Abstract
Erythrocyte pyruvate kinase deficiency is the most common cause of hereditary nonspherocytic hemolytic anemia. We present 6 previously undescribed mutations of the PKLR gene associated with enzyme deficiency located at cDNA nt 476 G→T (159Gly→Val), 884 C→T (295Ala→Val), 943 G→A (315Glu→Lys), 1022 G→A (341Gly→Asp), 1511 G→T (504Arg→Leu), and 1528 C→T (510Arg→Ter). Two of these mutations are near the substrate binding site: the 315Glu→Lys (943A) mutation may be involved in Mg2+ binding and159Gly→Val (476T) mutation has a possible effect on ADP binding. Four of six mutations produce deduced changes in the shape of the molecule. Two of these mutations,504Arg→Leu (1511T) and510Arg→Ter (1528T), are located at the interface of domains A and C. One of them (510Arg→Ter) is a deletion of the C-terminal residues affecting the integrity of the protein. The 504Arg→Leu mutation eliminates a stabilizing interaction between domains A and C. Changes in amino acid 341(nt 1022) from Gly to Asp cause local perturbations. The mutation295Ala→Val (884T) might affect the way pyruvate kinase interacts with other molecules. We review previously described mutations and conclude that there is not yet sufficient data to allow us to draw conclusions regarding genotype/phenotype relationship.
PYRUVATE KINASE (PK) is the glycolytic enzyme that catalyzes the formation of Mg-ATP by transfer of a phosphate group from phosphoenol pyruvate to Mg-ADP in the presence of potassium:
Four PK isoenzymes are present in mammalian tissues: M1 (in skeletal muscle), M2 (in kidney, adipose tissue, and lungs), L (in liver), and R (in red blood cells). Each of these isoenzymes have different kinetic properties. The M1 isoenzyme has predominantly hyperbolic Michaelis-Menten kinetics and isoenzymes M2, L, and R are all allosterically regulated. The four different types of PK enzyme are products of only two PK genes in humans and rats (PKLR andPKM). The PKM1 and PKM2 enzymes are formed from the PKM gene by alternative splicing. PKL and PKR are products of the other pyruvate kinase gene (PKLR), transcribed by two different, tissue-specific promoters. In the case of the liver enzyme (PKL), exon 2 but not exon 1 is present in the processed transcript; in the red blood cell enzyme (PKR), exon 1, but not 2, is represented.1 Because the PKLR gene is expressed in liver and in erythrocytes, whereas the PKM gene is expressed in muscle, it is mutations in the PKLR gene that lead to erythrocyte pyruvate kinase deficiency and hemolytic anemia.
PATIENTS AND METHODS
Seven unrelated subjects were studied. Five had hemolytic anemia, whereas the other two (no. 2 and 6) had low PK activity, as documented by the assay of the erythrocyte enzyme. Subject no. 2 is the husband of a woman with pyruvate kinase deficiency hemolytic anemia, and his red blood cells were found to have low PK activity in the course of genetic counseling. Subject no. 6 is the mother of a child who is transfusion dependent. The mutation found in this subject, 884T, is known to lower PK activity, but, because this couple have no children, it is not certain whether it is capable of causing hemolytic anemia. Clinical data, where available, are presented under the Results.
Genomic DNA was isolated from the peripheral blood using an Applied Biosystems DNA extracter (Foster City, CA). The oligonucleotides used for amplification of exons are shown in Table 1. The polymerase chain reaction was performed using the following conditions: denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds, and extension at 72°C for 30 seconds. Amplified DNA samples were sequenced using the 375A sequencer model (Applied Biosystems, Perkin Elmer, Foster City, CA). All new mutations were confirmed by restriction endonuclease digestion (Table 2).
Residual Enzyme Activity and Clinical Manifestations of PK-Deficient Patients
| Subject . | Genotype . | PK Vmax (IU/g Hb) . | Clinical Data . |
|---|---|---|---|
| 1 | 476T/1529A (G159V/R510Q) | 5.15 | Hemolytic anemia |
| 2 | 884T/+ (A295V) | 10.32 | Heterozygote |
| 3 | 943A/1456T (E315K/R486W) | 2.71 | Hemolytic anemia, neonatal jaundice |
| 4 | 1022A/993A (G341D/D331Q) | 8.9 | Hydrops Fatalis |
| 5 | 1022A/? (G341D) Older Younger | 2.3 3.5 | Severe hemolylic anemia, neonatal jaundice (older sister) |
| 6-151 | 1511T/1511T (R504L) | 9.96 | Severe hemolytic anemia |
| 7 | 1528T/+ (R510Ter) | 6.05 | The mother of a child with severe hemolytic anemia |
| Normal values | 11.1-18.9 | ||
| Subject . | Genotype . | PK Vmax (IU/g Hb) . | Clinical Data . |
|---|---|---|---|
| 1 | 476T/1529A (G159V/R510Q) | 5.15 | Hemolytic anemia |
| 2 | 884T/+ (A295V) | 10.32 | Heterozygote |
| 3 | 943A/1456T (E315K/R486W) | 2.71 | Hemolytic anemia, neonatal jaundice |
| 4 | 1022A/993A (G341D/D331Q) | 8.9 | Hydrops Fatalis |
| 5 | 1022A/? (G341D) Older Younger | 2.3 3.5 | Severe hemolylic anemia, neonatal jaundice (older sister) |
| 6-151 | 1511T/1511T (R504L) | 9.96 | Severe hemolytic anemia |
| 7 | 1528T/+ (R510Ter) | 6.05 | The mother of a child with severe hemolytic anemia |
| Normal values | 11.1-18.9 | ||
Abbreviation: IU, International units.
Transfused monthly.
Previously Undescribed Pyruvate Kinase Mutations
| Patient No. . | cDNA Nucleotide . | Genomic Nucleotide* . | Exon . | Base Substitution . | Amino Acid No. . | Amino Acid Substitution . | Detection Method . | Other Allele . |
|---|---|---|---|---|---|---|---|---|
| 1 | 476 | 2757 | 4 | G → T | 159 | Gly → Val | +Kpn I | 1529A |
| 2 | 884 | 3660 | 6 | C → T | 295 | Ala → Val | −Tse I | Normal |
| 3 | 943 | 3719 | 6 | G → A | 315 | Glu → Lys | +MseI | 1456T |
| 4 | 1022 | 3894 | 7 | G → A | 341 | Gly → Asp | −SfaNI | 993A |
| 5 | 1022 | 3894 | 7 | G → A | 341 | Gly → Asp | −SfaNI | ? |
| 6 | 1511 | 6360 | 10 | G → T | 504 | Arg → Leu | −Aci I | 1511T |
| 7 | 1528 | 6377 | 10 | C → T | 510 | Arg → Ter | +Dde I | Normal |
| Patient No. . | cDNA Nucleotide . | Genomic Nucleotide* . | Exon . | Base Substitution . | Amino Acid No. . | Amino Acid Substitution . | Detection Method . | Other Allele . |
|---|---|---|---|---|---|---|---|---|
| 1 | 476 | 2757 | 4 | G → T | 159 | Gly → Val | +Kpn I | 1529A |
| 2 | 884 | 3660 | 6 | C → T | 295 | Ala → Val | −Tse I | Normal |
| 3 | 943 | 3719 | 6 | G → A | 315 | Glu → Lys | +MseI | 1456T |
| 4 | 1022 | 3894 | 7 | G → A | 341 | Gly → Asp | −SfaNI | 993A |
| 5 | 1022 | 3894 | 7 | G → A | 341 | Gly → Asp | −SfaNI | ? |
| 6 | 1511 | 6360 | 10 | G → T | 504 | Arg → Leu | −Aci I | 1511T |
| 7 | 1528 | 6377 | 10 | C → T | 510 | Arg → Ter | +Dde I | Normal |
*cDNA and genomic nucleotide are numbered from the initiator ATG of the PK-R gene.
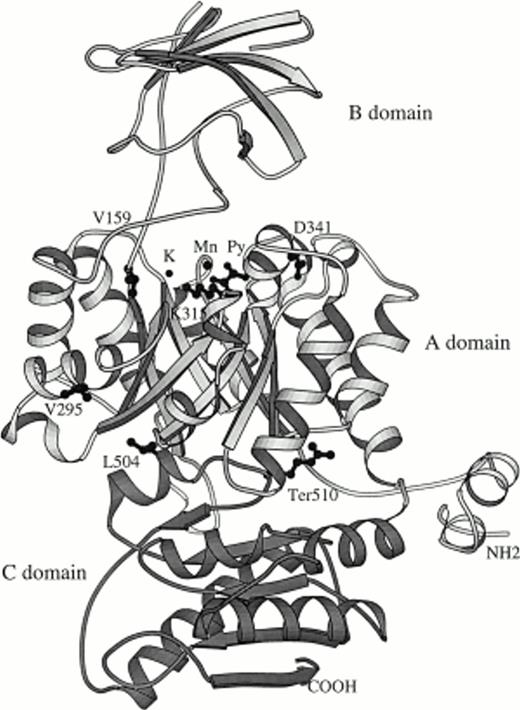
The scientific visualization package Insight (Biosym Inc, San Diego, CA) was used to view the molecule on the Silicon Graphics workstation and also to study possible interactions inside and outside of the PK molecule. Figure 1 was produced by using the Molscript software package.2 New human mutations were placed at the corresponding positions in rabbit muscle PK by using the computer graphics program of Jones et al.3
RESULTS
The residual enzyme activity and clinical findings are summarized in Table 1 and previously undescribed PK mutations are shown in Table 2. Primers used for amplification of the new mutation shown in Table 3. The location of all 6 new human mutations are shown on the structure of rabbit muscle pyruvate kinase in Fig 1.
Primers Used for Amplification of the New Mutations
| cDNA nt . | Sense Primer . | Antisense Primer . | Detection Method . |
|---|---|---|---|
| 476 | 5′-cgaggtcctggccaccttcc-3′ | 5′-gcgccgcctttccggccctg-3′ | +Kpn I |
| 884 | 5′-caactgtgccccgtcctca-3′ | 5′-gtgatggggaatagcgacag-3′ | −Tse I |
| 943 | 5′-caactgtgccccgtcctca-3′ | 5′-gtgatggggaatagcgacag-3′ | +Mse I |
| 1022 | 5′-cacctttcttctcctgcctg-3′ | 5′-caggtgtccctaaaacccac-3′ | −SfaNI |
| 1511 | 5′-ctcgttcaccactttcttgc-3′ | 5′-gggaagctgggttggggggc-3′ | −Aci I |
| 1528 | 5′-ctcgttcaccactttcttgc-3′ | 5′-gggaagctgggttggggggc-3′ | +Dde I |
| cDNA nt . | Sense Primer . | Antisense Primer . | Detection Method . |
|---|---|---|---|
| 476 | 5′-cgaggtcctggccaccttcc-3′ | 5′-gcgccgcctttccggccctg-3′ | +Kpn I |
| 884 | 5′-caactgtgccccgtcctca-3′ | 5′-gtgatggggaatagcgacag-3′ | −Tse I |
| 943 | 5′-caactgtgccccgtcctca-3′ | 5′-gtgatggggaatagcgacag-3′ | +Mse I |
| 1022 | 5′-cacctttcttctcctgcctg-3′ | 5′-caggtgtccctaaaacccac-3′ | −SfaNI |
| 1511 | 5′-ctcgttcaccactttcttgc-3′ | 5′-gggaagctgggttggggggc-3′ | −Aci I |
| 1528 | 5′-ctcgttcaccactttcttgc-3′ | 5′-gggaagctgggttggggggc-3′ | +Dde I |
Ribbon representation of the rabbit pyruvate kinase structure. The shown residue corresponds to human mutations observed in patients. Py is the catalytic site that binds pyruvate. The letters K and Mg denote the position of potassium and magnesium ions, respectively.
Ribbon representation of the rabbit pyruvate kinase structure. The shown residue corresponds to human mutations observed in patients. Py is the catalytic site that binds pyruvate. The letters K and Mg denote the position of potassium and magnesium ions, respectively.
Subject no. 1. 476T mutation (159Gly→Val, corresponding to rabbit muscle 115Gly).
The patient is a 4-year-old Caucasian female with hemolytic anemia. The mutation at nt 476 represents a G→T transition and159Gly→Val amino acid substitution. This mutation was confirmed by showing that it produced a Kpn I restriction site. From the structural point of view, such a change could affect the ADP binding site. ADP and ATP binding sites are glycine-rich. The small size of the glycine residues enables the ADP and ATP to approach the peptide at a range sufficiently short for strong interactions. The Gly→Val mutation can weaken or eliminate ADP binding, which results in reduction or elimination of the enzyme activity. The second mutation in this patient is 1529A (510Arg→Gln). It is the most common mutation in the European population, producing moderate enzyme deficiency with residual activity of about 10% to 25% of normal and no obvious change in kinetic properties.4 The amino acid changed corresponds to rabbit muscle amino acid 466. The mutation is located in the C domain, but interacts with the A domain carbonyl of Val 395 (human). A possible effect of this mutation can be a change in disposition of domain C with respect to domain A.
Subject no. 2. 884T mutation (295Ala→Val corresponding to rabbit muscle 251His).
This subject did not have hemolytic anemia. The mutation was found fortuitously in the course of genetic counseling. The patient has only 1 defective allele and his other allele is normal. The mutation in this patient was found to be at cDNA nucleotide 884 with a G→T substitution (295Ala→Val). This mutation destroys a Tse I restriction site. Because the red blood cell enzyme activity was reduced, the mutation must affect either the catalytic activity or stability of enzyme. In the crystal structure, the side chain of this residue projects externally from domain A, and it appears unlikely for this mutation to have a significant effect on the structure. However, the location of the Val at this position may have an effect on the interaction of pyruvate kinase with other molecules. Also, because of the surface location of this amino acid, the increase in hydrophobicity caused by Ala→Val change is thermodynamically less favorable for protein folding.
Subject no. 3. 943A mutation (315Glu→Lys corresponding to rabbit muscle 271Glu).
Two siblings with hemolytic anemia were jaundiced in the neonatal period, but neither had required an exchange transfusion. Both have splenomegaly. As expected, the patient and her brother were found to have the same genotype. The mutation was a G→A transition at cDNA nt 943 (315Glu→Lys). This mutation creates a Mse I restriction site. The amino acid at this position is involved in Mg2+ binding, so the change from Glu to Lys may lead to reduction in catalysis. The other allele has a previously described 1456T (486Arg→Trp) mutation. The rabbit muscle enzyme amino acid at this position is 442. This mutation is located in the C domain, but points to a space between A and C domains. Again, this could adversely influence the orientation of domain C relative to domain A.
Subjects no. 4 and 5. 1022A mutation (341Gly→Asp, corresponding to rabbit muscle297Gly).
These 2 patients, unrelated but coming from the same region in Portugal, were found to have the same mutation. One patient, a hydropic infant, was found to have a 1022A/993A genotype. She is 9 months old and requires red blood cell transfusions every 3 weeks. The activity of pyruvate kinase in the blood sample before exchange transfusion was 8.9 IU/g hemoglobin (Hb; normal value, 12.6 IU/g Hb), with a reticulocyte count of 45%. There was no family history of hydrops or of hemolytic anemia. The other patient and her sister with the 1022A mutation carry an as yet unidentified second allele. The older patient had jaundice at 24 hours of life, requiring phototherapy. Her mean Hb values are approximately 8.8 g/dL (range, 8.1 to 9.4 g/dL), with persistent reticulocytosis (mean, 8%). She has no liver or spleen enlargement. At 14 months, during a febrile episode, her Hb level decreased to 6.3 g/dL. The PK activity was 2.3 IU/g Hb (normal value, 9.7 IU/g Hb) and the low substrate activity was 1.3 IU/g Hb (normal value 8.2 IU/g Hb). The sister, who is 3 years younger, had no neonatal jaundice. Her anemia was diagnosed at 7 months. She had two crises, at 7 and at 22 months, when her Hb value reached 5 mg/dL and she was transfused. Her baseline Hb values are approximately 7.5 g/dL (range, 7.2 to 7.9 g/dL) and her reticulocyte counts ranged between 15% and 20%. PK activity was 3.5 IU/g Hb (normal value, 9.7 IU/g Hb) and the low substrate activity of PK is 1.2%. The mutation at nt 1022 represents a G→A transition predicting a341Gly→Asp amino acid substitution. This mutation was confirmed with the SfaNI restriction enzyme. The mutation is located in the A domain on the surface of the molecule.
The previously described 993A (331Asp→Gln) mutation found in the first patient is also in the A domain and has a strong salt bridge interaction with Arg 298 within a distance of 2.73 Å. Amino acid 331 corresponds to amino acid 287 in rabbit muscle pyruvate kinase.
Subject no. 6. 1511T mutation (504Arg→Leu, corresponding to rabbit muscle 460Arg).
This patient is a 3-year-old white female with hereditary nonspherocytic hemolytic anemia. She has hepatosplenomegaly and requires transfusion monthly. This patient was found to be homozygous for a G→T substitution at cDNA nucleotide 1511 (504Arg→Leu). The presence of this mutation was confirmed by the loss of an Aci I restriction site. The mutation is in the C domain, but is located at the interface between the A and C domains. It forms a salt bridge Asp 281 of domain A. The mutation to Leu eliminates this strong interaction that holds domain C to domain A. It is interesting to note that this mutation is located far away from the active binding site, but is clinically associated with severe hemolytic anemia.
Subject no. 7. 1528T mutation (510Arg→Ter, corresponding to rabbit muscle 466Arg).
This sample was obtained from the mother of a child with severe hemolytic anemia. The mutation was found to be a C→T transition at cDNA 1528 nt (510Arg→Ter) and was confirmed by demonstrating the formation of a Dde I restriction site. The predicted result of this mutation is the formation of a truncated enzyme with deletion of 63 residues at the C-terminal. As expected, the other allele of this carrier was normal. Unfortunately, we have been unable to obtain DNA from either the child or the father.
DISCUSSION
A considerable number of different red blood cell enzyme defects result in nonspherocytic hemolytic anemia.5,6 The most common of these defects are glucose-6-phosphate dehydrogenase (G6PD) deficiency and pyruvate kinase deficiency. In the case of each of these enzyme deficiencies, a broad range of clinical severity is observed. The intensity of hemolysis cannot be accurately predicted from the amount of residual enzyme activity, although complete deficiency of G6PD appears to be incompatible with life.7 8
In the case of pyruvate kinase deficiency, the situation is even more complex than in G6PD deficiency. Because the mutation is autosomal, most of the patients are compound heterozygotes. As a result, five different tetramers are potentially present in each cell. Moreover, red blood cell pyruvate kinase has, in addition to the active sites for ADP and PEP, an allosteric site that binds fructose diphosphate. The clinical effects of a mutation are probably also modulated by the extent to which activity of the other pyruvate kinase gene PKMpersists in deficient erythrocytes. The very drastic Gypsy mutation of PK, in which the penultimate exon is deleted resulting in a frameshift, is not uncommonly found in the homozygous state with only moderately severe anemia, but it is not clear how much residual enzyme is present in the red blood cells of patients with this mutation. Although such a truncated enzyme might be quite sensitive to proteolysis and lacks the subunit contact region,9 it might still have some intracellular activity. On the other hand, the recently described variant PK Kowloon10 has lost almost one-half of the polypeptide, and the twin patients who were homozygous for this defect were transfusion dependent. However, it is not clear in the latter case whether the normal mRNA that was found in the peripheral blood may have been derived from transfused reticulocytes.
The clinically most severe manifestation of PK deficiency is hydrops fetalis, a rare manifestation11 12 that was found in one of our patients who did, however, survive. This patient was heterozygous for the 1022A (341Gly→Asp) and the 993A (331Asp→Gln) mutations. The341Gly→Asp mutation and the331Asp→Gln mutation are in the A domain. But other patients with the 943A (315Glu→Lys) mutation in the A domain were not nearly as severely affected.
Pyruvate kinase enzymes are tetramers of identical subunits with about 570 amino acid residues. There is significant homology between the human red blood cell and the rabbit muscle enzyme, with 68% identity in a region containing 469 amino acids. The crystal structure of the human PK has not been solved. However, the coordinates of the cat and rabbit muscle enzyme are available.13,14 Each subunit is composed of N-terminal domain and A, B, and C domains. The A domain is the most highly conserved and domain B is the most variable.13 Three of the six new mutations (884T, 943A, and 1022A) we describe are located in domain A, one in domain B (476T), and two in domain C (1511T and 1528T). The three-dimensional structure of rabbit muscle pyruvate kinase was used to analyze the possible changes in the molecular structure of the mutant PK enzymes. Based on the crystal structure of the rabbit enzyme, we are able to make some deductions regarding the effect of the mutations on the enzyme activity. However, due to the difference in structure between muscle and erythrocyte pyruvate kinase, we were able to analyze only the pyruvate and ADP binding site and not the fructose-1,6-diphosphate allosteric site. The location of the pyruvate binding site and an approximate location of the ADP-binding site were deduced previously.13
The 56 missense point mutations summarized in Table 4change 50 different codons. Sequence alignment of the human PK with cat muscle PK and rabbit muscle PK show that the sequence identity among them is 68%. In this context, it is significant that 45 of the 50 amino acids changed (90%) show identity among the three mammalian PKs. Twelve of the changed amino acids are arginines. Comparison of human amino acid sequences with the new mutations described here and sequences of four other species shows them to be largely conserved at the amino acid 159, 315, and 341 positions (Fig 2). At amino acid 504 and 510, they have the same sequence, but regions around these positions are quite variable. The sequence around the mutation at human red blood cell amino acid 295 has a different sequence than the rabbit muscle PK.
Sites of PK Point Mutations That Cause Enzyme Deficiency
| cDNA nt Substitution . | Amino Acid Substitution . | Domain . | Reference . |
|---|---|---|---|
| 238 T → C | Ser 80 Pro | A | 15 |
| 320 T → C | Met 107 Thr | A | 15 |
| 343 G → C | Ala 115 Pro | A | 16 |
| 359 C → T | Ser 120 Phe | A | 16 |
| 401 T → A | Val 134 Asp | A | 15 |
| 476 G → T | Gly 159 Val | A | This study |
| 464 T → C | Leu 155 Pro | A | 15 |
| 487 C → T | Arg 163 Cys | B | 15 |
| 514 G → C | 3-150Glu 172 Gln | B | 17 |
| 787 G → A | 3-150Gly 263 Arg | B | 4 |
| 787 G → T | 3-150Gly 263 Trp | B | 17 |
| 823 G → C | Gly 275 Arg | A | 15 |
| 823 G → A | Gly 275 Arg | A | 17 |
| 841 G → A | Asp 281 Asn | A | 15 |
| 859 T → G | Phe 287 Val | A | 15 |
| 884 G → T | Ala 295 Val | A | This study |
| 941 T → C | Ile 314 Thr | A | 15 |
| 943 G → A | Glu 315 Lys | A | This study |
| 993 C → A | Asp 331 Glu | A | 15 |
| 994 G → A | Gly 332 Ser | A | 4, 15 |
| 1006 G → T | Ala 336 Ser | A | 4, 15 |
| 1010 G → C | Arg 337 Pro | A | 15 |
| 1010 G → A | Arg 337 Gln | A | 4 |
| 1022 G → A | Gly 341 Asp | A | This study |
| 1022 G → C | Gly 341 Ala | A | 15 |
| 1075 C → T | Arg 359 Cys | A | 15 |
| 1076 G → A | Arg 359 His | A | 15 |
| 1081 A → G | Asn 361 Asp | A | 4, 15 |
| 1091 G → A | Gly 364 Asp | A | 18 |
| 1102 G → T | Val 368 Phe | A | 15 |
| 1127 G → T | Ser 376 Ile | A | 4 |
| 1151 C → T | Thr 384 Met | A | 15 |
| 1168 G → A | Asp 390 Asn | A | 15, 17 |
| 1174 G → A | Ala 392 Thr | A | 4, 15 |
| 1178 A → G | Asn 393 Ser | A | 15, 16 |
| 1179 T → A | Asn 393 Lys | A | 15 |
| 1261 C → A | Gln 421 Lys | A | 15 |
| 1276 C → T | Arg 426 Trp | A | 15 |
| 1277 G → A | Arg 426 Gln | A | 15 |
| 1281 G → T | Glu 427 Asp | A | 4 |
| 1373 G → A | Gly 458 Asp | C | 15 |
| 1376 C → T | 3-150Ala 459 Val | C | 15 |
| 1378 G → A | Val 460 Met | C | 15 |
| 1403 C → T | Ala 468 Val | C | 15 |
| 1454 C → T | 3-150Ser 485 Phe | C | 4 |
| 1456 C → T | Arg 486 Trp | C | 4, 15 |
| 1468 C → T | Arg 490 Trp | C | 15 |
| 1484 C → T | Ala 495 Val | C | 15 |
| 1493 G → A | Arg 498 His | C | 4, 15 |
| 1511 G → T | Arg 504 Leu | C | This study |
| 1529 G → A | Arg 510 Gln | C | 4, 15, 16, 18 |
| 1552 C → A | 3-150Arg 518 Ser | C | 15, 17 |
| 1594 C → T | Arg 532 Trp | C | 4, 15 |
| 1654 G → A | Val 552 Met | C | 15 |
| 1675 C → G | Arg 559 Gly | C | 15 |
| 1698 C → A | Asn 566 Lys | C | 15 |
| cDNA nt Substitution . | Amino Acid Substitution . | Domain . | Reference . |
|---|---|---|---|
| 238 T → C | Ser 80 Pro | A | 15 |
| 320 T → C | Met 107 Thr | A | 15 |
| 343 G → C | Ala 115 Pro | A | 16 |
| 359 C → T | Ser 120 Phe | A | 16 |
| 401 T → A | Val 134 Asp | A | 15 |
| 476 G → T | Gly 159 Val | A | This study |
| 464 T → C | Leu 155 Pro | A | 15 |
| 487 C → T | Arg 163 Cys | B | 15 |
| 514 G → C | 3-150Glu 172 Gln | B | 17 |
| 787 G → A | 3-150Gly 263 Arg | B | 4 |
| 787 G → T | 3-150Gly 263 Trp | B | 17 |
| 823 G → C | Gly 275 Arg | A | 15 |
| 823 G → A | Gly 275 Arg | A | 17 |
| 841 G → A | Asp 281 Asn | A | 15 |
| 859 T → G | Phe 287 Val | A | 15 |
| 884 G → T | Ala 295 Val | A | This study |
| 941 T → C | Ile 314 Thr | A | 15 |
| 943 G → A | Glu 315 Lys | A | This study |
| 993 C → A | Asp 331 Glu | A | 15 |
| 994 G → A | Gly 332 Ser | A | 4, 15 |
| 1006 G → T | Ala 336 Ser | A | 4, 15 |
| 1010 G → C | Arg 337 Pro | A | 15 |
| 1010 G → A | Arg 337 Gln | A | 4 |
| 1022 G → A | Gly 341 Asp | A | This study |
| 1022 G → C | Gly 341 Ala | A | 15 |
| 1075 C → T | Arg 359 Cys | A | 15 |
| 1076 G → A | Arg 359 His | A | 15 |
| 1081 A → G | Asn 361 Asp | A | 4, 15 |
| 1091 G → A | Gly 364 Asp | A | 18 |
| 1102 G → T | Val 368 Phe | A | 15 |
| 1127 G → T | Ser 376 Ile | A | 4 |
| 1151 C → T | Thr 384 Met | A | 15 |
| 1168 G → A | Asp 390 Asn | A | 15, 17 |
| 1174 G → A | Ala 392 Thr | A | 4, 15 |
| 1178 A → G | Asn 393 Ser | A | 15, 16 |
| 1179 T → A | Asn 393 Lys | A | 15 |
| 1261 C → A | Gln 421 Lys | A | 15 |
| 1276 C → T | Arg 426 Trp | A | 15 |
| 1277 G → A | Arg 426 Gln | A | 15 |
| 1281 G → T | Glu 427 Asp | A | 4 |
| 1373 G → A | Gly 458 Asp | C | 15 |
| 1376 C → T | 3-150Ala 459 Val | C | 15 |
| 1378 G → A | Val 460 Met | C | 15 |
| 1403 C → T | Ala 468 Val | C | 15 |
| 1454 C → T | 3-150Ser 485 Phe | C | 4 |
| 1456 C → T | Arg 486 Trp | C | 4, 15 |
| 1468 C → T | Arg 490 Trp | C | 15 |
| 1484 C → T | Ala 495 Val | C | 15 |
| 1493 G → A | Arg 498 His | C | 4, 15 |
| 1511 G → T | Arg 504 Leu | C | This study |
| 1529 G → A | Arg 510 Gln | C | 4, 15, 16, 18 |
| 1552 C → A | 3-150Arg 518 Ser | C | 15, 17 |
| 1594 C → T | Arg 532 Trp | C | 4, 15 |
| 1654 G → A | Val 552 Met | C | 15 |
| 1675 C → G | Arg 559 Gly | C | 15 |
| 1698 C → A | Asn 566 Lys | C | 15 |
Nucleotides (and amino acids) are numbered from the upstream ATG (and methionine). The numbering from Rouger et al16 has been adjusted to conform to this convention.
Positions that are not conserved in cat and rabbit PKs.
Comparison of amino acid sequences in 4 different species at the locations of 6 new mutations.
Comparison of amino acid sequences in 4 different species at the locations of 6 new mutations.
Knowing the locations of PK mutations together with the analysis of the crystal structure may help us in the future to predict more specifically the clinical manifestation of the pyruvate kinase disease. Table 5 summarizes the as yet scanty information relating the clinical phenotype to the mutations that have been identified. At present, although the number of cases in which the mutations are known has expanded rapidly, the number of cases in which clinical manifestations are also described is still quite limited. The fact that most patients are compound heterozygotes for two differentPKLR mutations poses further difficulties in interpretation of the effects of specific mutations. Accumulation of further data regarding PK mutations and clinical phenotypes may allow a clearer picture to emerge.
The Ethnic Origin, Location of Mutations, and Clinical Manifestation of Patients Previously Reported and From This Study
| Ethnic Origin . | Genotype (amino acid substitution) . | Domains . | Clinical Information . | Reference . |
|---|---|---|---|---|
| US white | 1022G → C/1456C → T (G341A/R486W) | A/C | Transfused | 9 |
| Irish | 823G → C/1484C → T (G275R/A495V) | A/C | Splenectomy | 19 |
| Irish | 391-393del/1529G → A (1131del/R510Q) | A/C | Splenectomy | 19 |
| Hispanic | 1501C → T/1529G → A (Q501Ter/R510Q) | A/C | Transfused | 19 |
| English | 1127G → T/1456C → T (S376I/R486W) | A/C | Splenectomy | 4 |
| Variable | 1529G → A/1529G → A (R510Q) | C/C | Was variable | 4 |
| Unknown | 721G → T/1529G → A (E241Ter/R510Q) | B/C | Transfused | 19 |
| Irish | 391-393del/603G → A (I131del/W201Ter) | A/B | Splenectomy | 19 |
| US white | 476G → T/1529G → A (G159V/R510Q) | B/C | Hemolytic anemia | This study |
| Indian | 884G → T/+ (A295V/+) | A/+ | Heterozygote | This study |
| Indian | 943G → A/1456C → T (E315K/R486W) | A/C | Hemolytic anemia, neonatal jaundice | This study |
| Portuguese | 1022G → A/993A (G341D/D331Q) | A/A | Hydrops | This study |
| Portuguese | 1022G → A/? (G341D/?) | A/? | Severe hemolytic anemia | This study |
| Brazilian white | 1511G → T/1511G → T (R504L) | C/C | Severe hemolytic anemia | This study |
| Romanian/Puerto Rican | 1528T/+ (R510Ter/+) | C/+ | Mother of a child with severe hemolytic anemia | This study |
| Ethnic Origin . | Genotype (amino acid substitution) . | Domains . | Clinical Information . | Reference . |
|---|---|---|---|---|
| US white | 1022G → C/1456C → T (G341A/R486W) | A/C | Transfused | 9 |
| Irish | 823G → C/1484C → T (G275R/A495V) | A/C | Splenectomy | 19 |
| Irish | 391-393del/1529G → A (1131del/R510Q) | A/C | Splenectomy | 19 |
| Hispanic | 1501C → T/1529G → A (Q501Ter/R510Q) | A/C | Transfused | 19 |
| English | 1127G → T/1456C → T (S376I/R486W) | A/C | Splenectomy | 4 |
| Variable | 1529G → A/1529G → A (R510Q) | C/C | Was variable | 4 |
| Unknown | 721G → T/1529G → A (E241Ter/R510Q) | B/C | Transfused | 19 |
| Irish | 391-393del/603G → A (I131del/W201Ter) | A/B | Splenectomy | 19 |
| US white | 476G → T/1529G → A (G159V/R510Q) | B/C | Hemolytic anemia | This study |
| Indian | 884G → T/+ (A295V/+) | A/+ | Heterozygote | This study |
| Indian | 943G → A/1456C → T (E315K/R486W) | A/C | Hemolytic anemia, neonatal jaundice | This study |
| Portuguese | 1022G → A/993A (G341D/D331Q) | A/A | Hydrops | This study |
| Portuguese | 1022G → A/? (G341D/?) | A/? | Severe hemolytic anemia | This study |
| Brazilian white | 1511G → T/1511G → T (R504L) | C/C | Severe hemolytic anemia | This study |
| Romanian/Puerto Rican | 1528T/+ (R510Ter/+) | C/+ | Mother of a child with severe hemolytic anemia | This study |
Supported by National Institutes of Health Grant No. RR00833 and the Stein Endowment Fund. This is manuscript number 11273-MEM.
Address reprint requests to Ernest Beutler, MD, Department of Molecular and Experimental Medicine, The Scripps Research Institute, 10550 N Torrey Pines Rd, La Jolla, CA 92037.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.
© 1998 by the American Society of Hematology.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal