Abstract
Rhnull disease, which includes the amorph and regulator types, is a rare genetic disorder characterized by stomatocytosis and chronic hemolytic anemia. We studied here a German family transmitting a putative amorph Rhnull disease gene and identified a rare mutation causing the loss-of-function phenotype. We analyzed the genomic and transcript structure of RH30, RH50, andCD47, the three loci thought to be most critical for expression of the Rh complex in the red blood cell membrane. We showed that in this family the Rh50 and CD47 transcripts were normal in primary sequence. However, the RH30 locus contained an unusual double mutation in exon 7 of the RhCe gene, in addition to a deletion of the RhD gene. The mutation targeted two adjacent codons in multiple arrangements probably via the mechanism of microgene conversion. One scheme entails a noncontiguous deletion of two nucleotides, [ATT(Ile322)→AT] and [CAC(His323)→CC], whereas the other involves a T→C transition [ATT(Ile322)→ATC] and a dinucleotide deletion [CAC(His323)→C]. They caused the same shift in open reading frame predicted to encode a shortened protein with 398 amino acids. The loss of two transmembrane domains and gain of a new C-terminal sequence are likely to alter the protein conformation and impair the Rh complex assembly. Our findings establish the molecular identity of an amorph Rhnull disease gene, showing that Rh30 and Rh50 are both essential for the functioning of the Rh structures as a multisubunit complex in the plasma membrane.
Rh DEFICIENCY SYNDROME is a rare genetic disorder of the red blood cell (RBC) transmitted via an autosomal recessive mode generally through consanguineous pedigrees.1Its occurrence is characterized by two phenotypic conditions: the absence of all Rh antigens defines Rhnull disease, whereas an extremely suppressed expression defines the Rhmodphenotype. Both Rhnull and Rhmod patients manifest a mild to moderate hemolytic anemia, and their RBCs show changes in morphology (stomatocytosis) and abnormalities in plasma membranes.2-5 Classic genetic studies suggest that Rhnull disease results from distinct mechanisms, depending on whether the mutations target the Rh antigen locus itself (amorph type) or some unlinked loci that modulate the Rh protein expression (regulator type).1 Biochemical analyses show that Rhnull cells not only lack the carrier antigens but are deficient in several membrane glycoproteins, including Rh50, CD47, LW, and GPB.2-5 This intricate relationship highlights the organization of Rh and its related proteins as a multisubunit complex in the RBC membrane and pinpoints the impaired protein-protein interaction as the hallmark of Rh deficiency syndrome.
Although multiple components are likely involved, the nonglycosylated Rh30 polypeptides (ie, RhD and RhCE) and Rh50 glycoprotein emerge as the most important ones in formation of the Rh complex.3-5Despite the localization of their loci to separate chromosomes, Rh30 and Rh50 belong to the same family and share a notable sequence homology, particularly in their putative α-helices that characterize a 12-transmembrane (TM) topology.6-11 Moreover, evidence is accumulating that Rh30 and Rh50 may interact directly with each other to form a structural core in the RBC membrane.12-14 The other potentially crucial component of the Rh complex may be CD47, an integrin-associated protein,15,16 whose expression is profoundly reduced in Rhnull cells,17,18regardless of the genetic mechanisms involved. The still others, namely LW and GPB, are likely to be dispensable,19 20 because their absence neither disrupts the Rh complex nor alters cell morphology and physiology.
The recent identification of mutations associated with Rh50 gene in regulator Rhnull patients defines the Rh50 glycoprotein as a critical coexpressor of Rh30 polypeptides.21 22Nevertheless, no mutations have been reported to date that target either the RH30 or CD47 locus in Rhnullpatients.
We report here the identification of the first molecular defect that occurs as an amorph Rhnull disease gene in a German family described 25 years ago.23 We show that in this family the genomic structure and transcript expression of RH50 andCD47 are apparently normal. However, the RH30 locus contains a novel noncontiguous deletion of two nucleotides in exon 7 of RhCe gene, in addition to a complete deletion of RhD gene. This novel double mutation has caused frameshift and premature chain termination, thereby leading to a loss-of-function phenotype.
MATERIALS AND METHODS
Blood samples, phenotyping, and immunoblotting.
Control blood samples were from D-positive (genotype, DCe/DCe) and D-negative (genotype, dce/dce) human blood donors. Blood samples under study were from 4 members of the Rhnullfamily, including 2 homozygotes (II-2, B.K., and II-3, D.R.) and 2 heterozygotes (III-1 and III-2; Fig 1). Members I-1 and I-2 are related23 but not available for molecular analysis. Retesting of the patients' RBCs confirmed the absence of all Rh antigens. Immunoblot of RBC membrane proteins was performed as described.24 Two antibodies, LOR-15C9 for RhD25 and 2D10 for Rh50,26 were used. Peroxidase-conjugated antihuman and antimouse Igs were used as the respective secondary antibodies for band visualization.
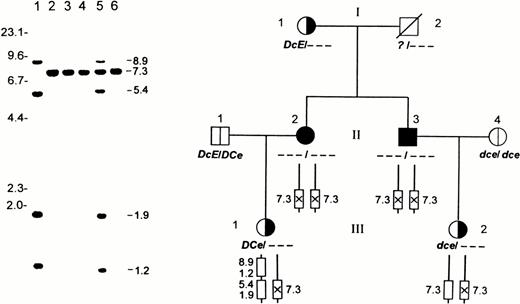
Gross structure of RH30 locus in the amorph Rhnull family as determined by Sph I RFLPs. (Left) Southern blot analysis of 4 members from the Rhnull family. Lane 1, D-positive; lane 2, D-negative; and lanes 3 through 6, family members II-2, II-3, III-1, and III-2, respectively. Genomic DNAs were digested with Sph I and the polymorphic regions spanning exons 4-7 are shown. Note that RhD gene bands (8.9 and 1.2 kb) are missing in II-2, II-3, and III-2, whereas the RH50 locus is grossly normal in all members (gels not shown). Size marker (in kilobases) of λ phage DNA cleaved by HindIII is indicated. (Right) Family pedigree transmitting a putative amorph Rhnull disease gene. I-1, I-2 (deceased), II-1, and II-4 were not available for molecular analyses. II-2 (propositus) and II-3 are Rhnullhomozygotes, whereas their children, III-1 and III-2, are heterozygotes. The genotype of each member was deduced from RFLP analysis and phenotyping. The amorph copy of RhCe gene on RhD deletion background is denoted by a crossed box with a vertical line.
Gross structure of RH30 locus in the amorph Rhnull family as determined by Sph I RFLPs. (Left) Southern blot analysis of 4 members from the Rhnull family. Lane 1, D-positive; lane 2, D-negative; and lanes 3 through 6, family members II-2, II-3, III-1, and III-2, respectively. Genomic DNAs were digested with Sph I and the polymorphic regions spanning exons 4-7 are shown. Note that RhD gene bands (8.9 and 1.2 kb) are missing in II-2, II-3, and III-2, whereas the RH50 locus is grossly normal in all members (gels not shown). Size marker (in kilobases) of λ phage DNA cleaved by HindIII is indicated. (Right) Family pedigree transmitting a putative amorph Rhnull disease gene. I-1, I-2 (deceased), II-1, and II-4 were not available for molecular analyses. II-2 (propositus) and II-3 are Rhnullhomozygotes, whereas their children, III-1 and III-2, are heterozygotes. The genotype of each member was deduced from RFLP analysis and phenotyping. The amorph copy of RhCe gene on RhD deletion background is denoted by a crossed box with a vertical line.
Nucleic acid isolation and Southern blot analysis.
RNA was isolated from hemolysates and genomic DNA from leukocytes, as described.27,28 The Rh cDNA probes were as described29; they span the 5′ (exon 1-3), middle (exon 4-7), and 3′ (exon 8-10 plus 3′-untranslated or UT) regions, respectively. The Rh50 and CD47 cDNA probes were as detailed previously.21 Southern blot was performed as described.28
Reverse transcriptase-polymerase chain reaction (RT-PCR).
The expression of transcripts was analyzed by RT-PCR, as described.21,29 Rh30-specific primers are shown (Table 1), and those for Rh50 and CD47 were detailed in our previous report.21 The mRNA was converted into cDNA using either a gene-specific 3′-UTa21 or anchored (dT)16 oligomer.30 Total RNA (2 μg) and 50 ng of primer were incubated at 65°C for 5 minutes and on ice for 5 minutes. Reverse transcription was incubated at 42°C for 75 minutes and inactivated at 72°C for 10 minutes. The cDNA was then amplified by different pairs of upstream primers spanning the entire coding region using AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, CT). PCR was repeated 35 cycles as follows: 94°C for 1 minute, 55°C for 45 seconds, and 72°C for 1 minute. The last step for chain extension at 72°C was for 7 to 10 minutes.
Primers Specific for Rh30 Genes
| Category . | Sequence (5′-3′)* . | Location . | Position-151 . |
|---|---|---|---|
| (I) Exon assay | |||
| 5′-UTs-152 | ATCGCTCCCTCAAGCCCTCAAGTA | 5′UT | −86-−63 |
| Ex-1a | CTTTTGATCCTCTAAGGAAGCGTCATA | Ex 1 | 99-123 |
| Ex-2s | GGCCAAGATCTGACCGTGATGGCG | Ex 2 | 151-174 |
| Ex-2a | CTGAACAGTGTGATGACCACCTTC | Ex 2 | 312-335 |
| Ex-4s | attgcagACAGACTACCACATGAAC | In 3/Ex 4 | |
| In-4a | tctgagccattctgctcagcccaa | In 4 | |
| In-4s | gacgtgacttccccatctaactct | In 4 | |
| Ex-5a | GCTGATCTTCCT(C)TTGGGGGTGAGC | Ex 5 | 775-798 |
| Ex-6a | ACTTATGTGCACAGTGCGGTGTTGG | Ex 6 | 802-826 |
| In-6a | tgtctagtttcttacCGGCAGG | In 6/Ex 6 | |
| Ex-7s | TGTTGTAACCGAGTGCTGGGGATT | Ex 7 | 943-966 |
| In-7a | agtgcacacgccccagctaagga | In 7 | |
| In-8s | cttgagattaaaaatcctgtgctc | In 8 | |
| Ex-9a | CCAGAAAACTTGGTCATCAAAATATTT | Ex 9 | 1198-1224 |
| Ex-10s | AGTTTCCTCATTTGGCTGTTGGATTT | Ex 10 | 1226-1250 |
| 3′-UTa(D) | GTATTCTACAGTGCATAATAAATGGTG | 3′UT, D | 1432-1458 |
| 3′-UTa(CE) | CTGTCTCTGACCTTGTTTCATTATAC | 3′UT, CE | 1363-1388 |
| (II) Amplification of exon 7 | |||
| In-6s | tttgaggtgagccttagtgcccat | In 6 | |
| Ex-7a | gtgacccacATGCCATTGCCGTTC | In 7/Ex 7 | |
| In-7a | Same as in (I) | ||
| (III) RT-PCR | |||
| 5′-UTs | CCTGCACAGAGACGGACACA | 5′UT | −33-−22 |
| Ex-5a | TGGCCAGAACATCCACAAGAAGAG | Ex 5 | 663-640 |
| Ex-4s | CCAAAATAGGCTGCGAACACGTAGA | Ex 4 | 515-539 |
| Ex-9s | GGTTTGCTCCTAAATCTCAAAATATGG | Ex 9 | 1153-1179 |
| 3′-UTa(D) | Same as in (I) | ||
| 3′-UTa(CE) | Same as in (I) |
| Category . | Sequence (5′-3′)* . | Location . | Position-151 . |
|---|---|---|---|
| (I) Exon assay | |||
| 5′-UTs-152 | ATCGCTCCCTCAAGCCCTCAAGTA | 5′UT | −86-−63 |
| Ex-1a | CTTTTGATCCTCTAAGGAAGCGTCATA | Ex 1 | 99-123 |
| Ex-2s | GGCCAAGATCTGACCGTGATGGCG | Ex 2 | 151-174 |
| Ex-2a | CTGAACAGTGTGATGACCACCTTC | Ex 2 | 312-335 |
| Ex-4s | attgcagACAGACTACCACATGAAC | In 3/Ex 4 | |
| In-4a | tctgagccattctgctcagcccaa | In 4 | |
| In-4s | gacgtgacttccccatctaactct | In 4 | |
| Ex-5a | GCTGATCTTCCT(C)TTGGGGGTGAGC | Ex 5 | 775-798 |
| Ex-6a | ACTTATGTGCACAGTGCGGTGTTGG | Ex 6 | 802-826 |
| In-6a | tgtctagtttcttacCGGCAGG | In 6/Ex 6 | |
| Ex-7s | TGTTGTAACCGAGTGCTGGGGATT | Ex 7 | 943-966 |
| In-7a | agtgcacacgccccagctaagga | In 7 | |
| In-8s | cttgagattaaaaatcctgtgctc | In 8 | |
| Ex-9a | CCAGAAAACTTGGTCATCAAAATATTT | Ex 9 | 1198-1224 |
| Ex-10s | AGTTTCCTCATTTGGCTGTTGGATTT | Ex 10 | 1226-1250 |
| 3′-UTa(D) | GTATTCTACAGTGCATAATAAATGGTG | 3′UT, D | 1432-1458 |
| 3′-UTa(CE) | CTGTCTCTGACCTTGTTTCATTATAC | 3′UT, CE | 1363-1388 |
| (II) Amplification of exon 7 | |||
| In-6s | tttgaggtgagccttagtgcccat | In 6 | |
| Ex-7a | gtgacccacATGCCATTGCCGTTC | In 7/Ex 7 | |
| In-7a | Same as in (I) | ||
| (III) RT-PCR | |||
| 5′-UTs | CCTGCACAGAGACGGACACA | 5′UT | −33-−22 |
| Ex-5a | TGGCCAGAACATCCACAAGAAGAG | Ex 5 | 663-640 |
| Ex-4s | CCAAAATAGGCTGCGAACACGTAGA | Ex 4 | 515-539 |
| Ex-9s | GGTTTGCTCCTAAATCTCAAAATATGG | Ex 9 | 1153-1179 |
| 3′-UTa(D) | Same as in (I) | ||
| 3′-UTa(CE) | Same as in (I) |
*Intronic sequences are denoted by lowercase letters and others by uppercase letters.
Nucleotide positions of exon primers are accounted from the first base of initiation codon ATG.
Sense (s) and antisense (a) primers to 3′-UT, exon (Ex), and intron (In) are denoted.
Amplification and analysis of RhCe amorph gene.
Sequences of Rh30 genes were amplified by PCR using total genomic DNA as a template. Exon sequences were analyzed by diagnostic restriction enzymes to distinguish their D or CEorigin.29 To retrieve unknown intron sequences for primer design (Table 1), genomic libraries generated by 6 restriction enzymes were amplified with Rh30 exon-specific primers, as described.21 Amplification was repeated 30 cycles as follows: 94°C for 1 minute, 60°C for 30 seconds, and 72°C for 30 seconds.
Nucleotide determination and sequence analysis.
Amplified cDNA and genomic products were purified by native 5% polyacrylamide gel electrophoresis (PAGE) and sequenced in both strands on a Model 373A automated sequencer (Applied Biosystems, Foster City, CA). The nucleotide and deduced amino acid sequences were analyzed using the DNASIS program (Hitachi, South San Francisco, CA).
RESULTS
Gross structure of Rh30 polypeptide genes.
Figure 1 shows a representative Sph1 blot of the Rhnull family hybridized with the cDNA probe spanning the polymorphic region of RH30.31 Notably, the homozygotes and one heterozygote each had a single 7.3-kb band retaining exons 4-7 whose intensity was comparable to that of D-negatives. This indicated that the Rhnull patients lacked RhD gene but had two copies of RhCE gene (Fig 1, left). The other heterozygote inherited from her father a DCe haplotype; thus, her D-specific (8.9 and 1.2 kb) and Ce-specific (5.4 and 1.9 kb) bands showed reduced intensity. Analysis of RH50and CD47 showed no difference between controls and Rhnull family members (gels not shown), indicating that all family members had two intact copies of Rh50 and CD47 genes. These results indicate that the amorph gene occurs on background ofRHD deletion and that both copies of RHCE are defective in the Rhnull patients (Fig 1, right).
To determine whether the RhD gene deletion is total or partial, we analyzed 8 of the 10 exons of Rh30 genes by PCR and diagnostic restriction cleavage (Fig 2). Whereas heterozygote III-1 had a pattern apparently identical with that of D-positives, other members lacked D-specific exons 1, 4, 5, 6, 7, 9, and 10. With regard to the cleavage of exons 1 and 2, the two homozygotes contained Ce- or CE-specific exons, whereas the heterozygotes exhibited a composite pattern. Taken together, these data demonstrate a total absence of RhD gene in Rhnullpatients and indicate that some subtle molecular defect(s) have silenced the phenotypic expression of their RhCE gene.
Assay of Rh30 exons in the Rhnull family by PCR and restriction analysis. Individual exons were amplified by PCR and their gene origin was determined by unique restriction site or primer specificity.29 The digested PCR products were separated by native 6% PAGE and stained with ethidium bromide. Enzymes used and exon numbers are indicated above gel panels. Lanes 1 through 6 are as in Fig 1. Below each panel, the size and gene origin of the respective fragments are shown (“+” indicates a cleavage). In the exon 10 panel, the expected RhD (233 bp) and RhCe (163 bp) bands were from coamplification of both gene fragments using a common 5′ primer, Ex-10s, coupled with two 3′-UTa primers (Table 1). Exons 3 and 8 were not analyzed, because the former lacks the unique site and the latter is identical between RhCE and RhD.6-10
Assay of Rh30 exons in the Rhnull family by PCR and restriction analysis. Individual exons were amplified by PCR and their gene origin was determined by unique restriction site or primer specificity.29 The digested PCR products were separated by native 6% PAGE and stained with ethidium bromide. Enzymes used and exon numbers are indicated above gel panels. Lanes 1 through 6 are as in Fig 1. Below each panel, the size and gene origin of the respective fragments are shown (“+” indicates a cleavage). In the exon 10 panel, the expected RhD (233 bp) and RhCe (163 bp) bands were from coamplification of both gene fragments using a common 5′ primer, Ex-10s, coupled with two 3′-UTa primers (Table 1). Exons 3 and 8 were not analyzed, because the former lacks the unique site and the latter is identical between RhCE and RhD.6-10
Identification of the novel mutation in RhCe gene by transcript analysis.
Although being a consanguineous pedigree,23 the inheritance pattern of this Rhnull family (Fig 1) did not rule out the occurrence of possible mutations in related suppressor loci. Therefore, we analyzed and sequenced the Rh50 and CD47 cDNAs from the Rhnull patients. No abnormality was found in the Rh50 (Fig 3A) or CD47 transcript (not shown), conforming to the transmission of an amorph gene.
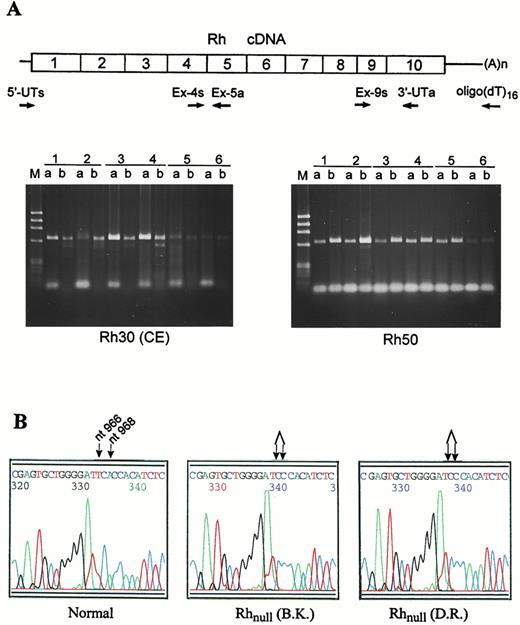
Identification of the novel mutation in RhCe gene by RT-PCR and sequencing. (A) Strategy for synthesis and amplification of Rh30 cDNA. Rh30 mRNA was reverse-transcribed into cDNA with either the gene-specific 3′-UTa primer or (dT)16 oligomer and then amplified with two pairs of upstream primers (Table 1). The cDNA products for Rh30 (left panel) and Rh50 (right panel) were separated on 1.8% agarose gels. Designations 1 through 6 are as in Fig 1. Lane a, 5′-UT/Ex-5a segment; lane b, Ex-4s/3′-UTa segment. Bands at bottom are primer dimers. (B) Nucleotide sequencing profiles for the novel double mutation identified in the amorph form of RhCe cDNAs from the 2 Rhnull patients (B.K. and D.R.). The mutation affects 2 codons (322 and 323) in exon 7, involving 2 single nucleotide deletions, ATT→AT (nt 965 or 966) and CAC→CC (nt 968) (indicated by arrow).
Identification of the novel mutation in RhCe gene by RT-PCR and sequencing. (A) Strategy for synthesis and amplification of Rh30 cDNA. Rh30 mRNA was reverse-transcribed into cDNA with either the gene-specific 3′-UTa primer or (dT)16 oligomer and then amplified with two pairs of upstream primers (Table 1). The cDNA products for Rh30 (left panel) and Rh50 (right panel) were separated on 1.8% agarose gels. Designations 1 through 6 are as in Fig 1. Lane a, 5′-UT/Ex-5a segment; lane b, Ex-4s/3′-UTa segment. Bands at bottom are primer dimers. (B) Nucleotide sequencing profiles for the novel double mutation identified in the amorph form of RhCe cDNAs from the 2 Rhnull patients (B.K. and D.R.). The mutation affects 2 codons (322 and 323) in exon 7, involving 2 single nucleotide deletions, ATT→AT (nt 965 or 966) and CAC→CC (nt 968) (indicated by arrow).
To define the nature of the amorph mutation, the Rh30 cDNA was amplified with different pairs of gene-specific primers. In both Rhnull patients, only RhCE but not RhD cDNA was detected, whose size and amount were comparable to that of controls (Fig 3A). Sequencing showed that the Rh30 cDNA was of Ce type, containing C- and e-specific polymorphisms.6,7 10 However, this cDNA harbored a very rare double mutation in exon 7, which targeted two adjacent codons (322 and 323) and caused a noncontiguous deletion of two nucleotides, 966T and 968A (Fig 3B). Significantly, the mutation resulted in both frameshift and premature termination. Thus, the cDNA was predicted to encode a shortened RhCe-like polypeptide of 398 amino acids (Fig 4A). The premature termination ablated the last 2 TM domains of the protein, whereas the frameshift gave rise to a new C-terminal sequence of 76 amino acids likely facing the cytoplasmic side (Fig 4B). We did not try to establish the status of the RhCe-like protein in the membrane, because no antibody can detect RhCE proteins by immunoblot. However, we showed an absence of RhD but a significant expression of Rh50 in the Rhnullpatients (Fig 4C). In light of a normal structure of Rh50 and CD47, we conclude that the double deletion is the primary defect underlying the amorph Rhnull disease gene in this family.
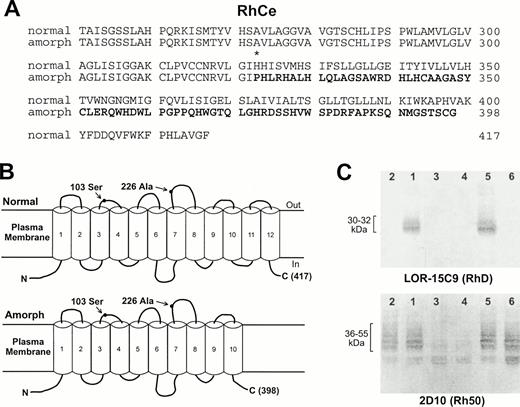
The deduced amino acid sequence and topology of amorph RhCe from the 2 Rhnull patients. (A) Comparison of the sequence between the normal (residues 251-417) and putative amorph RhCe protein (residues 251-398). The frameshift starts from Pro323 (marked by a star) and the new stretch of 76 amino acids terminates at Gly398 (bold). (B) Topology and organization of the putative amorph protein in the membrane. The amorph protein lacks the last 2 TM domains and does not express any Rh antigens, although it contains C and e antigen-specific polymorphisms, Ser103 and Ala226, on the second and fourth exofacial loops. (C) Immunoblot analysis of Rh30(D) and Rh50 proteins in the Rhnull membrane. The upper panel was probed with antibody LOR-15C9 and the lower one with 2D10. Lane designations are as in Fig 1. Note that no RhD protein was detected in D-negative and Rhnull patients, as expected. In contrast, a significant amount of Rh50 protein was present in the Rhnull cells.
The deduced amino acid sequence and topology of amorph RhCe from the 2 Rhnull patients. (A) Comparison of the sequence between the normal (residues 251-417) and putative amorph RhCe protein (residues 251-398). The frameshift starts from Pro323 (marked by a star) and the new stretch of 76 amino acids terminates at Gly398 (bold). (B) Topology and organization of the putative amorph protein in the membrane. The amorph protein lacks the last 2 TM domains and does not express any Rh antigens, although it contains C and e antigen-specific polymorphisms, Ser103 and Ala226, on the second and fourth exofacial loops. (C) Immunoblot analysis of Rh30(D) and Rh50 proteins in the Rhnull membrane. The upper panel was probed with antibody LOR-15C9 and the lower one with 2D10. Lane designations are as in Fig 1. Note that no RhD protein was detected in D-negative and Rhnull patients, as expected. In contrast, a significant amount of Rh50 protein was present in the Rhnull cells.
Inheritance of amorph gene and confirmation of genomic mutation.
The detection of a unique RhCe-like transcript (Fig 3) indicated that the patients are homozygous, whereas their children are heterozygous, for the mutation. As a new BamHI site was created, we performed a diagnostic assay in the 4 members. Whereas no cleavage was seen in controls, a total digestion in the 2 patients and a partial digestion in their children were observed (Fig 5A). This pattern is entirely consistent with the transmission of the amorph gene in an autosomal recessive fashion. Direct sequencing of the exon 7 containing fragment showed the same noncontiguous deletion (Fig 3B), confirming homozygosity of the mutation in the 2 patients.
Family inheritance of the amorph Rhnull gene. (A) The genomic region encompassing exon 7 of RhCE gene was amplified by 2 pairs of primers In-6s/Ex-7a and In-6s/In-7a (Table 1). The In-6s/Ex-7a fragments were cut by BamHI (G↓GATCC), which is diagnostic of the mutation, and separated by native 6% PAGE. Although no cleavage is seen in control lanes, the respective fragments of 264 bp (denoted by a star) were cut completely in the Rhnullpatient and partially in the heterozygotes. (B) Sequencing of the genomic fragments In-6s/In-7a. The 2 patients showed the same mutation, 2 deleted nucleotides (966T and 968A, boxed), in this fragment, which was sequenced on both strands. Intronic nucleotides are denoted by lowercase letters and exonic ones by uppercase letters. Dashes indicate identical nucleotides. The 2 primers for PCR amplification are overlined.
Family inheritance of the amorph Rhnull gene. (A) The genomic region encompassing exon 7 of RhCE gene was amplified by 2 pairs of primers In-6s/Ex-7a and In-6s/In-7a (Table 1). The In-6s/Ex-7a fragments were cut by BamHI (G↓GATCC), which is diagnostic of the mutation, and separated by native 6% PAGE. Although no cleavage is seen in control lanes, the respective fragments of 264 bp (denoted by a star) were cut completely in the Rhnullpatient and partially in the heterozygotes. (B) Sequencing of the genomic fragments In-6s/In-7a. The 2 patients showed the same mutation, 2 deleted nucleotides (966T and 968A, boxed), in this fragment, which was sequenced on both strands. Intronic nucleotides are denoted by lowercase letters and exonic ones by uppercase letters. Dashes indicate identical nucleotides. The 2 primers for PCR amplification are overlined.
DISCUSSION
In this study, we examined a German family in which a putative amorph Rhnull disease gene is transmitted on a consanguineous background.23 Molecular analysis of three genetic loci relating to the expression of the Rh complex, RH30, RH50, andCD47, has led to the detection of a rare double mutation in the Rh30 gene and exclusion of possible candidate mutations in Rh50 and CD47 genes. The mutation is defined as a noncontiguous deletion of 2 nucleotides targeting 2 adjacent codons in exon 7 of the RhCe gene and thus becomes the first known amorph defect at RH30. With a deleted RhD gene positioned in cis and trans, the mutation has silenced the antigen expression of the remaining RhCe gene, causing a loss-of-function phenotype in the two Rhnull patients.
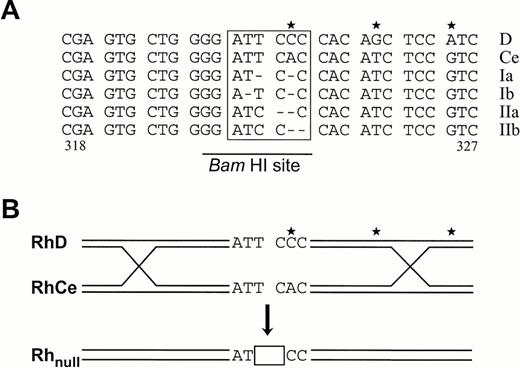
Of the double deletions identified, one hit the second or third position of codon ATT(Ile322) and the other the second position of codon CAC(His323). Based on the alignment of deletion forms, two models may be postulated to account for their origin. In spontaneous model, two hypothetical schemes could be proposed, each evoking two molecular events. In scheme I, the deletion of 966T (or 965T) and 968A is assumed to occur separately (Fig 6A). In scheme II, one event causes a transition [ATT→ATC] and the other a dinucleotide deletion [CAC→C] (Fig 6A). In the microgene conversion model,32the same end product may arise by a single event in which a heteroduplex of RhD and RhCe could be formed via homologous pairing; failure in repair synthesis involving codons 322 and 323 would result in patched transfer of the RhD-specific C residue and a contiguous deletion of TC dinucleotide (Fig 6B). Although microgene conversion might be infrequent in the Rh system,33 it is implicated as an important mechanism for the diversity of Ig and MHC gene families and provides a plausible explanation to the origin of some rare spontaneous mutations.34 35
Two models accounting for origin of the double mutation in the amorph Rhnull disease gene. (A) Spontaneous mutation model. Sequences of codons 318-327 in RhD and RhCe genes are shown on top. Three mismatches at center right are marked by stars. Possible arrangements of the mutated region (boxed) in the amorph gene are depicted in two hypothetical schemes (see Discussion for details). a and b denote alternatives of the same scheme. Scheme I shows a noncontiguous deletion of two nucleotides, whereas scheme II shows a contiguous deletion of 2 nucleotides in association with a T→C transition. The BamHI site is shown. (B) Microgene conversion model. A heteroduplex is formed between RhD and RhCe genes via homologous pairing and strand synapsis. A failure in repair synthesis involving codons 322 and 323 would result in A→C transition and contiguous deletion of 2 nucleotides (boxed). This model is compatible with scheme II shown above but accommodates the latter in a single event.
Two models accounting for origin of the double mutation in the amorph Rhnull disease gene. (A) Spontaneous mutation model. Sequences of codons 318-327 in RhD and RhCe genes are shown on top. Three mismatches at center right are marked by stars. Possible arrangements of the mutated region (boxed) in the amorph gene are depicted in two hypothetical schemes (see Discussion for details). a and b denote alternatives of the same scheme. Scheme I shows a noncontiguous deletion of two nucleotides, whereas scheme II shows a contiguous deletion of 2 nucleotides in association with a T→C transition. The BamHI site is shown. (B) Microgene conversion model. A heteroduplex is formed between RhD and RhCe genes via homologous pairing and strand synapsis. A failure in repair synthesis involving codons 322 and 323 would result in A→C transition and contiguous deletion of 2 nucleotides (boxed). This model is compatible with scheme II shown above but accommodates the latter in a single event.
The amorph mutation has targeted a nonconserved region, possibly the fifth endofacial loop, in the Rh30 polypeptides.6-10 Thus, its result as a loss-of-function phenotype is most likely due to the inevitable shift in open reading frame. The status of the putative RhCe-like protein in the Rhnull cells remains to be established. Nevertheless, several lines of evidence suggest that the mutant protein may be inserted into the membrane but not accessible to the antibodies that are strictly conformation-dependent. (1) The RhCe-like transcript was abundantly expressed, suggesting that the steady-state level of mRNA is not affected by the mutation. (2) Albeit at a reduced level, Rh50 was readily detectable by immunoblot. As a critical interaction partner of Rh30,12-14 this expression implies a possible association of Rh50 with the RhCe-like protein. (3) Prior surface-labeling studies detected a structurally similar Rh30 polypeptide in the Rhnull membrane,36indicating that the absence of Rh antigens does not necessarily mean the absence of Rh proteins. As predicted, the primary sequence of the RhCe-like protein indicates a loss of 2 TM domains and a gain of 76 new amino acids in the C-terminal region. Such structural changes would disturb the conformation of the RhCe-like protein and affect its interaction with Rh50 and other related glycoproteins in the RBC membrane.
Identification of the first amorph Rh30 gene establishes that Rh30, akin to its interacting partner Rh50, is an essential member of the Rh membrane structures. However, the molecular details regarding this protein-protein interaction remain to be elucidated. Limited proteolysis and antibody probing suggested that Rh30 and Rh50 are each composed of two 6-TM domains, forming a tetramer through N-terminal contacts.13 However, additional contact sites are likely present to further dictate the assembly of Rh membrane structures. The amorph protein has a change in the C-terminal portion after Pro323, but its N-terminal sequence is identical with the wild-type one. Its association with the dysfunction of the Rh complex suggests a possible interaction of Rh30 with Rh50 through their C-terminal regions. Notably, the C-termini of Rh50 mutants in regulator Rhnull disease resulting from a splicing donor mutation21 or a point deletion22 are also abnormal because of the inherent frameshift and premature termination. In the former, the only difference from the Rh50 protein lies C-terminal to the 10th TM domain, whereas the latter is altered after the 11th TM domain. Missense mutations targeting the conserved C-terminal TM domains of Rh50 are also associated with regulator Rhnull (our unpublished data). These coincidental observations suggest that the C-terminal regions of Rh30 and Rh50 play a crucial role in forming the multisubunit Rh structures in the RBC membrane.
ACKNOWLEDGMENT
The authors are grateful to Dr A. Blancher for his monoclonal antibody LOR-15C9 against RhD and to Dr A.E.G.Kr. von dem Borne for his monoclonal antibody 2D10 against Rh50. We thank G. Halverson for immunoblot analysis and T. Huima-Byron, Y. Oskov, and R. Ratner for photoprints and computer graphics.
Supported by National Institutes of Health Grant No. HL 54459.
Address reprint requests to Cheng-Han Huang, MD, PhD, Laboratory of Biochemistry and Molecular Genetics, Lindsley F. Kimball Research Institute, New York Blood Center, 310 E 67th St, New York, NY 10021; e-mail: chuang@nybc.org.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
© 1998 by the American Society of Hematology.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal