data were normalized to the β-galactosidase activities measured by the method described previously.26 All transfection experiments were performed at least three times and similar results were obtained.
RESULTS
AML1/Evi-1 inhibits TGF-β–mediated transcriptional activation of the target promoter.
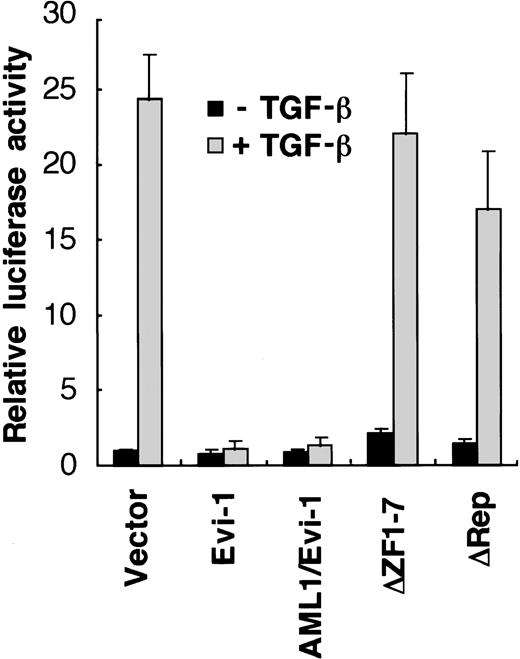
We have reported that Evi-1 represses TGF-β signaling in epithelial cells.46 Because AML1/Evi-1 fusion protein contains the whole Evi-1 sequence (Fig 1), it is plausible that AML1/Evi-1 can also affect TGF-β signaling. To examine potential roles of AML1/Evi-1 in the TGF-β signaling pathway, we evaluated the effects of AML1/Evi-1 on the TGF-β–mediated transcriptional response using transient cotransfection assays. We made use of p3TP-Lux, a luciferase reporter plasmid that contains three repeats of a 12-O-tetradecanoylphorbol 13-acetate (TPA) response element and a fragment from positions −636 to −740 of the human plasminogen activator inhibitor-1 (PAI-1) promoter.52 This construct has been shown to be efficiently stimulated by TGF-β through its receptors in a variety of cell lines.52-55 p3TP-Lux was transiently transfected either alone or together with Evi-1 expression vector (pME-Evi-1), and luciferase activity was measured in the extracts from untreated cells or cells treated with 5 ng of TGF-β per mL for 24 hours. As a model for these experiments, we used HepG2 cells, which are frequently used for studies on TGF-β–induced transactivation because they express TGF-β receptors and are highly responsive to TGF-β.56When p3TP-Lux alone was transfected into HepG2 cells, a 25-fold increase in luciferase activity was observed in the presence of TGF-β, similar to the previous report40 (Fig2). These transactivations were substantially repressed when pME-Evi-1 was cotransfected as we have reported previously.46 When AML1/Evi-1 was transfected, TGF-β activation of the PAI-1 promoter was also repressed back to the control level. We have identified the two separate regions that are responsible for the Evi-1 inhibition of TGF-β signaling: one is the first zinc finger domain and the other is a 125-amino acid contact region located N-terminally to the second zinc finger domain,46 which we refer to as the repression domain in this report. As shown in Fig 2, the mutant AML1/Evi-1 protein that lacks the first zinc finger domain of Evi-1 failed to suppress TGF-β activation of the PAI-1 promoter. Deletion of the repression domain also abolished the repressor activity of AML1/Evi-1. These results indicate that AML1/Evi-1 can interfere with TGF-β signaling depending on the two separate region of the Evi-1 portion.
TGF-β–mediated transcriptional responses are suppressed by AML1/Evi-1. p3TP-Lux was cotransfected into HepG2 cells along with either the empty pME18S, Evi-1, AML1/Evi-1, or its derivatives in pME18S. Cells were incubated for 24 hours in the presence or absence of 5 ng of TGF-β per mL. Relative luciferase activities were measured in cell extracts, normalized to the β-galactosidase activity. Values and error bars depict the means and the standard deviations, respectively, of four separate experiments.
TGF-β–mediated transcriptional responses are suppressed by AML1/Evi-1. p3TP-Lux was cotransfected into HepG2 cells along with either the empty pME18S, Evi-1, AML1/Evi-1, or its derivatives in pME18S. Cells were incubated for 24 hours in the presence or absence of 5 ng of TGF-β per mL. Relative luciferase activities were measured in cell extracts, normalized to the β-galactosidase activity. Values and error bars depict the means and the standard deviations, respectively, of four separate experiments.
Constitutive expression of Evi-1 or AML1/Evi-1 in 32Dcl3 cells overcomes TGF-β–mediated inhibition of cellular growth.
Given the data from the cotransfection experiments indicating that AML1/Evi-1, as Evi-1, can perturb TGF-β signaling, we examined whether AML1/Evi-1 and Evi-1 affect the antiproliferative effects of TGF-β in hematopoietic cells. To this end, we used 32Dcl3 cells, a murine IL-3–dependent immature myeloid cell line, which undergo growth arrests in response to TGF-β.57
We introduced the Evi-1 expression vector that enables concomitant expression of the neomycin resistant gene (pMV-Evi-1) into 32Dcl3 cells and selected them for neomycin resistance in the presence of G418. Individual G418-resistant clones were screened for expression of Evi-1, and several stable 32Dcl3 cell lines that express Evi-1 were isolated. Of these clones, Western blot analyses using the antiserum against Evi-126 showed that E1 and E11 are representative clones expressing high levels of Evi-1 (Fig 3A, lanes 3 and 4). We had established elsewhere the 32Dcl3 clones that stably express the AML1/Evi-1 protein, which were designated as A51 and A53.47 Expression of AML1/Evi-1 in A51 and A53 was also replicated (Fig 3A, lanes 3 and 4) as presented previously.47 Two independent clones, P1 and P2, which were transfected with the empty pMV7 vector, were used as controls. In these experiments, intrinsic Evi-1 was under the detectable level in the native 32Dcl3 cells. When cultured in complete medium without TGF-β treatment, all these clones showed comparable viabilities and proliferative abilities with each other (data not shown). In the absence of TGF-β, all the Evi-1- or AML1/Evi-1–expressing clones required IL-3 for proliferation and viability, as well as the mock-transfected 32Dcl3 cells. We evaluated growth-inhibitory effects of TGF-β on these 32Dcl3 clones using [3H]thymidine incorporation assays. Shown in Fig 3B are the effects on [3H]thymidine uptake when the 32Dcl3 clones were exposed to increasing amounts of TGF-β. The growth of the two control clones, P1 and P2, was inhibited by 1 ng of TGF-β per mL. In contrast, E1 and E11, which express high levels of Evi-1, showed diminished responsiveness to the TGF-β growth-inhibitory signaling. The AML1/Evi-1–expressing clones, A51 and A53, were also resistant to TGF-β–mediated growth inhibition to a similar extent as E1 and E11. These data indicate that AML1/Evi-1 can interrupt growth-inhibitory signals triggered by TGF-β and that both AML1/Evi-1 and Evi-1 can release hematopoietic cells from growth-arrested states induced by TGF-β.
Constitutive expression of AML1/Evi-1 in 32Dcl3 cells overcomes TGF-β–mediated inhibition of cell growth. (A) Expression of the Evi-1 and the AML1/Evi-1 proteins in stable 32Dcl3 transfectants. Clones P1 (lane 1) and P2 (lane 2) are control lines obtained from 32Dcl3 cells transfected with pMV7 followed by G418 selection. Clones E1 (lane 3) and E11 (lane 4) were established from cells transfected with pMV-Evi-1, while clones A51 (lane 5) and A53 (lane 6) were from cells transfected with pME-AML1/Evi-1. The arrows indicate the location of Evi-1 and AML1/Evi-1, and the positions of molecular-weight standards are shown at left. (B) Analysis of growth inhibition in response to TGF-β. The 32Dcl3 clones stably transfected with Evi-1 (E1 and E11) or AML1/Evi-1 (A51 and A53), and control clones (P1 and P2) were subjected to a [3H]thymidine incorporation assay in the presence of different concentrations of TGF-β. Results are expressed as percentages relative to values observed in control cultures that did not receive TGF-β. Representative values from four independent experiments are shown.
Constitutive expression of AML1/Evi-1 in 32Dcl3 cells overcomes TGF-β–mediated inhibition of cell growth. (A) Expression of the Evi-1 and the AML1/Evi-1 proteins in stable 32Dcl3 transfectants. Clones P1 (lane 1) and P2 (lane 2) are control lines obtained from 32Dcl3 cells transfected with pMV7 followed by G418 selection. Clones E1 (lane 3) and E11 (lane 4) were established from cells transfected with pMV-Evi-1, while clones A51 (lane 5) and A53 (lane 6) were from cells transfected with pME-AML1/Evi-1. The arrows indicate the location of Evi-1 and AML1/Evi-1, and the positions of molecular-weight standards are shown at left. (B) Analysis of growth inhibition in response to TGF-β. The 32Dcl3 clones stably transfected with Evi-1 (E1 and E11) or AML1/Evi-1 (A51 and A53), and control clones (P1 and P2) were subjected to a [3H]thymidine incorporation assay in the presence of different concentrations of TGF-β. Results are expressed as percentages relative to values observed in control cultures that did not receive TGF-β. Representative values from four independent experiments are shown.
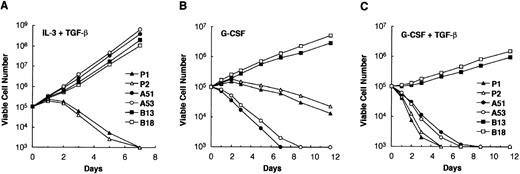
32Dcl3 cells are known to differentiate into mature granulocytes when cultured in the presence of G-CSF.58 We previously reported that AML1/Evi-1 can affect granulocytic differentiation of 32Dcl3 cells with dependence of the second zinc finger domain of the Evi-1 portion.47 When treated with G-CSF, AML1/Evi-1–expressing 32Dcl3 cells rapidly die within a week without obvious granulocytic differentiation. In contrast, 32Dcl3 cells that express the mutant AML1/Evi-1, which lacks the second zinc finger domain of the Evi-1 portion (AML1/Evi-1ΔZF8-10), continuously proliferate without maturation in response to G-CSF. We examined effects of TGF-β on growth and differentiation of these stable transfectants of 32Dcl3 cells. In the presence of IL-3, P1, P2, A51, A53, and the clones that stably express AML1/Evi-1ΔZF8-10 (B13 and B18) proliferated exponentially in a comparable manner (data not shown), as described previously.47 When deprived of IL-3, they died completely within 3 days without granulocytic differentiation, either in the absence or the presence of TGF-β (data not shown). When cultured with a combination of IL-3 and TGF-β, A51, A53, B13, and B18 clones exponentially proliferated without losing viability and did not show any morphological change (Fig 4A). These results are compatible with the fact that the first zinc finger and the repression domains of Evi-1 are responsible for AML1/Evi-1 repression of TGF-β signaling. In contrast, control P1 and P2 clones showed decline in viable cell number in 2 days and mostly died within a week (Fig 4A). No morphological differentiation into granulocytes was seen in P1 and P2 clones (data not shown), suggesting that TGF-β does not affect differentiation stages of 32Dcl3 cells in the presence of IL-3. When cultured with G-CSF instead of IL-3, P1 and P2 underwent terminal differentiation into morphologically mature granulocytes and showed gradual decline in viable cell number (Fig 4B), as described previously.14,47 In contrast, A51 and A53 lost viability and rapidly died without maturation, whereas B13 and B18 proliferated continuously in an immature state.47 When both G-CSF and TGF-β were added to these clones, control P1 and P2 clones immediately lost viability and completely died within 5 days, showing granulocytic differentiation as seen in the presence of G-CSF alone (Fig 4C). A51 and A53 also died promptly in the presence of G-CSF and TGF-β (Fig 4C). In contrast to P1 and P11, however, they showed no morphological differentiation into granulocytes, as in the presence of G-CSF alone (data not shown). B13 and B18 clones showed continuous proliferation without differentiation, reflecting unresponsiveness of B13 and B18 to the antiproliferative effect of TGF-β and the maturation signal of G-CSF. These results indicate that TGF-β does not significantly influence G-CSF–induced granulocytic differentiation but may simply inhibit growth of 32Dcl3 cells at any differentiation stage.
Growth curve of 32Dcl3 cells in response to TGF-β in the presence IL-3 or G-CSF. (A) The indicated cells were cultured with 0.25 ng of IL-3 per mL, 10 ng of TGF-β per mL, and 10% FCS. Cultures were diluted when the cell number reached 1 × 106 cells per mL. Viable cells were counted by the trypan blue exclusion method at each time point. Clones B13 and B18 are established from cells transfected with pMV-AML1/Evi-1▵ZF8-10. Two independent experiments were performed and similar results were obtained. Representative data are shown. (B and C) The indicated cells were washed twice with phosphate-buffered saline and subsequently cultured with 5 ng of G-CSF per mL either alone (B) or together with 10 ng of TGF-β per mL (C) in the presence of 10% FCS. Cultures were diluted when the cell number reached 1 × 106 cells per mL.
Growth curve of 32Dcl3 cells in response to TGF-β in the presence IL-3 or G-CSF. (A) The indicated cells were cultured with 0.25 ng of IL-3 per mL, 10 ng of TGF-β per mL, and 10% FCS. Cultures were diluted when the cell number reached 1 × 106 cells per mL. Viable cells were counted by the trypan blue exclusion method at each time point. Clones B13 and B18 are established from cells transfected with pMV-AML1/Evi-1▵ZF8-10. Two independent experiments were performed and similar results were obtained. Representative data are shown. (B and C) The indicated cells were washed twice with phosphate-buffered saline and subsequently cultured with 5 ng of G-CSF per mL either alone (B) or together with 10 ng of TGF-β per mL (C) in the presence of 10% FCS. Cultures were diluted when the cell number reached 1 × 106 cells per mL.
Suppression of endogenous Evi-1 expression in inv(3) leukemic cells recovers their responsiveness to TGF-β.
From the data above, AML1/Evi-1 and Evi-1 can potentially block growth inhibition of hematopoietic cells mediated by TGF-β. We further investigated whether the naturally expressed Evi-1 can also act as an inhibitor of TGF-β signaling. MOLM-1 cells are a human megakaryoblastoid cell line carrying the inv(3)(q21q26) chromosomal aberration. As a result of inv(3), MOLM-1 cells uniquely express the truncated from of the Evi-1 protein in which the C-terminal 44 amino acids of wild-type Evi-1 were replaced by five amino acids.20 The Evi-1 protein in MOLM-1 cells retains both the first zinc finger domain and the repression domain, which are the requirements for repression of TGF-β signaling. It was also shown to increase AP-1 activity when expressed in NIH3T3 cells as wild-type Evi-1.20 These facts allow us to expect it to possess the ability to inhibit TGF-β signaling. To modulate endogenous gene expression of Evi-1 in MOLM-1 cells, we used the antisense gene inhibition by oligonucleotides.59 As shown in Fig5A, the endogenous Evi-1 expression in MOLM-1 cells was effectively diminished by treatment with the antisense oligonucleotide that is complementary to the sequence encoding the N terminus of the first zinc finger domain of Evi-1, compared with cells that received the corresponding sense oligonucleotide or no oligonucleotide (Fig 5A). We determined TGF-β responsiveness using [3H]thymidine incorporation assays in these cells. In the presence of TGF-β, [3H]thymidine uptake was significantly reduced in the MOLM-1 cells in which Evi-1 expression has been suppressed by the antisense oligonucleotide treatment, compared with those of cells treated with no or the sense oligonucleotide (Fig5B). These results indicate that Evi-1 that is endogenously expressed in leukemic cells acts as an antagonist for TGF-β signaling and strongly support a model that Evi-1 contributes to leukemic transformation of hematopoietic cells by inhibiting TGF-β signaling.
Inhibition of endogenous Evi-1 restores responsiveness of MOLM-1 cells to TGF-β. (A) MOLM-1 cells were treated with no (−), the sense (S), or the antisense (AS) oligonucleotide for Evi-1. Whole-cell extracts containing 100 μg of proteins were subjected to SDS-PAGE and immunoblotting with the anti–Evi-1 antibody. The amount of the Evi-1 protein expressed in these cells is shown. The arrow indicates the migration of the Evi-1 protein, and the positions of molecular-weight standards are shown at left. (B) Analysis of growth inhibition in response to TGF-β. MOLM-1 cells treated with the indicated oligonucleotide were subjected to a [3H]thymidine incorporation assay in the presence of different concentrations of TGF-β. Results are expressed as percentages relative to values observed in control cultures that did not receive TGF-β. Representative values from four independent experiments are shown.
Inhibition of endogenous Evi-1 restores responsiveness of MOLM-1 cells to TGF-β. (A) MOLM-1 cells were treated with no (−), the sense (S), or the antisense (AS) oligonucleotide for Evi-1. Whole-cell extracts containing 100 μg of proteins were subjected to SDS-PAGE and immunoblotting with the anti–Evi-1 antibody. The amount of the Evi-1 protein expressed in these cells is shown. The arrow indicates the migration of the Evi-1 protein, and the positions of molecular-weight standards are shown at left. (B) Analysis of growth inhibition in response to TGF-β. MOLM-1 cells treated with the indicated oligonucleotide were subjected to a [3H]thymidine incorporation assay in the presence of different concentrations of TGF-β. Results are expressed as percentages relative to values observed in control cultures that did not receive TGF-β. Representative values from four independent experiments are shown.
AML1/Evi-1 physically interacts with Smad3 through the first zinc finger domain of the Evi-1 portion.
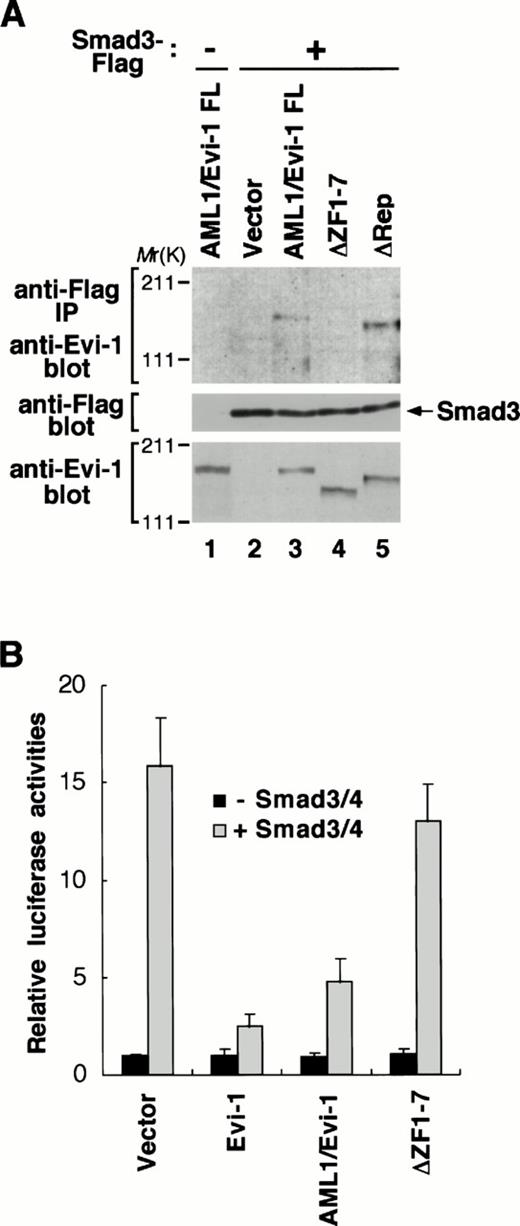
To explore a potential role of the ability of Evi-1 to interact with Smad3 in the AML1/Evi-1 repression of TGF-β signaling, we examined whether AML1/Evi-1 is also able to associate with Smad3. To this end, we transfected Smad3 tagged C-terminally with the Flag peptide (Smad3-Flag) into COS7 cells in the absence or the presence of the AML1/Evi-1 expression plasmid. Smad3-Flag was expressed efficiently along with AML1/Evi-1 in the transfected cells, as can be seen by immunoblotting with the anti-Flag or the anti–Evi-1 antibody (Fig6A, middle and bottom, lanes 1 through 3). Whole extracts from these cells were immunoprecipitated with the anti-Flag antibody and the precipitates were analyzed by immunoblotting with the anti–Evi-1 antibody. As shown in Fig 6A, we observed that AML1/Evi-1 and Smad3 were specifically coimmunoprecipitated. We have reported elsewhere that the Evi-1-Smad3 association is mediated by the first zinc finger domain.46On the other hand, the first zinc finger domain and the repression domain of AML1/Evi-1 are required for TGF-β signaling, as shown in Fig 2. To confirm roles of these domains in the interaction between AML1/Evi-1 and Smad3, we tested the Smad3 binding ability of the AML1/Evi-1 mutants that lack these domains. These mutants were coexpressed with Smad3-Flag in COS7 cells, and the whole cell extracts were subjected to immunoprecipitation using the anti-Flag antibody. By immunoblotting with the anti–Evi-1 antibody, the mutants are recognized to be expressed at the levels comparable to each other at the predicted sizes (Fig 6A, bottom, lanes 4 and 5). As seen in Fig 6A, AML1/Evi-1ΔRep was coimmunoprecipitated with Smad3 (Fig 6A, top, lane 5), whereas AML1/Evi-1ΔZF1-7 could not (Fig 6A, top, lane 4). Thus, deletion extending the entire of the first zinc finger domain of the Evi-1 portion completely abolishes the AML1/Evi-1-Smad3 association, as is the case with Evi-1.46 These results indicate that the first zinc finger of the Evi-1 portion is essential for the interaction of AML1/Evi-1 and Smad3.
AML1/Evi-1 physically interacts with Smad3 and inhibits the Smad3 activity. (A) Association between AML1/Evi-1 and Smad3 in vivo. Twenty micrograms of pME18S (lane 2), AML1/Evi-1 (lanes 1 and 3), AML1/Evi-1▵ZF1-7 (lane 4), or AML1/Evi-1▵Rep (lane 5) in pME18S was transfected into 4 × 106 of COS7 cells with 20 μg of the empty pCMV5 (lane 1) or Smad3-Flag in pCMV5 (lanes 2 to 5). Cells were obtained 48 hours later and subjected to the immunoprecipitation procedure with the anti-Flag antibody. Immunoprecipitates were resolved by SDS-PAGE, and detected by the anti–Evi-1 antiserum (top). Expression of Smad3-Flag and Evi-1 is monitored with the anti-Flag (middle) and the anti–Evi-1 (bottom) antibodies, respectively. The location of Smad3 is shown at right, and the positions of molecular-weight standard at left. (B) AML1/Evi-1 inhibits Smad3/4-induced TGF-β responses. Either the empty pME18S, Evi-1, AML1/Evi-1, or AML1/Evi-1▵ZF1-7 in pME18S in combination with p3TP-Lux was cotransfected into HepG2 cells together with pCMV5 or Smad3 plus Smad4 (Smad3/4). Relative luciferase activities were measured in cell extracts, normalized to the β-galactosidase activity. Values and error bars depict the means and the standard deviations, respectively, of four separate experiments.
AML1/Evi-1 physically interacts with Smad3 and inhibits the Smad3 activity. (A) Association between AML1/Evi-1 and Smad3 in vivo. Twenty micrograms of pME18S (lane 2), AML1/Evi-1 (lanes 1 and 3), AML1/Evi-1▵ZF1-7 (lane 4), or AML1/Evi-1▵Rep (lane 5) in pME18S was transfected into 4 × 106 of COS7 cells with 20 μg of the empty pCMV5 (lane 1) or Smad3-Flag in pCMV5 (lanes 2 to 5). Cells were obtained 48 hours later and subjected to the immunoprecipitation procedure with the anti-Flag antibody. Immunoprecipitates were resolved by SDS-PAGE, and detected by the anti–Evi-1 antiserum (top). Expression of Smad3-Flag and Evi-1 is monitored with the anti-Flag (middle) and the anti–Evi-1 (bottom) antibodies, respectively. The location of Smad3 is shown at right, and the positions of molecular-weight standard at left. (B) AML1/Evi-1 inhibits Smad3/4-induced TGF-β responses. Either the empty pME18S, Evi-1, AML1/Evi-1, or AML1/Evi-1▵ZF1-7 in pME18S in combination with p3TP-Lux was cotransfected into HepG2 cells together with pCMV5 or Smad3 plus Smad4 (Smad3/4). Relative luciferase activities were measured in cell extracts, normalized to the β-galactosidase activity. Values and error bars depict the means and the standard deviations, respectively, of four separate experiments.
To evaluate the functional consequence of the interaction between AML1/Evi-1 and Smad3, we examined the AML1/Evi-1 effect on the Smad3 activity. It is known that Smad3 can activate the PAI-1 promoter by itself and that this activation is potentiated when Smad4 is cotransfected.41 Coexpressed Smad3 and Smad4 (Smad3/4) synergized to induce about a 16-fold increase of the PAI-1 promoter activity without TGF-β, and this transactivation was effectively repressed in the presence of Evi-1, as is consistent with the previous studies.41 46 We examined whether AML1/Evi-1 can inhibit these Smad3/4-mediated promoter activation. As shown in Fig 6B, cotransfection of AML1/Evi-1 fully inhibited Smad3/4-dependent transcriptional activation. In contrast to full-length AML1/Evi-1 and Evi-1, AML1/Evi-1ΔZF1-7, which has lost the ability to interact with Smad3, was unable to inhibit Smad3/4-induced activation of the PAI-1 promoter (Fig 6B). These findings indicate that AML1/Evi-1 blocks TGF-β responses by inactivating Smad3 functions through the interaction with Smad3.
DISCUSSION
In the present study, we have found that the AML1/Evi-1 functions as a negative regulator of TGF-β signaling. We have also shown that both Evi-1 and AML1/Evi-1 can repress TGF-β–mediated growth inhibition in hematopoietic cells. AML1/Evi-1 physically interacts with Smad3, a mediator of TGF-β signaling, as Evi-1 does, and this interaction contributes to the AML1/Evi-1 repression of TGF-β signaling. These observations will provide a novel insight into a molecular basis for the t(3;21) leukemia.
In this study, we identified a relationship between the two distinct transcription regulators, AML1/Evi-1 and Smad3. Recent studies on transcriptional networks have shown a growing number of instances of functional antagonism between transcriptional regulators. For example, it was reported that c-Jun inhibits myogenesis by interfering with the MyoD function through the physical interaction.60 There are several ways by which transcriptional repressors can block gene expression.61 The simplest one is competitive DNA binding, where binding of the repressor to the promoter prevents binding of other activators. Indeed, sequence-specific transcriptional repression by Evi-1 has been documented in transient cotransfection experiments with reporter constructs containing the artificial binding sequences for Evi-1.25 However, it seems unlikely that AML1/Evi-1 inhibits TGF-β–mediated gene expression by binding directly to the target promoters, because DNA consensus sequences for Evi-1 binding are not found in the PAI-1 promoter used in this study. Recent studies have suggested that Smad3 can be a DNA-binding protein and potentially regulates gene expression through DNA binding.62-64Therefore, it is highly likely that AML1/Evi-1 blocks TGF-β signaling by preventing Smad3 from interacting with DNA, as Evi-1 does. These findings define a novel function of AML1/Evi-1 to regulate transcription by the protein-protein interaction. The association of AML1/Evi-1 with Smad3 is mediated by the first zinc finger domain of the Evi-1 portion. Remarkably, the repression domain of Evi-1 is dispensable for the association, indicating that the minimal Smad3-binding domain of Evi-1 is not sufficient for inhibition of TGF-β signaling. These findings suggest several possibilities: the repression domain may contribute to masking a domain of Smad3 responsible for interaction with DNA or transcriptional partners, and AML1/Evi-1 needs to interact with corepressors through this region to efficiently block the Smad3 activity.
Members of the TGF-β superfamily are potent regulators of growth and differentiation in various types of cells. The spatial and temporal controls of their activities are thus important in the normal cellular proliferation. A variety of mechanisms exist to affect the activities of these proteins, including the cytoplasmic protein FKBP12 that inhibits TGF-β signaling by interacting with its receptor,65,66 and the viral oncoprotein E1A that circumvents ligand-induced growth inhibition by blocking negative regulators of cell proliferation.67 Recently, it was reported that Smad6 and Smad7 associate with the TGF-β receptor and function as an antagonist for TGF-β signaling, which suggests the existence of intracellular control mechanisms that may contribute to cell-type specific responses to ligand stimuli.68-70 In this report, we showed that AML1/Evi-1 can block the two distinct responses induced by TGF-β; activation of the PAI-1 promoter and inhibition of hematopoietic cell proliferation. Together with the fact that AML1/Evi-1 interacts with Smad3, AML1/Evi-1 is now shown to belong to a new class of regulators of TGF-β signaling: a chimeric nuclear oncoprotein48 derived from a chromosomal translocation that inhibits the intracellular signaling component of TGF-β signaling. TGF-β is one of the most studied cytokines that can negatively regulate hematopoietic cell proliferation. It is also a potent inhibitor of the number and ploidy of megakaryocytes.71,72It is suggested that elevated expression of the Evi-1 proteins is associated with dysmegakaryopoiesis in some myeloid malignancies,73 as is typically seen in the 3q21q26 syndrome.74 Hence, disturbance of TGF-β signaling by AML1/Evi-1 may account for one of the mechanisms of AML1/Evi-1–induced leukemogenesis and dysmegakaryopoiesis.
The TGF-β receptor and its downstream components can be targets for mutations in some types of cancer. For instance, the gene encoding the TGF-β receptor type II is commonly inactivated in colon cancer.75 Smad2 and Smad4 genes have also been found inactivated or deleted in colon cancer, suggesting their roles as a tumor suppressor.38,76,77 Smad4 has been shown to be a candidate tumor suppressor gene of human pancreatic cancer78 and its mutations have been reported in various types of tumors including head and neck, lung, and esophageal cancers.79-84 Some of these naturally occurring mutations are proved to impair the activities of Smad proteins,38,85,86 suggesting that alteration of Smad functions will contribute to oncogenesis. Recently it was also reported that Smad5 is involved in the TGF-β–mediated inhibition of primitive human hematopoietic progenitor cell proliferation.87Although definite involvement of Smad proteins in hematological malignancies remains yet to be determined, disintegration of the TGF-β signaling pathways may contribute to the progression toward certain types of leukemias. Our findings also provide an important clue for a role of Smad proteins in leukemogenesis.
ACKNOWLEDGMENT
The authors thank L. Wrana for the pCMV5 vector, R. Derynck for providing Flag-tagged Smad3 and Smad4, and K. Miyazono for p3TP-Lux.
Supported in part by Grants-in-Aid for Cancer Research from the Ministry of Health and Welfare and from the Ministry of Education, Science, and Culture of Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hisamaru Hirai, MD, Department of Hematology & Oncology, Graduate School of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8655, Japan.


![Fig. 3. Constitutive expression of AML1/Evi-1 in 32Dcl3 cells overcomes TGF-β–mediated inhibition of cell growth. (A) Expression of the Evi-1 and the AML1/Evi-1 proteins in stable 32Dcl3 transfectants. Clones P1 (lane 1) and P2 (lane 2) are control lines obtained from 32Dcl3 cells transfected with pMV7 followed by G418 selection. Clones E1 (lane 3) and E11 (lane 4) were established from cells transfected with pMV-Evi-1, while clones A51 (lane 5) and A53 (lane 6) were from cells transfected with pME-AML1/Evi-1. The arrows indicate the location of Evi-1 and AML1/Evi-1, and the positions of molecular-weight standards are shown at left. (B) Analysis of growth inhibition in response to TGF-β. The 32Dcl3 clones stably transfected with Evi-1 (E1 and E11) or AML1/Evi-1 (A51 and A53), and control clones (P1 and P2) were subjected to a [3H]thymidine incorporation assay in the presence of different concentrations of TGF-β. Results are expressed as percentages relative to values observed in control cultures that did not receive TGF-β. Representative values from four independent experiments are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/11/10.1182_blood.v92.11.4003/5/m_blod42356003w.jpeg?Expires=1765888258&Signature=1TEXhdH0ovRg0zn9Ewd-dx8Ul2KDiDQyTyKBmP-v5xsyxprvYzLCiW9c1at0sfwpHwwTfrGerUNjSEP9GbHx5BKwiwYPxG5H9BAUKgrfH~8nAyU4i76L6TJRGQ5~xmtZjXDzCneNlDOgcS9owIDoOV~4GiQqY7ZEJyRq4wD~qwARWGCn9NAnX818Q2jSDvgkbCAw6AcoIqssSt51cuYLz2IIGXp036SzGaK~eJcBt3dMsHPUvqJihosMQnTFtZF5uypWAEvAg7zmO0VZSS9iu9sE~NDo4U1lZhhAUWc8i~fJKPpmNpSFkHPlRdsx1y18TEOGSEvkdswWwlyDT4634Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Inhibition of endogenous Evi-1 restores responsiveness of MOLM-1 cells to TGF-β. (A) MOLM-1 cells were treated with no (−), the sense (S), or the antisense (AS) oligonucleotide for Evi-1. Whole-cell extracts containing 100 μg of proteins were subjected to SDS-PAGE and immunoblotting with the anti–Evi-1 antibody. The amount of the Evi-1 protein expressed in these cells is shown. The arrow indicates the migration of the Evi-1 protein, and the positions of molecular-weight standards are shown at left. (B) Analysis of growth inhibition in response to TGF-β. MOLM-1 cells treated with the indicated oligonucleotide were subjected to a [3H]thymidine incorporation assay in the presence of different concentrations of TGF-β. Results are expressed as percentages relative to values observed in control cultures that did not receive TGF-β. Representative values from four independent experiments are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/11/10.1182_blood.v92.11.4003/5/m_blod42356005w.jpeg?Expires=1765888258&Signature=BN81NduaUy7Y0aZ6Jgh5GClbJgNnI~S385EGUWsLX9JrMKWgGh~SbyOyKGEvEzsBbccicSLTW5uM8mMo3emMTWzi36uHIQOmBY-6fQU1qO8dendcs5~SPlzjDcF3nbcPitJiFDpzGN9P4V-q3rXg2WZX008FqCewUI-5tfh5669Jub7AEF7V91PUbwBvZlzk9Hr4Qq3Fq25PUwGqROnZnG9lrnPXTAXuRtNPDMl~ACgYJmR28npCY6xbrsjqvof44vrOdXvaY77q2yKSToKJJfFY82W~1vPySNjjuHgJnr1zgt5wUhl2B5sHsd~jziPXChsjm4RLcBLUjnxffQLIkA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal