Abstract
Loss of chromosome 7 (−7) or deletion of the long arm (7q−) are recurring chromosome abnormalities in myeloid leukemias. The association of −7/7q− with myeloid leukemia suggests that these regions contain novel tumor suppressor gene(s), whose loss of function contribute to leukemic transformation or tumor progression. Based on chromosome banding analysis, two critical regions have been identified, one in band q22 and another in bands q32-q35. Presently there are no data available on the molecular delineation of the distal critical region. In this study we analyzed bone marrow and blood samples from 13 patients with myeloid leukemia (de novo myelodysplastic syndrome [MDS] , n = 3; de novo acute myeloid leukemia [AML], n = 9; therapy-related (t-) AML, n = 1) which, on chromosome banding analysis, exhibited deletions (n = 12) or in one case a balanced translocation involving bands 7q31-qter using fluorescence in situ hybridization (FISH). As probes we used representative clones from a contig map of yeast artificial chromosome (YAC) clones that spans chromosome bands 7q31.1-qter. In the 12 cases with loss of 7q material, we identified a commonly deleted region of approximately 4 to 5 megabasepairs in size encompassing the distal part of 7q35 and the proximal part of 7q36. Furthermore, the breakpoint of the reciprocal translocation from the patient with t-AML was localized to a 1,300-kb sized YAC clone that maps to the proximal boundary of the commonly deleted region. Interestingly, in this case both homologs of chromosome 7 were affected: one was lost (−7) and the second exhibited the t(7q35). The identification and delineation of translocation and deletion breakpoints provides the first step toward the identification of the gene(s) involved in the pathogenesis of 7q35-q36 aberrations in myeloid disorders.
CHROMOSOME 7 has been a focus of attention as a site harboring tumor suppressor genes since cytogenetic studies have shown deletions of its long arm (7q) in various tumor types.1,2 In myeloid disorders, loss of chromosome 7 (−7) or deletion of the long arm (7q−) are among the most common recurrent chromosome abnormalities. These aberrations are associated with myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), in particular with therapy-related MDS/AML (t-MDS/t-AML) following therapy with alkylating agents or secondary MDS/AML after occupational exposure to chemical mutagens.3-5 Furthermore, −7/7q− occur in MDS and AML that develop in patients with constitutional disorders (eg, Fanconi’s anemia, Kostmann’s syndrome, neurofibromatosis type 1, familial monosomy 7).6 Clinically, myeloid leukemias exhibiting −7/7q− have been associated with high susceptibility to infections, poor response to chemotherapy, and short survival times.3 7
By chromosome banding analysis two critical regions on the long arm of chromosome 7 have been identified, one in band 7q22 and another in bands 7q32-q35.5,8-11 The recurrent loss of genetic material suggests that these regions contain as yet unidentified tumor suppressor gene(s) which contribute to myeloid leukemogenesis. The molecular delineation of the proximal critical region in 7q22 has been the focus of several investigations.11-16 Using fluorescence in situ hybridization (FISH) and loss of heterozygosity (LOH) studies, distinct critical regions in bands 7q22-q31.1 have been identified which are shown to be commonly deleted or to contain translocation/inversion breakpoints in myeloid disorders.11-16
The distal critical region in 7q has so far only been delineated by chromosome banding analysis. In a study by Rodrigues Pereira Velloso et al10 of 54 patients with MDS and AML, bands 7q22 and 7q32 were most commonly deleted in the proximal and distal critical region, respectively. Le Beau et al11 reported on 16 cases with de novo MDS/AML and t-MDS/t-AML exhibiting deletions involving the distal part of 7q. All deletions were interstitial with the proximal and the majority of distal breakpoints localized to 7q31 or 7q32 and 7q36, respectively. The commonly deleted segment in this study was delineated to bands 7q32-q33.
In the present study, we analyzed samples from 13 patients with myeloid leukemia exhibiting deletions or a translocation affecting bands 7q31-qter by FISH. As probes we selected representative yeast artificial chromosome (YAC) clones from a physical map encompassing bands 7q31-qter.17-19 Because overlapping YACs were used, it was possible to systematically delineate the extent of the deletions and to locate the breakpoint of one reciprocal translocation at the molecular level.
MATERIALS AND METHODS
Patients.
Bone marrow and/or blood samples from 13 patients with myeloid disorders (de novo MDS, n = 3 [nos. 1-3]; de novo AML, n = 9 [nos. 4-12]; t-AML, n = 1 [no. 13]) were studied which, on chromosome banding analysis and/or FISH, exhibited deletions or translocations of bands 7q31-qter (Table1). Chromosome banding analysis was performed using standard methods, and the karyotypes were designated according to the International System for Cytogenetic Nomenclature.20
Karyotypes of the 13 Myeloid Leukemias
| Case No. . | Karyotype . |
|---|---|
| De novo MDS | |
| 1 | 46,XX,del(7)(q35) |
| 47,XX,del(7)(q35),+8 | |
| 2 | 43-45,XX,add(7)(q33-36),inc |
| 3 | 41-46,XX,add(7)(q32),inc |
| De novo AML | |
| 4 | 48,XX,−7,inc |
| 5 | 46,XY,der(7)t(7;13)(q32;q12) |
| 6 | 45,XY,del(7)(q31),−17 |
| 7 | 46,XY,del(7)(q3?4),−17,+r |
| 8 | 46,XY,del(7)(q?22),inc |
| 46,XY,del(7)(q?22),−7,+mar,inc | |
| 9 | 46,XY,der(7)t(1;7)(q11;q31) |
| 10 | 46,XY,t(8;21)(q22;q22) |
| 46,XY,del(7)(q32),t(8;21)(q22;q22) | |
| 11 | 47,XX,add(7)(q31),+13 |
| 12 | 46,XX,add(7)(q3?1),t(15;17)(q22;q21) |
| t-AML | |
| 13* | 45,X,−X,t(3;7)(p13;q34 or q35),−7,−13,+mar |
| Case No. . | Karyotype . |
|---|---|
| De novo MDS | |
| 1 | 46,XX,del(7)(q35) |
| 47,XX,del(7)(q35),+8 | |
| 2 | 43-45,XX,add(7)(q33-36),inc |
| 3 | 41-46,XX,add(7)(q32),inc |
| De novo AML | |
| 4 | 48,XX,−7,inc |
| 5 | 46,XY,der(7)t(7;13)(q32;q12) |
| 6 | 45,XY,del(7)(q31),−17 |
| 7 | 46,XY,del(7)(q3?4),−17,+r |
| 8 | 46,XY,del(7)(q?22),inc |
| 46,XY,del(7)(q?22),−7,+mar,inc | |
| 9 | 46,XY,der(7)t(1;7)(q11;q31) |
| 10 | 46,XY,t(8;21)(q22;q22) |
| 46,XY,del(7)(q32),t(8;21)(q22;q22) | |
| 11 | 47,XX,add(7)(q31),+13 |
| 12 | 46,XX,add(7)(q3?1),t(15;17)(q22;q21) |
| t-AML | |
| 13* | 45,X,−X,t(3;7)(p13;q34 or q35),−7,−13,+mar |
46,XX in PHA-stimulated blood.
DNA probes.
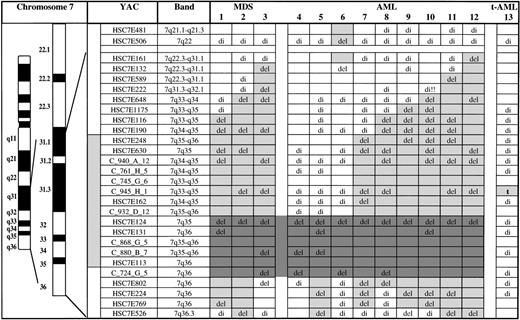
For the metaphase and interphase FISH experiments, we selected representative clones from a panel of YAC clones (Fig 1) that were previously mapped to chromosome bands 7q31.1 to 7qter (genes which are present on each YAC are given in parentheses)17-19 21: HSC7E161 (PDS, PRKAR2B, DRA), HSC7E132, HSC7E589, HSC7E222, HSC7E648, HSC7E1175 (ALDR1, BPGM,CALD1), HSC7E116 (CHRM2), HSC7E190, HSC7E248 (CLCN1, NEDD2), HSC7E630 (CLCN1,NEDD2), C_940_a_12 (NEDD2, TRCB, PRSS1,KEL, PIP1, CLCN1), C_761_H_5 (TIM1), C_745_G_6, C_945_H_1, HSC7E162, C_932_D_12, HSC7E124, HSC7E131, C_868_G_5, C_880_B_7, HSC7E113, C_724_G_5 (RHEB), HSC7E802 (XRCC2), HSC7E224, HSC7E769 (DPP6, HTR5A,APCR), and HSC7E526 (VIPR2). Clones HSC7E248 through HSC7E113 recognize a contiguous genomic fragment in 7q35-q36. Detailed information on the YACs is available at:http://www.genet.sickkids.on.ca/chromosome7/ andhttp://www.cephb.fr/ceph-genethon-map.html.
Mapping of deletions and a translocation involving chromosome bands 7q31-qter in 13 myeloid leukemias by FISH. HSC7E-YACs are from the chromosome 7–specific YAC library17 and the C_row_plate_ column YACs are from the CEPH-Généthon collection.21 Clones HSC7E248 to HSC7E113 recognize a contiguous genomic fragment in chromosome bands 7q35-q36. del, deletion (only one fluorescence signal); t, translocation breakpoint; di, disomy (two fluorescence signals indicating retention of both alleles); empty boxes, not done; light gray boxes, extent of the deletion; dark gray boxes, commonly deleted segment.
Mapping of deletions and a translocation involving chromosome bands 7q31-qter in 13 myeloid leukemias by FISH. HSC7E-YACs are from the chromosome 7–specific YAC library17 and the C_row_plate_ column YACs are from the CEPH-Généthon collection.21 Clones HSC7E248 to HSC7E113 recognize a contiguous genomic fragment in chromosome bands 7q35-q36. del, deletion (only one fluorescence signal); t, translocation breakpoint; di, disomy (two fluorescence signals indicating retention of both alleles); empty boxes, not done; light gray boxes, extent of the deletion; dark gray boxes, commonly deleted segment.
To ensure that the distal 7q deletions did not contiguously involve the proximal critical region in 7q22-q31, all cases were also hybridized with YAC clones HSC7E481 (7q21.1-q21.3) and HSC7E506 (7q22). We previously showed that YAC HSC7E506 recognizes a genomic fragment that is contained in the proximal critical region.12
Human sequences from the YAC clones were generated by a polymerase chain reaction (PCR) protocol using primers directed against Alu-sequences.22 Amplification was performed in a 100-μL reaction mixture containing approximately 160 ng YAC-DNA, 100 mmol/L of the four dNTPs (Boehringer Mannheim, Mannheim, Germany), 10 μL PCR-buffer (Boehringer Mannheim), and 2.0 mmol/L MgCl2(Boehringer Mannheim). Three Alu-PCR reactions were performed using either the primers CL1, CL2, or a combination of both (0.5 μmol/L). The products of all three reactions were combined for use in the FISH experiments. The Alu-PCR products were labeled by nick translation with biotin-16-dUTP or digoxigenin-11-dUTP (Boehringer Mannheim).
FISH.
FISH was performed as described.23 24 The hybridization mixture contained approximately 250 ng labeled Alu-PCR product, 10 μg Cot-1 DNA fraction (BRL/Life-Technologies, Gaithersburg, MD), and 10 μg salmon sperm DNA (Sigma Deisenhofen, Germany).
Interphase cytogenetic analysis was performed for deletion mapping. To monitor the hybridization efficiency we cohybridized with a YAC clone from 7q that mapped outside the region of interest. By analogy to our previous studies on deletion analyses, the cut-off level was defined by the mean + 3 SD of the frequency of control cells exhibiting only one fluorescence signal.12 23 The cut-off levels were determined for five representative YACs from the YAC map (range, 4.5% to 7.6%). Signal numbers were enumerated in 200 to 300 nuclei. The breakpoint of the balanced translocation t(3;7)(p13;q34 or q35) was additionally identified by metaphase FISH.
RESULTS
Chromosome banding analysis.
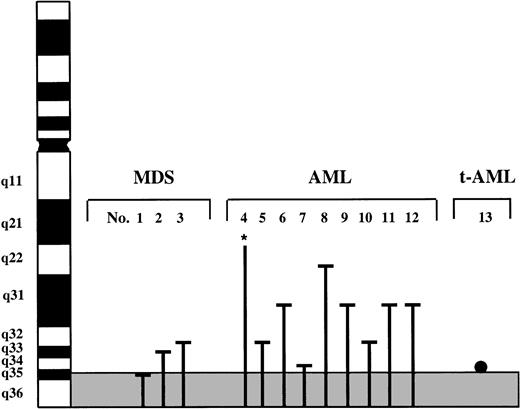
As determined by chromosome banding analysis, 11 of the 13 cases had aberrations involving bands 7q31-qter. The remaining two cases had monosomy 7 (no. 4) and a deletion del(7)(q?22) (no. 8) within a complex karyotype including unidentified marker chromosomes. The loss of 7q material resulted from terminal deletions (cases 1, 6, 7, 8, and 10) and unbalanced translocations (cases 2, 3, 5, 9, 11, and 12). The commonly deleted segment was defined as 7q35-qter (see Fig 2). Case 13 exhibited a balanced translocation t(3;7) with a breakpoint in band 7q34 or q35. Interestingly, this case also had monosomy 7. This patient had received total body irradiation and high-dose cyclophosphamide followed by autologous bone marrow transplantation for stage IV follicular lymphoma. Two years after transplantation the patient developed t-MDS which rapidly progressed to t-AML. Cytogenetic analysis of phytohemagglutinin (PHA)-stimulated blood at the time of diagnosis of the t-AML showed a normal karyotype. The karyotypes of the 13 leukemias are given in Table 1.
Deletions or translocations involving chromosome bands 7q31-qter in 13 patients with MDS, AML, and t-AML identified by chromosome banding analysis. The commonly deleted segment is delineated by the grey area. Line, deletion; point, translocation breakpoint; *, monosomy 7 within a complex karyotype.
Deletions or translocations involving chromosome bands 7q31-qter in 13 patients with MDS, AML, and t-AML identified by chromosome banding analysis. The commonly deleted segment is delineated by the grey area. Line, deletion; point, translocation breakpoint; *, monosomy 7 within a complex karyotype.
Deletion and translocation mapping by FISH.
The results of the deletion/translocation mapping by FISH are given in Fig 1. To delineate the commonly deleted segment in 7q31-qter at the molecular level, we selected a panel of YACs distributed along 7q31.1-qter, including a set of contiguously mapped YACs in bands 7q35-q36. In the 12 MDS/AML cases exhibiting 7q deletions, unbalanced 7q translocations, or monosomy 7 within a complex karyotype, we identified a commonly deleted region in bands 7q35-q36. In the case of t-AML with the t(3;7), we localized the 7q breakpoint to a 1,300-kb sized genomic segment in 7q35.
The commonly deleted segment in the 12 MDS/AML cases extended from YAC HSC7E124 in the distal part of band 7q35 to YAC C_724_G_5 in the proximal part of band 7q36 (Fig 1). This segment comprises approximately 4 to 5 megabasepairs (Mb). The proximal boundary of this segment was defined by cases 4 and 5. As illustrated in Fig 1, there is marked heterogeneity of the proximal deletion breakpoints among the cases: within bands 7q31.1-q35 the breakpoints were scattered along a large region extending from the genomic segments recognized by YAC HSC7E132 (case 3) to YAC HSC7E124 (cases 4 and 5). In case 12 the proximal deletion breakpoint was located between YAC HSC7E161 (7q31.1) and YAC HSC7E506 (7q22). For case 6 we could not determine the exact proximal deletion breakpoint because of the limited amount of material available. The distal boundary of the commonly deleted segment was defined by cases 4 and 6. In the remaining cases the deletions extended distal to the genomic region identified by YAC C_724_G_5 in band 7q36. In 5 (nos. 2, 5, 9, 11, and 12) of the 12 deletion cases, there was loss of the telomeric sequences (detected by YAC HSC7E526). Interestingly, case 6 had a noncontiguous deletion involving a genomic fragment in the proximal critical region in 7q22-q31 (identified by YAC HSC7E506) and in the distal critical region 7q35-q36 (identified by YACs HSC7E124 to C_724_G_5). In contrast to the interpretation of chromosome banding analysis, bands 7q32-q35 were retained. None of the other cases had deletion within the proximal critical region in 7q22-q31.
The breakpoint of the t(3;7)(p13;q34 or q35) (case 13) was mapped to a 1,300-kb genomic segment encompassed by YAC C_945_H_1 in band 7q35. This genomic fragment is located near the proximal boundary of the commonly deleted region. Based on the available physical map from this region, the distance between the translocation breakpoint and the commonly deleted region is estimated to be in the range of 2 to 3 Mb. There was no deletion detectable in the homolog involved in the translocation. FISH using YAC C_945_H_1 and a centromere 7 specific probe (Oncor Inc, Gaithersburg, MD) confirmed the results from chromosome banding analysis that the tumor also had monosomy 7. FISH using the centromere 7 probe and YAC C_945_H_1 on PHA-stimulated blood from this patient that was obtained at the time of the development of t-AML showed two hybridization signals for both probes, indicating that the translocation t(3;7) was not constitutional but somatically acquired. These data are supported by FISH with the same probes performed on mononuclear cells from the autologous bone marrow graft obtained 2 years before the development of t-AML. Hybridization again showed two fluorescence signals for the centromere 7 probe and YAC C_945_H_1.
DISCUSSION
Using FISH with YAC clones of defined physical position, we identified a commonly deleted genomic segment in 12 cases of myeloid leukemias exhibiting deletions of the distal part of 7q. Furthermore, we mapped the translocation breakpoint of a reciprocal translocation to a 1,300-kb sized genomic segment that is located at the proximal boundary of the commonly deleted region.
In the 12 MDS/AML cases with loss of 7q material we delineated a commonly deleted genomic segment of approximately 4 to 5 Mb in size encompassing the terminal part of band 7q35 and the proximal part of band 7q36. The proximal breakpoints in the deletion cases scattered along a large region extending from bands 7q31.1 to 7q35. The distal boundary of all the deletions extended into band 7q36, with 5 of the 12 cases involving the telomeric sequences detected with YAC HSC7E526. With respect to the chromosomal location of the critical region, our data are at variance to those obtained by chromosome banding analysis. In two recent chromosome banding studies, the commonly deleted region was assigned to bands 7q32-q33, whereas in our study the critical segment was delineated to a more distal region in 7q35-q36.10 11 FISH using probes from a defined physical map is the more sensitive technique for the delineation and chromosomal mapping of a critical segment, particularly in a region where chromosomal assignment of breakpoints may be difficult. In one of our cases (no. 6) we were able to identify a noncontiguous deletion by FISH involving the proximal (7q22-q31) and the distal (7q35-q36) critical region. Such high resolution cannot be achieved by the chromosome banding techniques.
In one case of t-AML, the translocation breakpoint of a reciprocal translocation t(3;7)(p13;q34 or q35) was identified and localized to a 1,300-kb sized genomic segment that is localized near the proximal boundary of the commonly deleted region. Interestingly, in this case chromosome 7 was homozygously affected by somatically acquired rearrangements, one homolog was lost (−7), and the second exhibited the t(7q34 or q35). One may speculate that the translocation breakpoint disrupts a gene relevant to the leukemogenic event. Translocations commonly lead to the activation of proto-oncogenes or to chimeric fusion genes; however, some translocations have been shown to be accompanied by submicroscopic deletions and have led to the identification of a genomic segment likely containing a novel tumor suppressor.25,26 Based on the physical map available, the estimated distance between the translocation breakpoint and the commonly deleted region is approximately 2 to 3 Mb. One possible interpretation of the significance of this translocation breakpoint is that—by analogy to the proximal critical region in 7q22-q31—there is heterogeneity of the translocation and deletion breakpoints.11-13 16 More than one gene in either critical region of chromosome 7 could be involved.
Only the gene encoding ras homologue enriched in brain 2 (RHEB)has been mapped to the commonly deleted region.27 The region has not yet been fully sequenced; thus, it is likely that other unidentified genes localize to this region. None of the genes that are located in the distal part of 7q and may be candidates based on their protein function such as NEDD2, XRCC2, and TIM map to the commonly deleted region or to the genomic segment containing the translocation breakpoint.28-30
In the present study, we defined a genomic fragment in chromosome bands 7q35-q36 that is commonly affected in malignant myeloid disorders. Refinement of the critical region by the analysis of additional tumor samples together with the growing data from the human chromosome 7 mapping and sequencing project will facilitate the identification of the relevant disease gene(s).
ACKNOWLEDGMENT
We gratefully acknowledge the members of the German multicenter AML HD Study Group and Dr B. Mohr for providing leukemia specimens.
Supported by the Forschungsförderungsprogramm of the Medical Faculty, University of Heidelberg, and the Medical Research Council (MRC) of Canada (to S.W.S. and L.C.T.). S.W.S. is a Scholar of the MRC and L.C.T. is a Senior Scientist of the MRC.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hartmut Döhner, MD, Medizinische Klinik and Poliklinik V, University of Heidelberg, Hospitalstraβe 3, 69115 Heidelberg, Germany; e-mail:hartmut_doehner@ukl.uni-heidelberg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal