Abstract
The receptor tyrosine kinase c-kit is necessary for normal hematopoiesis, the development of germ cells and melanocytes, and the pathogenesis of certain hematologic and nonhematologic malignancies. To better understand the regulation of the c-kit gene, a detailed analysis of the core promoter was performed. Rapid amplification of cDNA ends (RACE) and RNase protection methods showed two major transcriptional initiation sites. Luciferase reporter assays using 5′ promoter deletion-reporter constructs containing up to 3 kb of 5′ sequence were performed in hematopoietic and small-cell lung cancer cell lines which either did or did not express the endogenous c-kit gene. This analysis showed the region 83 to 124 bp upstream of the 5′ transcription initiation site was crucial for maximal core promoter activity. Sequence analysis showed several potential Sp1 binding sites within this highly GC-rich region. Gel shift and DNase footprinting showed that Sp1 selectively bound to a single site within this region. Supershift studies using an anti-Sp1 antibody confirmed specific Sp1 binding. Site-directed mutagenesis of the −93/−84 Sp1 binding site reduced promoter-reporter activity to basal levels in c-kit–expressing cells. Cotransfection into DrosophilaSL2 cells of a c-kit promoter-reporter construct with an Sp1 expression vector showed an Sp1 dose-dependent enhancement of expression that was markedly attenuated by mutation of the −93/−84 site. These results indicate that despite the fact that the human c-kit promoter contains multiple potential Sp1 sites, Sp1 binding is a selective process that is essential for core promoter activity.
C-KIT IS THE HUMAN cellular homologue of v-kit1 which was isolated from the Hardy-Zuckerman 4 feline sarcoma virus.2 c-kitencodes a receptor tyrosine kinase of the type III family which includes platelet-derived growth factor (PDGF) and colony-stimulating factor 1 receptors and is characterized by an intracellular split kinase domain as well as a cysteine-rich extracellular region. c-kit maps to the murine White Spotting (W) locus,3,4 and its ligand, known as mast cell growth factor, Kit ligand, or stem cell factor5 (SCF) maps to the mouseSteel (Sl) locus.6-8 Kit and SCF interaction is involved in the development of melanocytes, germ cells, and hematopoietic cells. Mutations of the mouse W locus and theSl locus lead to severe defects in hematopoiesis and deficiencies in mast cells, as well as unpigmented coats and sterility.9 SCF in combination with other hematopoietic growth factors supports the proliferation and differentiation of multiple hematopoietic cell lineages from early precursors.6,10,11 In addition to playing a preeminent role in normal hematopoiesis, Kit may be important in regulating the growth of some hematopoietic malignancies.12 Kit and SCF are coexpressed in a variety of solid tumors including gynecologic tumors,13 colon tumors,14 breast tumor specimens and tumor cell lines,15neuroblastomas,16 and over 70% of small-cell lung cancer (SCLC) lines and tumors.17-20 Coexpression in SCLC results in a functional autocrine growth loop.21
Sp1 is a ubiquitous transcription factor mostly associated with TATA-less GC-rich promoters and is mainly thought to be involved in basal promoter activity by interacting with transcription activation factors, which may stabilize components of the transcriptional machinery.22 Sp1 consists of three contiguous Cys2His2 Zinc-finger domains that bind to the decanucleotide consensus sequence 5′ GGGGCGGGGC 3′23,24 and similar sequences that are referred to as GC boxes. Family members Sp3 and Sp4 have similar structural features, and the DNA binding domains of all three proteins are highly conserved.25 Another family member, Sp2, seems to have somewhat different binding specificities.26 Functional analyses have shown that Sp4, like Sp1, is a transcriptional activator, whereas Sp3 has been shown to both repress27 and activate transcription.28,29 Studies have shown that Sp1-responsive promoters usually contain multiple binding sites of differing DNA sequence and Sp1 binding affinity,30-32 although Sp1 binding to a single site seems to be sufficient for promoter activation.
Previous studies of the regulation of c-kit transcription have characterized the mouse c-kit33 and human c-kit34-36 5′ flanking sequence and have identified potential Sp1 binding sites in the proximal promoter region. In studies of the mouse promoter, important cis-acting elements required for cell type–specific expression were localized within 105 bp of the transcription initiation site (TIS), and no additional elements in over 5 kb of 5′ sequence significantly influenced activity.33 The human c-kit promoter was found to lack a typical TATA box and contained several potential Sp1 binding sites, as well as putative binding sites for AP-2, Ets-domain proteins, Myb, and GATA-1.35 This study also showed that sequences mediating cell type–specific expression were contained within 120 bp of the TIS, which was different from the TIS identified in the mouse promoter, and that possible negative regulatory elements were located in the region between −992 and −604.35 A more recent study found that sequences up to 184 bp 5′ of the translational initiation codon were important for promoter activity, but the −4100 to −5500 region contained cell type–specific negative regulatory elements.36 With the possible exception of the microphthalmia transcription factor identified by genetic studies and specifically regulating c-kit expression in mast cells,37 and Myb36,38 and Ets38 proteins, which seem to have effects on expression only when they are overexpressed, no definitive binding of a transcription factor to the c-kit promoter has been shown. With this fact in mind along with the discrepancies over the sequences necessary for appropriating c-kit expression, we sought to further characterize the human c-kit promoter and specifically determine the role of Sp1 in its regulation.
MATERIALS AND METHODS
Cell cultures.
Cell lines used were as follows: c-kit–expressing SCLC cell lines H526, H510, and WB19 and the HEL leukemia cell line33,35; and the nonexpressing SCLC cell lines H146 and H8219 and the Jurkat T-cell line.36 Cultures were grown in RPMI 1640 (Bio-Whitaker, Walkersville, MD) medium supplemented with 10% fetal calf serum (Life Technologies, Bethesda, MD), 2 mmol/L L-glutamine, and 50 U/mL penicillin-50 U/mL streptomycin. Drosophila SL2 cells were cultured at 26°C in Schneider’s Drosophila medium (Life Technologies) containing 10% fetal calf serum (FCS).
Cloning of the c-kit promoter region.
A 6-kb NotI fragment containing 5′ c-kit promoter sequence was isolated from a human lambda (FIXII) placental library (Stratagene, La Jolla, CA) by screening with a radiolabelled 369-bpSstI-HindIII fragment of a 1.2-kb 5′ partial cDNA clone (American Type Culture Collection clone #594931). After primary screening, the positive plaques were purified through quaternary platings using two different T4 polynucleotide kinase-labeled oligonucleotides (complementary to bases 41-61 and 62-81 in exon 11) as probes. The genomic NotI fragment hybridizing to the exon 1 probes was excised and subcloned into the pGEM5Zf(+) plasmid vector (Promega, Madison, WI).
Cloning and characterization of c-kit mRNA primer extension products.
Cloning of c-kit mRNA primer extension products was accomplished by using a modification of the rapid amplification of cDNA ends (RACE) protocol for polymerase chain reaction (PCR) amplification of the 5′ ends of cDNAs.39 Two oligonucleotide primers (0.6 μg) specific for portions of the 5′ sequence of c-kit (complementary to bases 62-81 and 358-376 1) were annealed to 25 μg of WB total RNA by incubation for 16 hours at 55°C in 10 to 20 μL of 0.4 mol/L NaCl/8 mmol/L PIPES (piperazine-N-N′-bis[2-ethansulfonic acid]) buffer (pH 6.7) in separate reactions. The primer extension was accomplished by diluting the annealing mixture to 100 to 200 μL with the addition of 1,000 U of Moloney murine leukemia virus reverse transcriptase (MMLV-RT) or MMLV-RT Superscript (Life Technologies) and 0.5 mmol/L deoxynucleoside triphosphate with the manufacturer-supplied buffer. The extension reaction was performed at 37°C for 40 minutes and terminated by heating to 90°C for 2 minutes. Ten micrograms of RNase A was added, and the incubation was continued for 15 minutes at 37°C. The mixture was then made 0.5% sodium dodecyl sulfate, 30 μg of proteinase K was added, and the incubation was continued for 15 minutes at 42°C. The primer-extended species were then phenol-chloroform extracted, ethanol precipitated, and purified by binding to GlassMAX (Life Technologies). The cDNA was eluted from the GlassMAX matrix in TE [10 mmol/L Tris(hydroxymethyl) aminomethane-Cl, pH 7.4, 0.1 mmol/L Ethylenediamine Tetracetic acid] buffer, tailed with dATP as described by Frohman et al,39and repurified using GlassMAX.
One-half of the cDNA was diluted to 100 μL total volume and used in the PCR reaction. The PCR reaction mixture contained a 0.6 to 0.8 mmol/L concentration of orientation-specific primer designed to hybridize 3′ of the primer used for cDNA synthesis (complementary to bases 41-61 and bases 62-81, respectively1); a 0.9-mmol/L concentration of a (dT)17 primer-adapter containing HindIII, SalI, and XhoI cloning sites (5′-GACTCGAGTCGACAAGCTTTTTTTTTTTTTTTTT-3′); 50 mmol/L KCl, 10 mmol/L Tris HCl (pH 8.8), 1.5 mmol/L MgCl2, 3 mmol/L dithiothreitol (DTT), 100 μg/mL bovine serum albumin; 0.2 mmol/L deoxynucleoside triphosphates; and 2.5 U of Taqpolymerase (AmpliTaq; Perkin Elmer, Norwalk, CT). Amplification was performed in a programmable thermal reactor by first heating the mixture to 95°C for 1 minute and then using a step program (95°C, 1 minute; 50°C, 2 minutes; 72°C, 2 minutes) for 35 cycles, with a final extension for 15 minutes at 72°C. The amplified cDNA products were then phenol-chloroform extracted, ethanol precipitated, resuspended in TE buffer, and cut at the NarI (within exon 1) and SalI (within primer-adapter) restriction sites. These restriction-cut cDNA products were then ligated into either pGEM7Zf(−) or pSP70 vectors (Promega) cut withClaI and XhoI restriction endonucleases. The cDNA inserts were sequenced using the dideoxynucleotide chain termination method as described in Davis et al.40
RNase protection.
A 3-kb BamHI-BamHI fragment containing the promoter region was subcloned into a pGEM7Zf(−) vector and then cut at aSalI restriction site 445 bases 5′ of the BamHI site within exon 1 to make a template for the generation of a riboprobe. RNase protection assays were performed using 30 μg of total RNA as previously described.41
Luciferase constructs.
The promoter deletion constructs were made by restriction endonuclease digestion of the 6-kb NotI clone subcloned into the promoterless luciferase reporter plasmid pGL2:Basic (Promega). The 3′ 3 kb and 935 bp of the NotI fragment were ligated asBamHI fragments into pGL2:Basic to make the −3-kb and the −935-bp constructs, respectively. A BglII-XhoI fragment was ligated into pGL2:Basic to make the −409-bp construct. A SmaI fragment was ligated into pGL2:Basic to make the −124-bp construct. A BamHI-NaeI fragment was ligated into pGL2:Basic to make the −83-bp construct. All plasmid constructs were purified by double cesium chloride gradient centrifugation and verified by sequence analysis.
Mutagenesis of the Sp1 binding site.
The 2-bp mutation of −93/−84 Sp1 site in the −124 construct (XN2mt) was performed using the QuikChange Mutagenesis protocol (Stratagene). The oligonucleotides used for this site-directed mutagenesis were 5′-GGGGAGGCGAGGAGGTTCGTGGCCGCGCG-3′ and 5′-CGCGCCGGCCACGAACCTCCTCGCCTCCCC-3′. Reinsertion of the −93/−84 Sp1 site upstream of the XN2mt construct was performed by annealing two oligonucleotides, 5′ CCGGGAGGGGCGTGGCCG 3′ and 5′ CCGGCGGCCACGCCCCTC 3′, which were then ligated into the −124/XN2mt construct cut withXmaI.
Luciferase reporter assays.
Transfection was initiated by adding DNA to 8 × 106cells suspended in 0.4 mL in 2-mm cuvettes that were incubated on ice for 10 minutes. H526 cells were electroporated in phosphate-buffered saline (PBS) (pH 7.4) at 300 V and 500 μF, H146 cells in PBS at 300 V/1,000 μF, HEL cells in PBS at 200 V/1,500 μF, and Jurkat cells in RPMI 1640 at 300 V/1,250 μF using a BTX Electro Cell Manipulator 600 (BTX Inc, San Diego, CA). The cells were transfected with 25 μg of the pGL2:−3-kb construct and equimolar amounts of the smaller reporter plasmids. pSP70 plasmid was used as a filler plasmid to maintain a final DNA concentration of 25 μg for each transfection. Ten micrograms of pCMV βgal plasmid was cotransfected to serve as a control for transfection efficiency. After electroporation, the cells were plated in 6 mL of RPMI 1640/10% FCS in 60-mm tissue culture dishes, cultured for 24 hours, and harvested according to the Reporter Lysis protocol (Promega) using 400 μL of reporter lysis buffer. Luciferase and β-galactosidase activities were assayed according to the manufacturer’s protocol (Promega). Luciferase activity was analyzed using 20 μL of cell extract mixed with 100 μL of luciferase substrate (Promega), which was quantitated for 30 seconds in a Lumat LB 9507 luminometer (EG&G Berthold, Bad Wildbad, Germany). β-galactosidase activity was used to normalize for transfection efficiency. SL2 cells were transfected using the calcium phosphate coprecipitation technique, as described in Davis et al.40 Luciferase and β-galactosidase activity in the SL2 cells were assayed as above.
Gel mobility shift analysis.
Whole cell extracts used in this assay were prepared in the following manner. Approximately 5 × 107 cells were obtained, washed with PBS (pH 7.4), and resuspended in 1 mL of extraction buffer (20 mmol/L Hepes [pH 7.8], 450 mmol/L NaCl, 0.4 mmol/L EDTA, 0.5 mmol/L DTT, 25% glycerol, 0.5 mmol/L phenylmethylsulfonyl fluoride). This suspension was frozen and thawed three times using a dry ice/ethanol bath and a 37°C water bath. After a 10-minute centrifugation at 13,000g at 4°C, the supernatant was aliquotted and stored at −70°C until use. Protein concentration was determined by BCA assay (Pierce, Cleveland, OH). Oligonucleotides used were as follows: Xma-Nae fragment (−124 to −83): 5′-CCGGGCGGGCGCGAGGGAGGGGAGGCGAGGAGGGGCGTGGCC-3′ and 5′-GGCCACGCCCCTCCTCGCCTCCCCTCCCTCGCGC-3′; XN (−125/−97): 5′-CCCGGGCGGGCGCGAGGGA-3′ and 5′-TCGCCTCCCCTCCCTCGCGCCCGC-3′; XN2 (−102/−82): 5′-GGGGAGGCGAGGAGGGGCGT-3′ and 5′-CGGCCACGCCCCTCCTCG-3′; the XN2 oligonucleotide with a 2-bp mutation in a putative Sp1 site, XN2mt: 5′-GGGGAGGCGAGGAGGTTCGT-3′ and 5′-CGGCCACGAACCTCCTCG-3′; and XN3 (−96/−82): 5′-GGAGGGGCGTGGC-3′ and 5′-CGGCCACGCCCCT-3′. Oligonucleotides were synthesized by the MCV-VCU Nucleic Acid Synthesis Core Facility (Richmond, VA), and Integrated DNA Technologies (Coralville, IA). Consensus and mutant Sp1 oligonucleotides were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The oligonucleotides were annealed; radiolabeled by fill-in reaction using Klenow fragment, dATP, dGTP, dTTP, and a32P-dCTP (3,000 Ci/mmol; NEN, Boston, MA) or T4 polynucleotide kinase and γ32P-ATP40; and unincorporated label was removed using a Sephadex G-50 column. Ten micrograms of H526 cell extract was incubated with 10,000 cpm of labeled DNA in 20 μL of binding buffer (20 mmol/L HEPES [pH 7.9], 1 mmol/L DTT, 1 mmol/L EDTA, 10% glycerol, 1 μg poly dI-dC, and 50 mmol/L NaCl) for 20 minutes at room temperature. Assays with recombinant human Sp1 (rhSp1; Promega) protein were performed in the same manner using 1 footprinting unit (fpu) of rhSp1 added to 1 μg ofDrosophila nuclear extract (Promega). For competition assays, unlabeled competitor oligonucleotides were added to the nuclear extracts 20 minutes before addition of radiolabeled probe. Reactions were electrophoresed at 100 V in a nondenaturing 4% polyacrylamide gel in 0.5 × Tris-borate, EDTA (TBE) buffer40at 4°C.
In supershift assays, 2 μg of an antibody specific for Sp1, Sp2, Sp3, Sp4 (Santa Cruz), or nonspecific rabbit IgG (Sigma Chemical Co, St Louis, MO) was incubated with the gel-shift mixture for 20 minutes at room temperature after normal gel-shift incubation. The 4% nondenaturing gel was run at 325 V at 4°C in 0.5 × TBE.
DNA footprinting.
Footprinting assays were performed according to the Core Footprinting system protocol (Promega). A fragment containing bases −124 to +37 was cut out of the pGL2:−409-bp construct usingHindIII and SmaI. It was subsequently subcloned into the HindIII/SmaI sites of the pSP64 plasmid vector (Promega). From this construct, a 172-bp fragment was cut out usingHindIII and EcoRI, kinased using γ32P-ATP, restriction enzyme digested with SmaI, leaving only the +37 end of the probe labeled. Cell extracts and binding buffer were prepared as described above. The labeled probe was incubated with 10 or 20 μg of H526 cell extract for 20 minutes at room temperature in a total volume of 20 μL. This mixture was digested with 1.2 U of DNase I for 1 or 2 minutes at room temperature and then run on a 6% acrylamide, 8 mol/L urea sequencing gel at 1,500 V/60 W until the bromophenol blue dye-front reached the bottom of the gel. For the sequencing ladder, a 17-bp oligonucleotide, 5′-AGCTTGGATCCGAGCTC-3′, corresponding to the exact 5′ end of the footprinting fragment was used as a primer for sequencing of the pSP64 plasmid containing theHindIII-SmaI insert.
RESULTS
Identification of the transcription initiation sites.
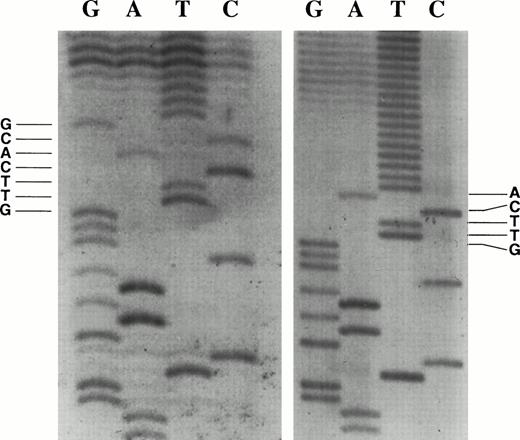
We initially used a modification of the RACE protocol to identify the transcriptional initiation sites using RNA isolated from the c-kit–expressing SCLC cell line WB. Oligonucleotide primers were annealed to WB total RNA, extended, tailed with dATP, amplified by PCR, subcloned, and inserts were sequenced. The transcription start site was identified as the base adjacent to the start of the terminal transferase-generated deoxynucleotide tail. We sequenced 11 independent clones generated from the RACE protocol. Seven of the 11 clones identified the major initiation sites at the nucleotide G, 58 bases upstream of the translational initiation codon, and the nucleotide A, 56 bases upstream (Fig 1). These TIS are designated as +1 and +3 on the proximal c-kit promoter sequence (Fig 2).
RACE clones show two major transcriptional initiation sites. The transcription initiation site was identified as the base adjacent to the start of the poly T tail. (Left panel) Transcription start site at nucleotide G, base +1 in Fig 2. (Right panel) Transcription start site at nucleotide A, base +3 in Fig 2.
RACE clones show two major transcriptional initiation sites. The transcription initiation site was identified as the base adjacent to the start of the poly T tail. (Left panel) Transcription start site at nucleotide G, base +1 in Fig 2. (Right panel) Transcription start site at nucleotide A, base +3 in Fig 2.
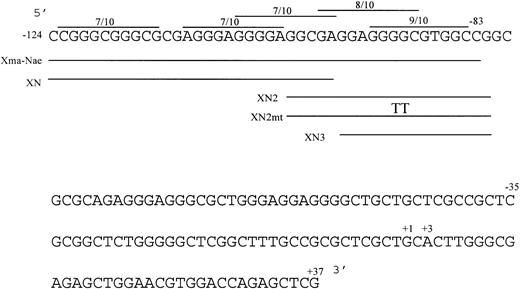
Nucleotide sequence of the human c-kit promoter from −124/+37. The sequence is numbered from the 5′ transcription initiation site. The locations of the double-stranded oligonucleotides (XmaI-NaeI, XN, XN2, XN3) used in gel shift analysis (below) are indicated. The locations of potential Sp1 binding sites are indicated as lines above the sequence showing degree of homology to the consensus decanucleotide. The TT in the XN2mt oligonucleotide marks the location of an inserted 2-bp mutation (below). The +37 nucleotide is the most 3′ base of c-kit promoter sequence before the pGL2 polylinker in the reporter plasmids.
Nucleotide sequence of the human c-kit promoter from −124/+37. The sequence is numbered from the 5′ transcription initiation site. The locations of the double-stranded oligonucleotides (XmaI-NaeI, XN, XN2, XN3) used in gel shift analysis (below) are indicated. The locations of potential Sp1 binding sites are indicated as lines above the sequence showing degree of homology to the consensus decanucleotide. The TT in the XN2mt oligonucleotide marks the location of an inserted 2-bp mutation (below). The +37 nucleotide is the most 3′ base of c-kit promoter sequence before the pGL2 polylinker in the reporter plasmids.
To confirm the RACE study and determine the relative usage of these transcriptional start sites, an RNase protection assay was performed. ABamHI-SalI fragment extending approximately 400 bases beyond the potential transcriptional start sites was used to transcribe a riboprobe. This riboprobe was annealed to total RNA isolated from cell lines that express c-kit mRNA (WB, H510, and HEL) as well as a cell line that does not express c-kit mRNA (H82), and an RNase protection assay was performed.
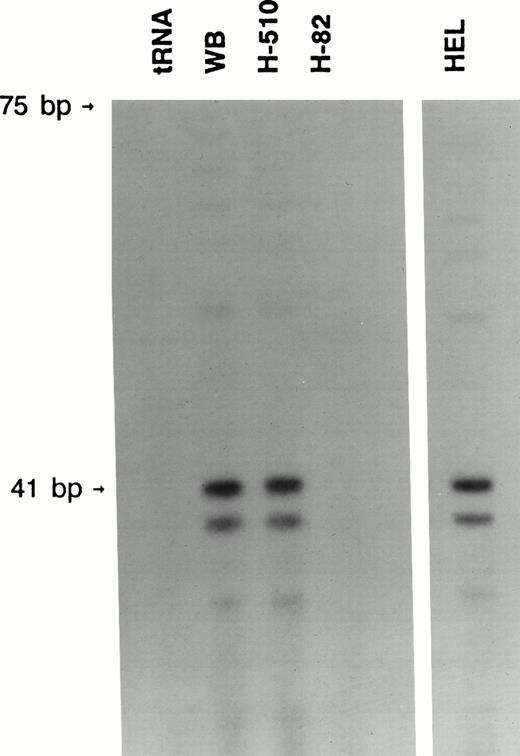
Both the WB and H510 SCLC cell lines, which express high levels of c-kit mRNA, were found to contain protected bands corresponding to at least three different transcriptional start sites (Fig 3). The two major bands correspond to the bases +1 and +3 identified by the RACE protocol (Figs 1 and 2). The third band corresponds to base +7, a base also identified by one of the RACE clones. We found no protected bands when our riboprobe was annealed to either RNA from the SCLC cell line H82, which does not express c-kit, or yeast tRNA (Fig 3). HEL cells were found to have the identical three bands, showing that these transcriptional start sites are common to SCLC and hematopoietic cells (Fig 3).
RNase protection analysis showed multiple transcriptional initiation sites. Two major and one minor protected bands, indicating the transcriptional start sites, were detected in the WB and H510 SCLC cell lines as well as the HEL leukemia cell line. These respectively correspond to base +1 (41 bp from 3′ BamHI site), +3, and +7 in Fig 2. The upper major band comigrated with a 41-base RNA size marker made by transcribing BamHI-cut pGEM3 with SP6 polymerase.
RNase protection analysis showed multiple transcriptional initiation sites. Two major and one minor protected bands, indicating the transcriptional start sites, were detected in the WB and H510 SCLC cell lines as well as the HEL leukemia cell line. These respectively correspond to base +1 (41 bp from 3′ BamHI site), +3, and +7 in Fig 2. The upper major band comigrated with a 41-base RNA size marker made by transcribing BamHI-cut pGEM3 with SP6 polymerase.
Scanning of the sequence immediately upstream of the transcription initiation sites (Fig 2) failed to reveal a TATA box. This region was also scanned for an initiator (Inr) promoter element, which can also mediate transcriptional initiation at unique sites.42 The Inr overlaps transcriptional start sites, and has the core consensus sequence Py Py A+1 N T/A Py Py.43 The c-kit transcriptional start site at base +1 corresponds to this sequence, except guanine is substituted for adenine at +1 (Fig 2). If the c-kit transcriptional start site at base +3 is considered, then the only differences are purines substituted for pyrimidines at both ends of the consensus sequence.
Functional analysis of the human c-kit promoter.
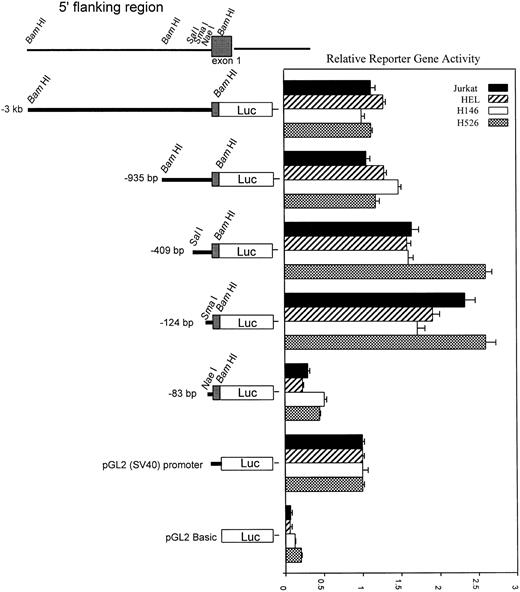
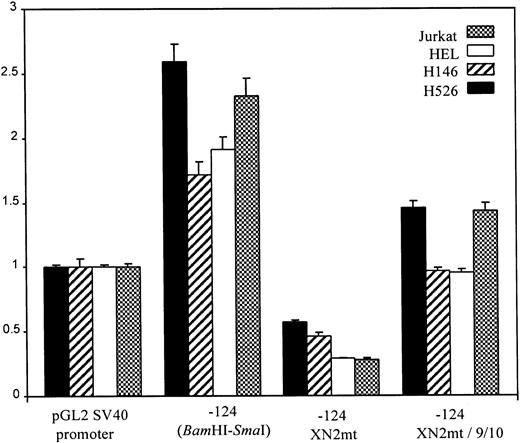
To characterize functional regulatory elements within the c-kitpromoter, we used a series of 5′-deletion fragments linked to the luciferase gene and tested them in transient transfection assays. The promoter fragments ranging from 83 bp up to 3 kb of 5′ sequence were cloned into the promoterless pGL2:Basic luciferase vector. The deletion construct plasmids were electroporated into the H526 SCLC and the HEL cell lines which express endogenous c-kit or the H146 SCLC and the Jurkat cell lines which do not express c-kit. Reporter assays showed the same level of expression independent of whether the cell lines expressed c-kit, indicating there are no cell type–specific regulatory elements within sequences 3 kb upstream of the TIS (Fig 4). The −83 construct showed a basal level of activity. There was a fourfold to fivefold increase in activity from the −83-bp construct to the −124-bp construct. Additional sequence beyond the −409 construct showed a decrease in reporter activity which may indicate negative regulatory sequences. Based on this analysis, regulatory elements responsible for maximal core promoter activity must lie within the −83 to −124 region.
Promoter-reporter activity of sequential 5′ deletion constructs. Promoter activity of all promoter constructs (diagrammed at left) was compared with that of pGL2:SV40 promoter with transient transfection of the constructs into H526, H146, HEL, and Jurkat cell lines. Cotransfection with pCMV βgal plasmid was used to control for transfection efficiency. After a 24-hour incubation, cell extracts were assayed, normalized for transfection efficiency, and relative reporter activity was determined. Error bars indicate standard error (SE) calculated from at least six separate electroporations. All activity values are relative to the pGL2:SV40 promoter, which was assigned a value of 1.0. Relative reporter gene activity was calculated as follows: luciferase activity divided by β-galactosidase activity, divided by the ratio obtained for the pGL2:SV40 promoter construct.
Promoter-reporter activity of sequential 5′ deletion constructs. Promoter activity of all promoter constructs (diagrammed at left) was compared with that of pGL2:SV40 promoter with transient transfection of the constructs into H526, H146, HEL, and Jurkat cell lines. Cotransfection with pCMV βgal plasmid was used to control for transfection efficiency. After a 24-hour incubation, cell extracts were assayed, normalized for transfection efficiency, and relative reporter activity was determined. Error bars indicate standard error (SE) calculated from at least six separate electroporations. All activity values are relative to the pGL2:SV40 promoter, which was assigned a value of 1.0. Relative reporter gene activity was calculated as follows: luciferase activity divided by β-galactosidase activity, divided by the ratio obtained for the pGL2:SV40 promoter construct.
Specific protein binding to the −93/−84 site in the core promoter.
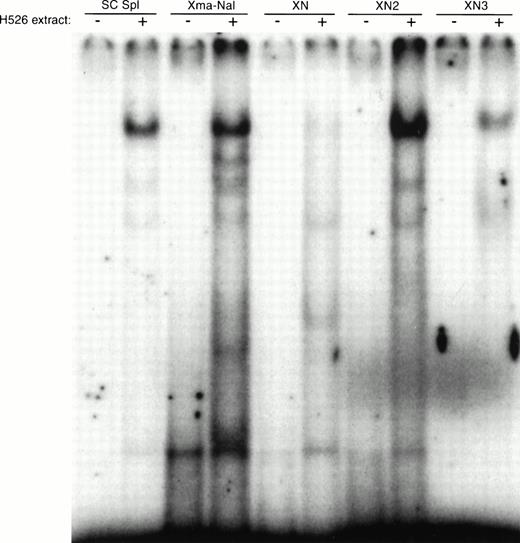
A search for transcription factor binding sites within this 40 bp yielded several potential consensus binding sites for Sp1 including a 9/10 consensus Sp1 site, GGGGCGTGGC (Fig 2). To characterize the proteins that bind to this region of DNA, we performed gel mobility shift analysis (Fig 5). When oligonucleotides corresponding to the 5′ and 3′ halves of the 40 bases were incubated with H526 cell extract, only the 3′ half oligonucleotide (XN2) produced a strong shifted band. A further gel shift using a smaller oligonucleotide corresponding to the 3′ 15 bp (XN3) showed that only a 15-bp fragment of DNA containing the −93/−84 consensus Sp1 site is necessary for a gel shift. This shifted band comigrates with the band obtained using a consensus Sp1 oligonucleotide.
Gel mobility shift analysis of protein binding to the −124 to −83 DNA fragment. Gel mobility shift analysis was performed with the double-stranded DNA fragment containing the −124 to −83 region alone and with 10 μg of cellular protein from H526 cells. The SC Sp1 oligonucleotide (Santa Cruz) contains a consensus Sp1 binding site. The oligonucleotides corresponding to c-kit sequences are: Xma-Nae (−124/−83), XN (−125/−97), XN2 (−102/−82), XN3 (−96/−82). The +/− indicates whether H526 extract was added to the gel shift mixture.
Gel mobility shift analysis of protein binding to the −124 to −83 DNA fragment. Gel mobility shift analysis was performed with the double-stranded DNA fragment containing the −124 to −83 region alone and with 10 μg of cellular protein from H526 cells. The SC Sp1 oligonucleotide (Santa Cruz) contains a consensus Sp1 binding site. The oligonucleotides corresponding to c-kit sequences are: Xma-Nae (−124/−83), XN (−125/−97), XN2 (−102/−82), XN3 (−96/−82). The +/− indicates whether H526 extract was added to the gel shift mixture.
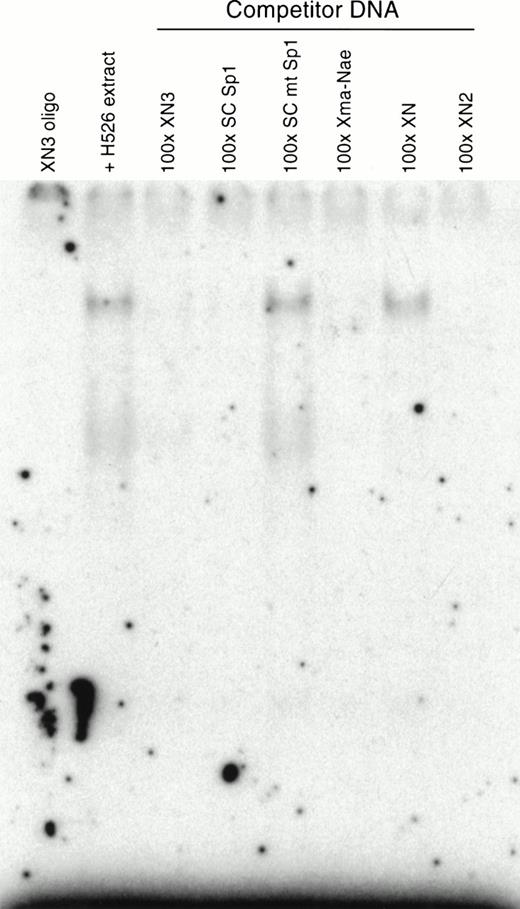
To show specificity, a competition assay was performed. Incubating the XN3 oligonucleotide with unlabeled competitor DNA containing a cognate binding site before addition of labeled probe should compete for protein binding and result in subsequent loss of a shifted band. Only the oligonucleotides containing the −93/−84 site and the consensus Sp1 oligonucleotide were able to successfully compete for binding (Fig 6). Neither the mutant consensus Sp1 oligonucleotide (5′-ATTCGATCGGTTCGGGGCGAGC-3′) nor the oligonucleotide corresponding to the 5′ half of the 40-bp (XN) fragment was able to compete for binding, which shows specific binding by the protein to the XN3 oligonucleotide.
Competition assays using specific and nonspecific competitor oligonucleotides. In the absence or presence of competing oligonucleotides, 0.1 ng of 32P-labeled XN3 oligonucleotide was incubated with H526 extract. Competing oligonucleotides were at 100-fold excess. The competing oligonucleotides corresponding to c-kit sequences were: Xma-Nae (−124/−83), XN (−125/−97), XN2 (−102/−82), XN3 (−96/−82). Labeled DNA:protein complexes were only formed in the presence of competitors that do not contain the −93/−84 Sp1 site. Competition using the consensus Sp1 oligonucleotide was used as a positive control, and the oligonucleotide containing a mutant Sp1 site (SC mt Sp1) was used as a negative control.
Competition assays using specific and nonspecific competitor oligonucleotides. In the absence or presence of competing oligonucleotides, 0.1 ng of 32P-labeled XN3 oligonucleotide was incubated with H526 extract. Competing oligonucleotides were at 100-fold excess. The competing oligonucleotides corresponding to c-kit sequences were: Xma-Nae (−124/−83), XN (−125/−97), XN2 (−102/−82), XN3 (−96/−82). Labeled DNA:protein complexes were only formed in the presence of competitors that do not contain the −93/−84 Sp1 site. Competition using the consensus Sp1 oligonucleotide was used as a positive control, and the oligonucleotide containing a mutant Sp1 site (SC mt Sp1) was used as a negative control.
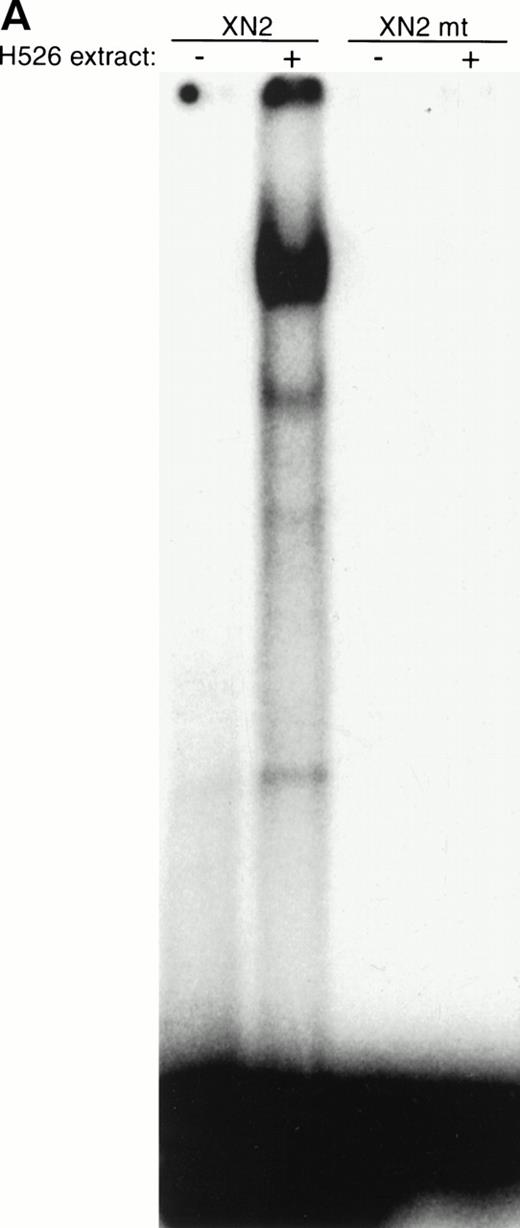
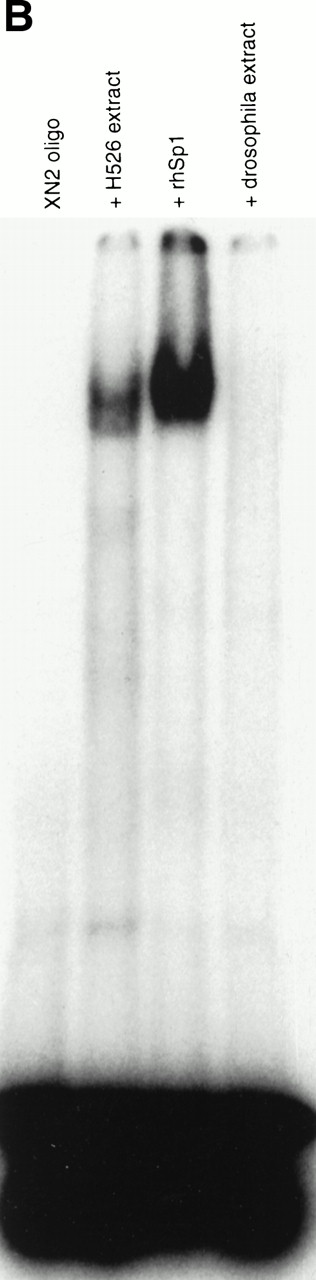
If the protein was bound to the −93/−84 site, mutation of this site should cause disruption of binding and loss of the shifted band. A 2-bp mutation (GGGGCGTGGC > GGTTCGTGGC) was introduced into the −93/−84 site in the XN2 oligonucleotide (Fig 2); gel shift analysis with this oligonucleotide failed to produce a shifted band (Fig 7A). This indicates that a protein from the H526 extract was specifically recognizing the 2-bp mutated sequence as part of its binding site. Furthermore, incubation of the XN2 oligonucleotide with H526 extract yielded a shifted complex with the same mobility as that of the XN2 oligonucleotide incubated with recombinant human Sp1 protein (Fig 7B). A Drosophila extract, which does not contain Sp1, failed to produce a shifted band. These data further suggest that the binding protein is Sp1.
The protein in the −93/−84 complex has the binding specificity and mobility of Sp1. (A) A 2-bp mutation (GGGGCGTGGC > GGTTCGTGGC) was introduced into the −93/−84 Sp1 site of the XN2 oligonucleotide (XN2mt), and binding ability was compared with the wild-type XN2 oligonucleotide. The −/+ indicates whether cellular extract was added to the gel shift mixture. Ten thousand cpm of the XN2 and XN2mt oligonucleotides were incubated with 10 μg of H526 extract. (B) Mobility shift of XN2 oligonucleotide with rhSp1. Mobility shift analysis using the XN2 oligonucleotide incubated with H526 extract (10 μg ) or with recombinant human Sp1 (rhSp1, 1 fpu) and Drosophila nuclear extract (1 μg). Incubation of the XN2 oligonucleotide with H526 extract (lane 2) yielded a band with a similar mobility pattern as that obtained by incubating XN2 oligonucleotide with rhSp1 andDrosophila extract (lane 3). Incubation of XN2 oligonucleotide with Drosophila extract alone was used as a negative control (lane 4).
The protein in the −93/−84 complex has the binding specificity and mobility of Sp1. (A) A 2-bp mutation (GGGGCGTGGC > GGTTCGTGGC) was introduced into the −93/−84 Sp1 site of the XN2 oligonucleotide (XN2mt), and binding ability was compared with the wild-type XN2 oligonucleotide. The −/+ indicates whether cellular extract was added to the gel shift mixture. Ten thousand cpm of the XN2 and XN2mt oligonucleotides were incubated with 10 μg of H526 extract. (B) Mobility shift of XN2 oligonucleotide with rhSp1. Mobility shift analysis using the XN2 oligonucleotide incubated with H526 extract (10 μg ) or with recombinant human Sp1 (rhSp1, 1 fpu) and Drosophila nuclear extract (1 μg). Incubation of the XN2 oligonucleotide with H526 extract (lane 2) yielded a band with a similar mobility pattern as that obtained by incubating XN2 oligonucleotide with rhSp1 andDrosophila extract (lane 3). Incubation of XN2 oligonucleotide with Drosophila extract alone was used as a negative control (lane 4).
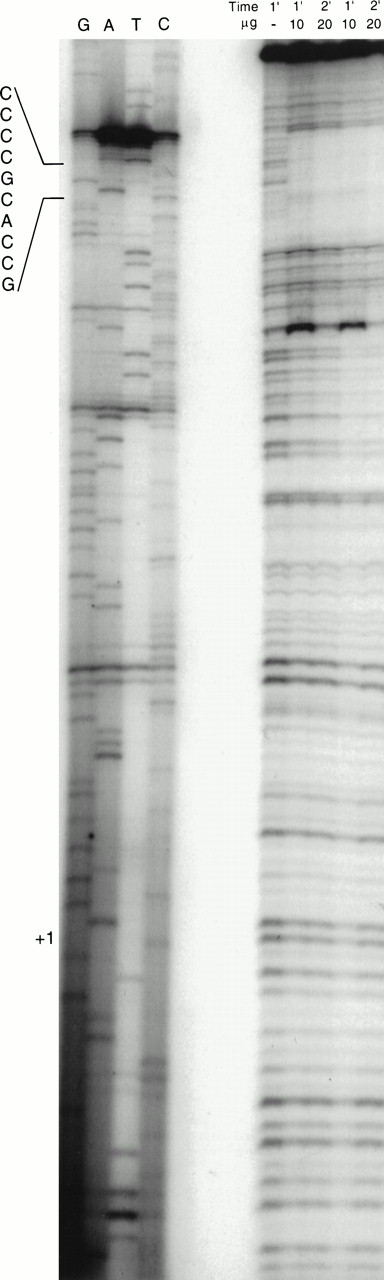
To confirm the location of protein binding and to screen for additional binding sites, a DNase footprint analysis was performed. A DNA fragment containing the −124/+37 region was 5′ end-labeled, incubated with H526 cell extract, and then digested with 1.2 U of DNase I. The digestion products were analyzed on a 6% sequencing gel. A sequencing reaction of the identical region was run on the same gel to identify the protected region. The region corresponding to the −93/−84 consensus Sp1 was protected from DNase I digestion (Fig 8). This was the only region shown to be protected under our assay conditions, but it is possible that there are other protected sites at the 5′ and 3′ ends of the fragment which cannot be well visualized.
DNase footprinting analysis of the −124 to +37 region. The ability of proteins to bind and protect the core promoter region of the c-kit promoter was analyzed. DNase I digestion for 1 and 2 minutes of the 32P-labeled DNA fragment was performed with no cell extract (lane 1) and in the presence of 10 and 20 μg of cell extract (right panel). The digested products were analyzed on a 6% sequencing gel. A sequencing reaction of the DNA region was run in parallel (left panel). The sequence of the protected region, corresponding to the −93/−84 SP1 site, is shown.
DNase footprinting analysis of the −124 to +37 region. The ability of proteins to bind and protect the core promoter region of the c-kit promoter was analyzed. DNase I digestion for 1 and 2 minutes of the 32P-labeled DNA fragment was performed with no cell extract (lane 1) and in the presence of 10 and 20 μg of cell extract (right panel). The digested products were analyzed on a 6% sequencing gel. A sequencing reaction of the DNA region was run in parallel (left panel). The sequence of the protected region, corresponding to the −93/−84 SP1 site, is shown.
Sp1 specifically binds the −93/−84 site.
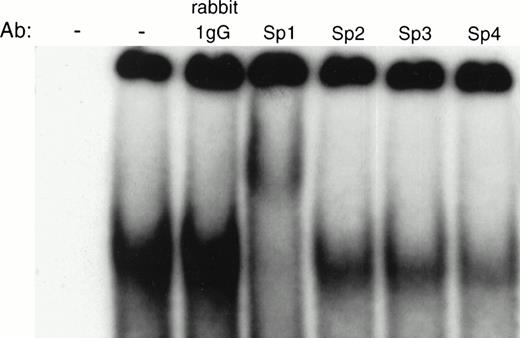
The above data strongly suggest that an Sp family member binds to the −93/−84 site based on the specificity of binding and mobility of gel-shifted species. To further determine if the protein binding to the −93/−84 site was indeed Sp1, a supershift assay was performed. If Sp1 was the binding protein, incubation of the protein:DNA complex with an anti-Sp1 antibody should result in a complex of slower mobility than the original shifted band. Because the other Sp family members also bind GC boxes, we also assayed the abilities of antibodies to Sp2, Sp3, and Sp4 to bind to the protein:DNA complex. Incubating 32P-labeled XN2 oligonucleotide with H526 cell extract and Sp1-specific antibody resulted in a supershifted band, whereas incubating with antibodies specific for Sp2, Sp3, Sp4, or nonspecific rabbit IgG did not (Fig 9), confirming that Sp1 specifically binds to this region of DNA.
Supershift analysis of Sp1 family members. Interaction of the XN2 fragment with H526 cell extracts in the absence and presence of antibodies specific for Sp1, Sp2, Sp3, Sp4, and nonspecific rabbit IgG. The DNA:protein complexes were analyzed on a 4% nondenaturing polyacrylamide gel. The first lane contains no extract and no antibody.
Supershift analysis of Sp1 family members. Interaction of the XN2 fragment with H526 cell extracts in the absence and presence of antibodies specific for Sp1, Sp2, Sp3, Sp4, and nonspecific rabbit IgG. The DNA:protein complexes were analyzed on a 4% nondenaturing polyacrylamide gel. The first lane contains no extract and no antibody.
Mutational analysis of the −93/−84 Sp1 site.
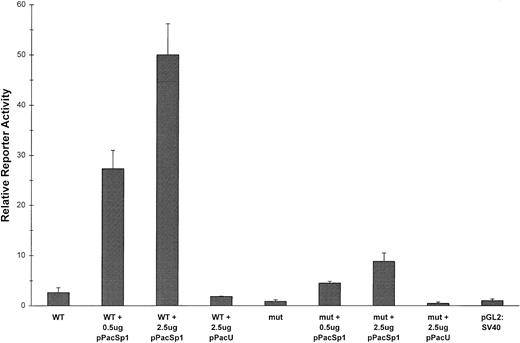
The reporter construct containing the −124/+37 region has maximal activity, likely representing the core promoter. To correlate Sp1 binding with the potency of this region to promote transcription, we introduced a 2-bp mutation into the −93/−84 Sp1 site (GGGGCGTGGC > GGTTCGTGGC) of the −124 construct (XN2mt). This mutation eliminated Sp1 binding by gel-shift analysis (Fig 7). Reporter activity of the mutant construct transfected into H526 cells was reduced to basal levels when compared with the wild-type construct (Fig 10). To study the effect of restoration of the Sp1 binding site, we inserted the wild-type −93/−84 consensus site directly upstream of the XN2mt construct (XN2mt/Sp1), which restored much of the activity, though it was less than wild-type. This lower activity may be related to the altered position of the Sp1 binding site in this construct.
Functional analysis of −93/−84 binding site. Reporter assays were performed as in Fig 4. A construct containing a 2-bp mutation of the −93/−84 Sp1 (GGGGCGTGGC > GGTTCGTGGC) site, −124 XN2mt, displayed basal reporter activity. The −124 XN2mt/9/10 construct, which has the −93/−84 Sp1 site inserted 40 bp upstream in the −124 XN2mt construct, restores reporter activity to near wild-type levels.
Functional analysis of −93/−84 binding site. Reporter assays were performed as in Fig 4. A construct containing a 2-bp mutation of the −93/−84 Sp1 (GGGGCGTGGC > GGTTCGTGGC) site, −124 XN2mt, displayed basal reporter activity. The −124 XN2mt/9/10 construct, which has the −93/−84 Sp1 site inserted 40 bp upstream in the −124 XN2mt construct, restores reporter activity to near wild-type levels.
The above data correlates the ability of Sp1 to uniquely bind to the −93/−84 site in vitro with the ability of this site to activate transcription in vivo. To correlate in vivo Sp1 binding with the ability of this site to activate transcription we turned to the Drosophila SL2 expression system.44 Courey and Tjian44 devised this system to analyze the functional domains of Sp1 because it is extremely difficult to observe the effects of exogenously expressed Sp1 on the background of the high constitutive levels of Sp1 in mammalian cells. Drosophila cells lack endogenous Sp1, yet transfected reporter plasmids containing Sp1 sites show a dose response to exogenously expressed Sp1, indicating that the conserved transcriptional machinery is capable of interacting with the transcriptional activation domain of Sp1.44 To correlate in vivo Sp1 binding with activation of c-kit transcription, we cotransfected into SL2 cells either the wild-type −124 reporter construct or the construct containing the mutant −93/−84 site with increasing amounts of a plasmid encoding Sp1 under the control of the Drosophila actin promoter (pPacSp1; kindly donated by R. Tjian).44Figure 11 illustrates that transcriptional activation of the wild-type −124 construct showed a dramatic dose response to Sp1 expression. Transfection of the emptyDrosophila expression vector, pPacU, had no effect on expression. The mutant construct also showed an Sp1 dose-dependent increase in transcription, but the levels of reporter expression approached only 10% to 20% of those of the wild-type construct. The Sp1-dependent transcription of the mutant construct suggests that low-level transcriptional activation may be obtained through Sp1 binding to alternative sites. However, it is clear from all the reporter assays that selective Sp1 binding to the −93/−84 site and not to the other potential binding sites is required for maximal promoter activity.
In vivo transcriptional activation mediated through the −93/−84 binding site correlates with Sp1 expression. Five micrograms of either the −124 promoter-reporter construct (WT) or the −124 XN2mt construct (mut) were cotransfected into SL2 cells in triplicate along with the indicated amounts of either the pPacSp1 expression plasmid or the empty pPacU expression vector. The pCMV βgal plasmid was also cotransfected to normalize for transfection efficiency. Relative reporter activity was calculated as in Fig 4, with the activity of the pGL2:SV40 promoter plasmid assigned a value of 1; error bars indicate ± SE.
In vivo transcriptional activation mediated through the −93/−84 binding site correlates with Sp1 expression. Five micrograms of either the −124 promoter-reporter construct (WT) or the −124 XN2mt construct (mut) were cotransfected into SL2 cells in triplicate along with the indicated amounts of either the pPacSp1 expression plasmid or the empty pPacU expression vector. The pCMV βgal plasmid was also cotransfected to normalize for transfection efficiency. Relative reporter activity was calculated as in Fig 4, with the activity of the pGL2:SV40 promoter plasmid assigned a value of 1; error bars indicate ± SE.
DISCUSSION
In this study, two major transcriptional initiation sites for the human c-kit gene were identified using the RACE protocol and RNase protection assays, and these sites were shown to be common to both hematopoietic and nonhematopoietic cell lines. The human c-kittranscriptional initiation site identified in this study as base +1 (Fig 2) is homologous to the mouse c-kit transcriptional start site33 and with one of the major start sites previously identified in the human promoter.35 In addition, Yamamoto et al35 identified a transcriptional initiation site 4 bases upstream of this site. We also identified multiple transcriptional initiation sites, but in addition to the site at +1, we localized the others to +3 as well as a minor site at +7 (Fig 3). Yamamoto et al35 used an M13mp18 sequencing ladder to determine the size of the primer-extended and S1 nuclease-protected bands. We believe the RACE method used in this study, which allows for direct sequencing of the primer-extended species, gives a more accurate localization of the transcription initiation sites. This accurate localization is a necessary prerequisite for the identification of elements that regulate transcriptional initiation.
Initiator elements are known to localize transcription initiation sites and mediate the action of some upstream activators in TATA-less promoters.43 The human c-kit promoter has sequences centered around bases +1 and +3 that are very similar to the initiator consensus sequence (Py Py A+1 N T/A Py Py).43Only one difference from the Inr consensus sequence is observed for transcription starting at base +1, substituting the guanine for adenine at the initiation site. For transcription starting at base +3, the pyrimidines at both ends of the consensus Inr sequence are replaced by purines. Although substituting a guanine for adenine at base +1 substantially weakens the Inr element,43 having two overlapping Inr sequences may compensate and allow transcriptional initiation at either the +1 or +3 position.
As mentioned earlier, there have been several studies performed on both the mouse and human c-kit promoter. The initial study of the mouse promoter found that as little as 105 bp upstream of the TIS was enough to show strong promoter activity in cell lines that express c-kit but not in nonexpressing cell lines.33 In a similar study of the human c-kit promoter, sequences 120 bp upstream of the TIS were shown to activate the c-kit promoter in a cell type–specific manner.35 In the study of the mouse promoter,33 the HL-60 cell line was used as the nonexpressing cell line. We also initially used HL-60 cells but found transfection efficiencies to be very low, making expression studies from any reporter plasmid difficult to interpret. Therefore, we switched to the H146 and Jurkat cell lines, which, in our hands, had much higher transfection efficiencies, and we found no cell type–specific elements in the proximal promoter. We suggest that some of the differences between our data and the previous studies may be caused by the use of c-kit nonexpressing cells with low transfection efficiencies.
In agreement with the studies mentioned above, our analysis of the human c-kit promoter has shown that a construct containing 124 bp of promoter sequence was able to maximally activate the c-kit promoter, albeit not in a tissue-specific manner. This finding agrees with Vandenbark et al,36 who concluded that the DNA region to −183 (from the translational initiation codon; equivalent to −125 in our numbering scheme) was important for c-kit promoter activation, but did not confer cell type–specific transcription. Their data suggested that a distal negative regulatory DNA segment between −4100 to −5500 was the main, though not the only, cis-acting DNA region controlling cell type–specific transcription. They also showed that Myb binding to a consensus site at approximately −1300 had a negative regulatory effect. This is in contrast to a recent study by Ratajczak et al,38 who showed that Myb and Ets-2 binding to consensus sites located between −179 and −471 cooperatively enhanced transcription. However, in the latter study overexpression of Myb and Ets-2 was required to observe these effects; deletion of all Myb and Ets consensus sequences had no significant effect on expression in the presence of endogenous levels of these two transcription factors. Thus, as has been previously shown,36 the tissue-specific regulation of c-kit is likely to be complex. We believe that the difficulty in reconciling all the published data indicates that regions of DNA containing major tissue-specific regulators of transcription have yet to be identified. Recently, two studies of theWsh mutation found an inversion disrupting tissue-specific positive regulatory elements controlling c-kitexpression,45 46 suggesting that potential c-kitupstream regulatory elements may lie near the breakpoint, located between the PDGFRα and c-kit loci, within 100 to 200 kb 5′ of c-kit. Our study did not show any cell type–specific elements within the proximal 3 kb of promoter sequence.
All proximal promoter-reporter studies performed thus far agree that deletion of the region between −124 and −83 from the TIS results in a drop in promoter function to basal levels in c-kit–expressing cells.33,35,36,38 This crucial region contains the −93/−84 Sp1 binding site we have characterized, which is completely conserved in the mouse promoter.33,37 Transfection studies of promoter-reporter plasmids containing a mutation which eliminated in vitro Sp1 binding resulted in a drop in transcriptional activity to basal levels, showing that the −93/−84 site is largely responsible for the activity of the whole −124/−83 fragment. However, overexpression studies in SL2 cells did demonstrate that Sp1 was still able to weakly transactivate in spite of mutation of the −93/−84 site, indicating that one of the upstream consensus sites may have some functional activity. In fact, gel-shift studies (Fig 5) have suggested weak in vitro binding of Sp1 to the XN oligonucleotide, which contains a perfect 6/6 Sp1 core consensus sequence (GGGCGG; −122/−117) that other investigators have identified as the likely site of Sp1 binding within the −124/−83 fragment based on computer homology algorithms.33 35 Mutation of this site in promoter-reporter constructs resulted in only a 20% to 25% decrease in transcriptional activity (not shown), consistent with our conclusion that the vast majority of Sp1-mediated transcriptional activation occurs as a result of binding to the −93/−84 site.
There are many examples where Sp1 plays an essential role in core promoter activity.47,48 In addition, in the human adenosine deaminase promoter, Sp1 not only controls basal level activation but is also involved in enhancer-mediated activation.49Furthermore, in some promoters Sp1 can discriminate between different consensus Sp1 binding sites,50 indicating that certain Sp1 binding sites contribute more to the overall activity than other sites. An example of a promoter that relies predominantly on only one of multiple potential Sp1 sites, in addition to the c-kitpromoter, is the promoter of the transforming growth factor β type I receptor51 (TGF-βRI). Deletion and site-specific mutation analyses explored the importance of multiple Sp1 sites throughout the TGF-βRI promoter and established that one downstream site at position −63 to −54 contributes heavily to basal promoter activity. In the nicotinamide adenine dincucleotide phosphate cytochrome P-450 oxidoreductase gene, promoter deletion studies indicated that loss of the seven distal GC boxes had minimal effect on transcriptional activity, but deletion of the two proximal Sp1 sites resulted in 90% loss of promoter activity.48 In addition, some cell type–specific promoters require cooperative activity of Sp1 with cell type–specific transcription factors, including Egr-1 for thrombospondin,52 AP2 for the acetylcholine receptor α3 gene,53 and GATA 1 for the γ-globin gene.54 Although a potential AP2 site exists on the −124 to −83 fragment, no evidence for binding of factors other than Sp1 was found by either gel-shift or DNase footprint analysis.
In conclusion, we have localized the major transcriptional initiation sites of the human c-kit gene to two bases 58 and 56 bases upstream of the translational initiation codon. Two overlapping weak Inr consensus sites may mediate transcriptional initiation at these specific sites. All the c-kit promoter studies to date33,35,36 38 agree that maximal promoter activity is contained within approximately 125 bp of the transcriptional initiation sites, and that deletion of the region between bases −124 and −83 results in a marked loss in activity. This region contains several possible binding sites for the transcription factor Sp1, but Sp1 seems to bind only to the −93/−84 site with high affinity, and mutation of this site results in a drop in promoter activity to basal levels. Taken together, the data presented here and previously published suggest that selective Sp1 binding to the −93/−84 site must cooperate with presumed upstream factors to control c-kit promoter activation in a cell type–specific manner. Characterization of the binding sites and identification of these upstream factors should be a subject for future investigation.
ACKNOWLEDGMENT
We thank Barbara Armstrong and Julie Litz for excellent technical assistance during the course of this study, Dr Robert Tjian for donating the pPacU and pPacSp1 plasmids, and Dr Richard Moran for donating the SL2 cells.
Supported by a Merit Review Award from the Department of Veterans Affairs.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Geoffrey W. Krystal, MD, PhD, Room 5A-128, McGuire VA Medical Center, 1201 Broad Rock Blvd, Richmond, VA 23249.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal