Abstract
Approximately 40% of Hodgkin's disease (HD) cases in Western countries carry Epstein-Barr virus (EBV) in the malignant Hodgkin-Reed-Sternberg (H-RS) cells. HLA class I–restricted cytotoxic T lymphocytes (CTLs) with specificity for viral antigens expressed in H-RS cells therefore have therapeutic potential. However, a prerequisite for CTL therapy is that the tumor target be capable of processing and presenting endogenously expressed antigens via the transporter associated with antigen processing (TAP)-dependent HLA class I pathway. We have assessed the antigen-presenting phenotype of H-RS cells in two ways. First, immunohistochemical analysis of 38 HD biopsies showed that H-RS cells were uniformly TAP1/TAP2-positive and expressed HLA class I in the majority (18 of 24, 75%) of EBV-positive cases compared with only 4 of 14 (29%) of EBV-negative cases. Second, using a panel of 5 H-RS cell lines, we showed that 4 of 5 could process and present EBV proteins to HLA class I–restricted EBV-specific CTL clones. Others have reported that human interleukin-10 (IL-10), which is expressed by H-RS cells in the majority of EBV-positive HD cases, can abrogate CTL recognition in some circumstances. However, IL-10 pretreatment of the H-RS lines or of the EBV-specific CTLs had no such effect in this system. These results support the possibility that EBV-specific CTLs may be used to treat virus-positive HD.
© 1998 by The American Society of Hematology.
THE ASSOCIATION BETWEEN Epstein-Barr virus (EBV) and Hodgkin's disease (HD) is now well established, with approximately 40% of HD cases in Western countries carrying EBV in the malignant Hodgkin-Reed Sternberg (H-RS) cell population.1In such cases, every malignant cell contains the viral genome, and where patients possess multiple lesions, the same unique viral episome is present at every site.2 The precise role of EBV in the origin of HD is unknown but the oncogenic potential of this virus is clear from its ability to transform resting B cells in vitro into permanently growing lymphoblastoid cell lines (LCLs) and from its association with several human tumors.3
Despite the oncogenic potential of EBV, this virus is widespread in the human population where it usually persists as an asymptomatic infection of B cells under the control of cytotoxic T lymphocytes (CTLs).3 EBV is a potent target for HLA class I–restricted CTLs, and such effectors are readily reactivated in vitro by stimulating peripheral blood mononuclear cells with the autologous EBV-transformed LCL. Within an LCL, EBV establishes a latent infection with the expression of six nuclear antigens (Epstein-Barr nuclear antigens [EBNAs] 1, 2, 3A, 3B, 3C, and -LP) and two latent membrane proteins (LMPs 1 and 2).3 Of these, the EBNA 3 family (EBNAs 3A, 3B, and 3C) represent the immunodominant target antigens for CTLs, although subdominant responses to many of the other EBV latent proteins have been detected.4-7 In clinical situations where such CTL control is ablated (for instance, in heavily immunosuppressed transplant patients), EBV-positive immunoblastic lymphomas arise in which the full spectrum of virus latent proteins are expressed. Importantly, these tumors remain highly susceptible to adoptive transfer of EBV-specific CTLs prepared by in vitro reactivation.8
Within H-RS cells, EBV also establishes a latent infection, but expression of viral proteins is restricted to just EBNA 1, LMP 1, and LMP 2.9-12 This alternative latent state may allow the tumor to evade virus-specific CTL surveillance because the immunodominant EBNA 3 proteins are not expressed. Nevertheless, LMP 1 and LMP 2 are known to be subdominant targets for CTL responses on at least some HLA backgrounds.4-6 Indeed, a number of CTL target epitope sequences restricted through common HLA alleles have been identified in LMP 2 and these sequences are conserved in viruses present within HD tumors.6 Accordingly, there is now considerable interest in the possibility of treating EBV-positive HD cases by boosting LMP-specific CTL responses through immunization or by adoptive transfer of appropriate CTL preparations.
In this context, LMP-specific memory CTLs have been detected in the blood of at least some HD patients,13,14 yet such reactivities have clearly failed to control the disease. The explanation may simply be quantitative in that LMP-specific CTLs almost always comprise a minor component of the EBV-induced CTL response5,7; in such circumstances, boosting these responses should prove an effective therapy for HD. However, there may be other factors which underlie the emergence of EBV-positive HD in a virus-immune individual. First, H-RS cells may be defective in the pathway whereby viral antigens are processed and presented to HLA class I–restricted T cells. In the normal HLA class I processing pathway, endogenously synthesized proteins are first cleaved by the proteasome complex into peptide fragments. These peptides are then delivered into the endoplasmic reticulum via the heterodimeric transporter associated with antigen processing (TAP1/TAP2) and compete for binding to nascent HLA class I molecules; the most stable peptide: HLA class I complexes are then transported to the cell surface for recognition by specific T-cell receptors.15 Defects in this pathway have been identified in several human malignancies including small cell lung carcinoma16 and the EBV-associated tumor, Burkitt's lymphoma (BL).17 In both cases, the defect involves downregulated expression of the TAP transporters with reduced expression of HLA class I on the cell surface. Immunohistochemical studies of HLA class I expression in HD have so far yielded conflicting results. Poppema and Visser18 reported that the majority of H-RS cells lack surface HLA class I expression, whereas Oudejans et al19 found that most EBV-positive H-RS cells are HLA class I-positive.
In addition to antigen-processing defects, it has been postulated that H-RS cells may evade T-cell recognition through the expression of interleukin-10 (IL-10). This cytokine is known to suppress cellular immune responses20-22 and a recent study has shown its expression by H-RS cells in the majority of EBV-positive HD cases.23 Furthermore, there is independent evidence to suggest that local suppression of the CTL response does occur in EBV-positive HD. Thus, by screening both the circulating and tumor-infiltrating T cells from HD patients, Frisan et al13found that EBV-specific CTLs could be isolated from the blood of a virus genome-positive HD patient but not from the biopsy itself. Not only might IL-10 suppress the CTL response, but it has also been reported in model systems to protect target cells from CTL recognition by downregulating surface HLA class I expression.24
In the present study we have sought to address these issues (1) by analyzing TAP1, TAP2, and surface HLA class I molecule expression in HD tumor biopsies, (2) by assaying a panel of H-RS–derived cell lines in vitro for their capacity to present antigen to EBV-specific CTLs, and (3) by determining whether exogenously added IL-10 can act either on the H-RS lines or on the CTL effectors to prevent CTL-mediated lysis of H-RS targets.
MATERIALS AND METHODS
HD biopsy samples and H-RS cell lines.
Lymph node biopsy samples from 38 cases of HD were obtained from Birmingham Heartlands Hospital and Good Hope NHS Trust Hospital (Birmingham, UK). Hematoxylin- and eosin-stained sections from each biopsy were examined and patients classified histologically according to the Rye convention.25 The H-RS cell lines used in this study were kindly provided by Dr J. Wolf and Dr V. Diehl (Cologne, Germany), and Dr H. Kamesaki (Kyoto, Japan) and have all been described previously.26-29 HDLM2, L540, and L428 were derived from advanced cases of nodular sclerosis subtype, whereas KMH2 and L1236 were derived from advanced cases of mixed cellularity subtype; all of the lines are EBV genome negative. H-RS lines were HLA typed by M. Bunce (The Oxford Transplant Centre, Churchill Hospital, Oxford, UK) using polymerase chain reaction–based DNA typing. Cells were cultured in RPMI 1640 (Life Technologies, Paisley, Scotland) containing 10% fetal calf serum, 2 mmol/L glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin (growth medium).
In situ hybridization.
The EBV status of HD biopsies was determined by in situ hybridization using a fluorescein-conjugated oligonucleotide probe specific for the abundantly expressed EBV-encoded EBER RNAs (Vector Laboratories, Inc, Burlingame, CA). Formalin-fixed paraffin-embedded sections were mounted on slides coated with 3-amino-propyl-triethoxy-saline (APES; Sigma-Aldrich Co Ltd, Poole, Dorset, UK), deparaffinized with xylene, hydrated in absolute alcohol, and immersed in phosphate-buffered saline (PBS). Sections were digested with pronase E (6.25 ng/μL) for 5 minutes, washed in PBS, dehydrated in absolute alcohol, and left to air dry. The probe hybridization solution was added to each section and incubated at 55°C overnight in a humidity chamber. Slides were then washed for 5 minutes each in 2 × standard saline citrate (SSC) buffer (twice), 0.1 × SSC, and then PBS. Sections were blocked using 10% normal rabbit serum and bound probe was detected using rabbit anti–fluorescein isothiocyanate (FITC) conjugated to alkaline phosphatase (diluted 1:100). Slides were incubated for 30 minutes at room temperature, then washed in PBS followed by alkaline phosphatase substrate buffer (pH 9.0) for 5 minutes. Alkaline phosphatase activity was demonstrated with chromogen overnight. Nuclei were counterstained with hematoxylin. The EBV-positive cell line B95.8 was used as a positive control.
Immunohistochemistry.
Tissues were cut to 5-μm thick sections and mounted on vectorbonded slides (Vectorbond reagent, Vector Laboratories Inc). The expression of LMP 1 was detected using the monoclonal antibody (MoAb) CS1-4 (kindly provided by M. Rowe, University of Cardiff, Cardiff, UK) as described previously.30 HLA class I levels were assessed using the antibody HC10 at a dilution of 1:300. This antibody, kindly provided by H. Ploegh (M.I.T., Cambridge, MA) preferentially recognizes HLA B/C locus products.31 TAP 1 and TAP 2 expression were measured using anti–C-terminal peptide immune rabbit sera32 (kindly supplied by J. Trowsdale, Cambridge University, UK) diluted 1:500 and 1:1,000, respectively. Bound antibody was detected using a standard ABC immunoperoxidase method (Universal VECTASTAIN Elite ABC kit, Vector Laboratories Inc) and visualized with the diaminobenzidine-based detection reaction (Vector Laboratories Inc). Nuclei were counterstained with hematoxylin.
Western blotting.
Cells for Western blot analysis were lysed by sonication in 8 mol/L urea; 10 mmol/L Tris buffer, pH 7.0; and the protein concentration determined using a Bio-Rad DC protein assay kit (Bio-Rad Laboratories, Richmond, CA). The lysate was diluted in electrophoresis sample buffer and 50 μg protein was loaded per track of a discontinuous sodium dodecyl sulfate (SDS)-polyacrylamide gel with a 7.5% acrylamide resolving gel. The electrophoresed proteins were transferred to a nitrocellulose membrane and TAP 1 and TAP 2 proteins were detected using anti–C-terminal peptide immune rabbit sera32 (anti-TAP 1 diluted 1:400; anti-TAP 2 diluted 1:500) and a chemiluminescence detection protocol.
Immunofluorescence.
Surface expression of HLA class I molecules on H-RS cell lines was measured by incubating viable cells with MoAb W6/3233 or BB7.234 for 30 minutes on ice. After washing with PBS containing 10% (vol/vol) normal goat serum (NGS), bound antibody was detected by incubation with FITC-labeled goat anti-mouse IgG (Sigma-Aldrich Co Ltd) for 20 minutes on ice. After four washes in PBS + 10% NGS, cells were fixed with 1% (wt/vol) paraformaldehyde and analyzed using a FACScan (Becton-Dickinson, Mountain View, CA). W6/32 (ascites) and BB7.2 (culture supernatant) were diluted 1:200 and 1:5, respectively, using PBS + 10% NGS, and FITC-labeled goat anti-mouse IgG was diluted 1:50 using PBS + 10% NGS + 10% EBV-seronegative human serum.
CTL clones and cytotoxicity assays.
LCLs were generated in vitro by EBV transformation of B cells from healthy laboratory donors of known HLA type and cultured in growth medium. EBV-specific CTLs were reactivated from the peripheral blood of healthy laboratory donors by in vitro stimulation with the autologous LCL (irradiated at 4,000 rads; responder to stimulator ratio = 40:1) in growth medium containing 1% human AB serum (Sigma-Aldrich Co Ltd). After 7 days, fresh medium and autologous LCL (irradiated) were added, and on day 14 cells were cloned by limiting dilution at 0.3 cells/well and maintained in IL-2–conditioned medium as described previously.6
In cytotoxicity assays, targets were incubated with [51Cr]O4 for 1 to 2 hours, washed, and then incubated in 96-well V-bottom plates (Life Technologies) at 2.5 × 103 cells/well in 100 μL of growth medium. CTLs in growth medium (100 μL/well) were then added at a known effector:target (E:T) ratio. Spontaneous and maximum levels of cell lysis were measured by culturing targets with growth medium alone or 1% (wt/vol) SDS solution, respectively (100 μL/well). After 5 hours incubation, 100 μL of culture supernatant was obtained from each well and levels of [51Cr]O4 measured using a gamma counter (Packard, Pangbourne, Berks, UK). All tests were conducted in triplicate and the percentage specific lysis was calculated as (release by CTL − spontaneous release) × 100/(total release in 1% SDS − spontaneous release). In some assays, BL cell lines were used as targets. The isolation and culture of the BL cell line DH has been described previously35; OKU is a group I BL cell line established in the same manner from an African BL patient (A.B.R., unpublished data). Where assays involved the use of recombinant vaccinia viruses, target cells were preinfected for 1 hour with virus at a multiplicity of infection of 10, followed by 4 to 15 hours incubation in growth medium. Targets were then labeled with [51Cr]O4 and used in the assay as described above. The vaccinia recombinant expressing HLA B*3501 was constructed using the protocol described by Blasco and Moss.36 The cDNA for B*3501 was cloned into the transfer plasmid pRB21 and then transfected into CV-1 cells. After infection of these cells with vRB12, a recombinant vaccinia expressing HLA B*3501 was rescued, and this was then plaque purified and titred on BSC-1 cells. Other vaccinia recombinants used in this study have all been described previously.7 37 Where assays involved the use of synthetic peptides, [51Cr]O4-labeled targets were incubated with peptide at 0.01 μg/mL for 1 hour at 37°C, washed, counted, and added to the assay plate as described above. None of the peptides used in this study mediated target cell lysis in the absence of CTLs. Peptides were synthesized using fluorenylmethoxycarbonyl chemistry by J. Fox (Alta Bioscience, University of Birmingham, Birmingham, UK). They were dissolved in dimethylsulphoxide (DMSO) and protein concentrations measured using a modified biuret assay.
IL-10.
Recombinant human IL-10 (hIL-10) was kindly provided by Schering-Plough Research Institute (Kenilworth, NJ). Biological activity was assessed using a method based on that described by Hsu et al.38Briefly, peripheral blood mononuclear cells (PBMCs) (106cells/mL) were cultured in AIM-V serum free medium (Life Technologies) containing phytohemagglutinin (PHA, 10 μg/mL) with or without hIL-10 (200 U/mL) for 3 days at 37°C in 5% CO2. Cultures were set up in triplicate in 96-well flat-bottomed plates (Life Technologies) at 200 μL/well. After 3 days, 50 μL of culture supernatant was obtained from each well and levels of interferon-γ (IFN-γ) measured using the Quantikine human IFN-γ assay (R & D Systems Europe Ltd, Abingdon, Oxford, UK) according to the manufacturer's instructions.
RESULTS
TAP 1, TAP 2, and HLA class I expression in H-RS cells within HD biopsies.
Biopsy material from 38 HD patients was studied for expression of HLA class I molecules and the peptide transporter proteins TAP 1 and TAP 2 by H-RS cells as markers of their antigen-processing phenotype. The results are shown in Table 1, in each case alongside the histological subtype of the tumor and its EBV status (based on in situ hybridization for EBERs and staining for the EBV protein LMP 1). Of the 24 EBV-positive tumors, 18 (75%) expressed detectable surface HLA class I on the H-RS cells, usually at medium to high levels. In contrast, of the 14 EBV-negative cases, only 4 (29%) showed clear HLA class I staining of H-RS cells and the majority of tumors appeared to be HLA class I negative. In three cases, one of which was an EBV-positive tumor, the biopsy contained a mixture of class I–positive and class I–negative H-RS cells. By comparison, immunohistochemical staining for the TAP proteins gave a much more uniform pattern of results. Thus TAP 1 and TAP 2 were detected in H-RS cells in all of the tumors, irrespective of their EBV genome status, although in three cases expression of one or other of these proteins was weak.
Expression of HLA Class I, TAP 1, and TAP 2 in H-RS Cells
| HD Case . | Subtype . | EBV Status . | HLA Class I . | TAP 1 . | TAP 2 . | |
|---|---|---|---|---|---|---|
| EBER in situ Hybridization . | LMP 1 Staining . | |||||
| 1 | MC | + | + | +++ | ++ | ++ |
| 2 | MC | + | + | +++ | ++ | ++ |
| 3 | MC | + | + | + | + | + |
| 4 | MC | + | + | +++ | +++ | +++ |
| 5 | MC | + | + | ++ | + | ++ |
| 6 | MC | + | + | ++ | ++ | ++ |
| 7 | NS | + | + | ++ | ++ | ++ |
| 8 | NS | + | + | ++ | ++ | ++ |
| 9 | NS | + | + | ++ | ++ | + |
| 10 | NS | + | + | ++ | ++ | + |
| 11 | NS | + | + | ++ | ++ | ++ |
| 12 | NS | + | + | ++ | ++ | ++ |
| 13 | NS | + | + | + | + | ++ |
| 14 | NS | + | + | ++ | ++ | + |
| 15 | NS | + | + | ++ | + | ++ |
| 16 | NS | + | + | + | +++ | ++ |
| 17 | NS | + | + | − | + | +/− |
| 18 | NS | + | + | − | ++ | +/− |
| 19 | NS | + | + | − | ++ | ++ |
| 20 | NS | + | + | − | ++ | ++ |
| 21 | LP | + | + | +/− | +++ | + |
| 22 | LP | + | + | − | ++ | ++ |
| 23 | IF | + | + | ++ | ++ | ++ |
| 24 | IF | + | + | ++ | ++ | ++ |
| 25 | MC | − | − | ++ | ++ | ++ |
| 26 | MC | − | − | +/− | +++ | +++ |
| 27 | NS | − | − | ++ | ++ | +++ |
| 28 | NS | − | − | − | +++ | ++ |
| 29 | NS | − | − | − | +++ | ++ |
| 30 | NS | − | − | − | ++ | ++ |
| 31 | NS | − | − | − | ++ | ++ |
| 32 | NS | − | − | − | ++ | ++ |
| 33 | NS | − | − | − | ++ | ++ |
| 34 | LP | − | − | +/− | ++ | ++ |
| 35 | LP | − | − | − | +++ | +++ |
| 36 | LP | − | − | − | +/− | ++ |
| 37 | ni | − | − | + | + | + |
| 38 | ni | − | − | + | ++ | ++ |
| HD Case . | Subtype . | EBV Status . | HLA Class I . | TAP 1 . | TAP 2 . | |
|---|---|---|---|---|---|---|
| EBER in situ Hybridization . | LMP 1 Staining . | |||||
| 1 | MC | + | + | +++ | ++ | ++ |
| 2 | MC | + | + | +++ | ++ | ++ |
| 3 | MC | + | + | + | + | + |
| 4 | MC | + | + | +++ | +++ | +++ |
| 5 | MC | + | + | ++ | + | ++ |
| 6 | MC | + | + | ++ | ++ | ++ |
| 7 | NS | + | + | ++ | ++ | ++ |
| 8 | NS | + | + | ++ | ++ | ++ |
| 9 | NS | + | + | ++ | ++ | + |
| 10 | NS | + | + | ++ | ++ | + |
| 11 | NS | + | + | ++ | ++ | ++ |
| 12 | NS | + | + | ++ | ++ | ++ |
| 13 | NS | + | + | + | + | ++ |
| 14 | NS | + | + | ++ | ++ | + |
| 15 | NS | + | + | ++ | + | ++ |
| 16 | NS | + | + | + | +++ | ++ |
| 17 | NS | + | + | − | + | +/− |
| 18 | NS | + | + | − | ++ | +/− |
| 19 | NS | + | + | − | ++ | ++ |
| 20 | NS | + | + | − | ++ | ++ |
| 21 | LP | + | + | +/− | +++ | + |
| 22 | LP | + | + | − | ++ | ++ |
| 23 | IF | + | + | ++ | ++ | ++ |
| 24 | IF | + | + | ++ | ++ | ++ |
| 25 | MC | − | − | ++ | ++ | ++ |
| 26 | MC | − | − | +/− | +++ | +++ |
| 27 | NS | − | − | ++ | ++ | +++ |
| 28 | NS | − | − | − | +++ | ++ |
| 29 | NS | − | − | − | +++ | ++ |
| 30 | NS | − | − | − | ++ | ++ |
| 31 | NS | − | − | − | ++ | ++ |
| 32 | NS | − | − | − | ++ | ++ |
| 33 | NS | − | − | − | ++ | ++ |
| 34 | LP | − | − | +/− | ++ | ++ |
| 35 | LP | − | − | − | +++ | +++ |
| 36 | LP | − | − | − | +/− | ++ |
| 37 | ni | − | − | + | + | + |
| 38 | ni | − | − | + | ++ | ++ |
Abbreviations: MC, mixed cellularity; NS, nodular sclerosing; LP, lymphocyte predominant; IF, interfollicular HD; ni, not interpretable; −, no staining; +/−, only some cells positive; +, weak staining; ++, medium staining; +++, strong staining.
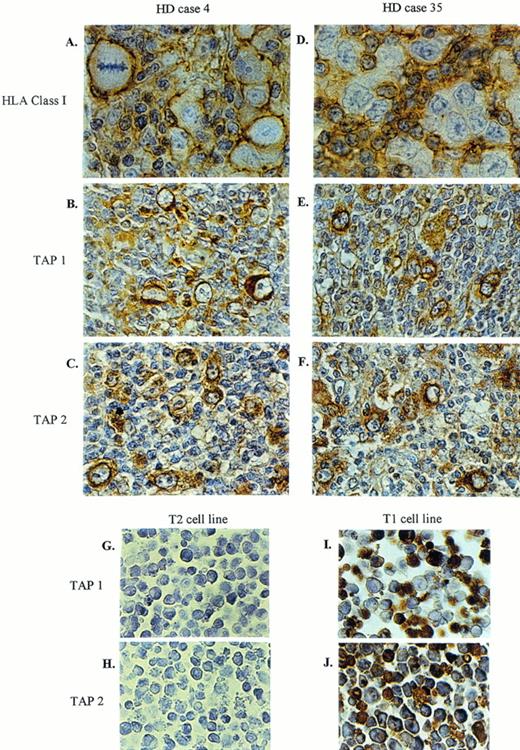
Figure 1shows staining for HLA class I and TAP 1 and 2 in two representative HD cases (Table 1, cases 4 and 35). Figure 1A shows H-RS cells from HD case 4 that are HLA class I positive (graded as “+++” in Table1); note that the infiltrating lymphocytes that surround the H-RS cells are also HLA class I positive, but that the H-RS cells stand out due to the increased level of expression of class I on these cells. The expression of TAP 1 and TAP 2 in H-RS cells from this same biopsy is shown in Fig 1B and C, respectively. Figure 1D shows the results of HLA class I staining of HD case 35 where the surrounding lymphocytes clearly express class I molecules but the H-RS cells do not. This same H-RS cell population did nevertheless express TAP 1 and TAP 2, as shown in Fig 1E and F, respectively. As controls for the TAP-specific reagents, we stained the T2 mutant LCL which lacks the genes coding for TAPs 1 and 2 and its TAP-positive parent LCL T1.39 40 T2 cells did not stain for TAPs (Fig 1G and H), whereas the T1 line shows positive staining with both the anti-TAP 1 and the anti-TAP 2 sera (Fig1I and J).
Two representative HD cases stained for HLA class I, TAP 1, and TAP 2. Biopsy material from HD case No. 4 expresses high levels of HLA class I (A), as well as TAP 1 (B) and TAP 2 (C). HD case No. 35 shows no detectable HLA class I (D), whereas TAP 1 (E) and TAP 2 (F) are expressed. Sections were counterstained with hematoxylin. T2 (TAP-negative cell line) and T1 (TAP-positive cell line) were used as controls for TAP 1 (G, I) and TAP 2 (H, J) staining. Original magnification × 672 (A, D) and × 420 (B, C, E-J).
Two representative HD cases stained for HLA class I, TAP 1, and TAP 2. Biopsy material from HD case No. 4 expresses high levels of HLA class I (A), as well as TAP 1 (B) and TAP 2 (C). HD case No. 35 shows no detectable HLA class I (D), whereas TAP 1 (E) and TAP 2 (F) are expressed. Sections were counterstained with hematoxylin. T2 (TAP-negative cell line) and T1 (TAP-positive cell line) were used as controls for TAP 1 (G, I) and TAP 2 (H, J) staining. Original magnification × 672 (A, D) and × 420 (B, C, E-J).
TAP 1, TAP 2, and HLA class I expression in H-RS cell lines.
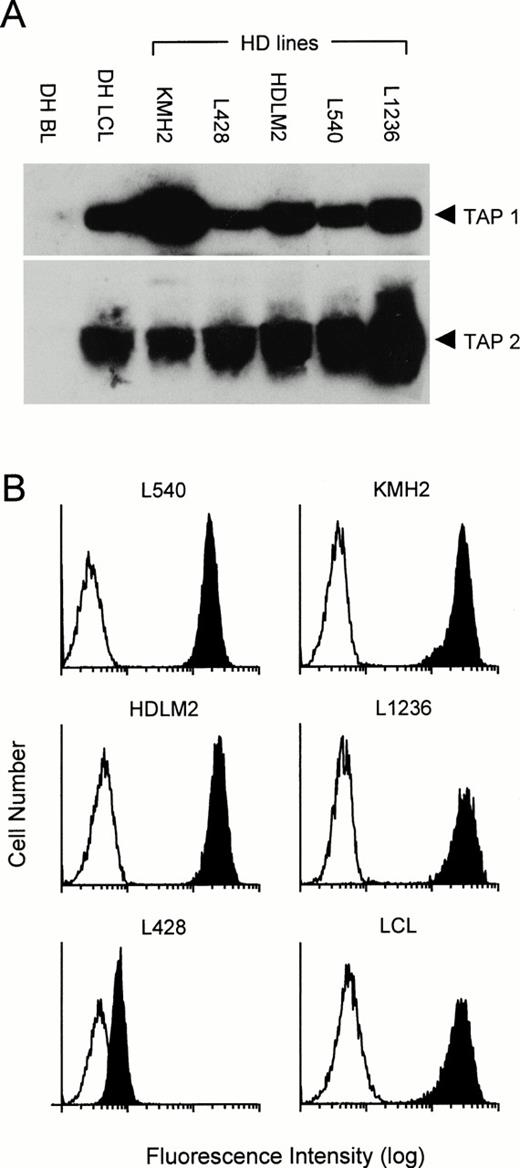
We then extended our analysis to study the class I processing pathway of five H-RS cell lines. Using Western blotting, we measured the levels of expression of TAP 1 and TAP 2 and compared them with a Burkitt tumor cell line DH and with a matched LCL established by EBV transformation of normal B cells from the same individual. As reported previously,17 41 and as shown here, BL cell lines express little or no TAP 1 and TAP 2, whereas both of these proteins are expressed at high level in LCLs (Fig2A). Levels of TAP expression in the H-RS lines showed some variation, but in general they were equivalent to or greater than that seen in the LCL. Using fluorescence-activated cell sorter (FACS) analysis we also compared expression of HLA class I molecules on the surface of the H-RS cell lines and an LCL. The results shown in Fig 2B show that cell lines KMH2, HDLM2, L1236, and L540 express high levels of HLA class I (comparable with that seen on an LCL), whereas L428 expresses very low levels.
Expression of TAP 1, TAP 2, and HLA class I in HD-derived cell lines. Five H-RS lines plus an LCL and BL cell line from donor DH were analyzed by Western blotting for expression of TAP 1 and TAP 2 (A). Viable cells from the H-RS cell lines plus an LCL were also analyzed by FACScan for surface expression of HLA class I using the MoAb W6/32 (B). Clear profile, staining with second step antibody alone; shaded profile, W6/32 staining. Results shown are representative of those observed in several repeated assays.
Expression of TAP 1, TAP 2, and HLA class I in HD-derived cell lines. Five H-RS lines plus an LCL and BL cell line from donor DH were analyzed by Western blotting for expression of TAP 1 and TAP 2 (A). Viable cells from the H-RS cell lines plus an LCL were also analyzed by FACScan for surface expression of HLA class I using the MoAb W6/32 (B). Clear profile, staining with second step antibody alone; shaded profile, W6/32 staining. Results shown are representative of those observed in several repeated assays.
CTL recognition of H-RS cell lines.
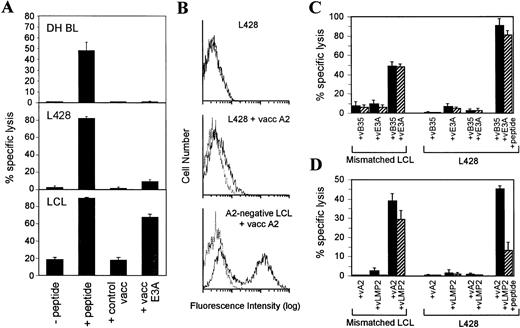
Studying the H-RS cell lines, we tested their ability to process and present EBV proteins to HLA class I–restricted CTL clones. All of the H-RS lines are EBV genome negative and therefore EBV proteins were expressed in the cells using recombinant vaccinia vectors and their processing monitored in standard cytotoxicity assays using as effectors HLA class I–restricted CTL clones appropriate to the class I type of the H-RS target line. As a positive control, H-RS cell lines were preloaded with synthetic peptides representing the relevant CTL target epitope. Once again, results were compared with those obtained using BL and LCL targets in the same assays. For example, HDLM2 (HLA A1,2 B8,44) was tested for its ability to present a B8-restricted epitope in EBNA 3A (amino acids 158-166). As shown in Fig3A, a B8-restricted CTL clone specific for this epitope mediated clear lysis of HDLM2 expressing EBNA 3A from a vaccinia vector, whereas HDLM2 infected with a control vaccinia vector was not lysed. Note that the level of lysis was comparable with that achieved by preloading HDLM2 directly with the target peptide epitope. The same pattern of results was observed when a B8-positive LCL was used as the target cell.* In contrast to both HDLM2 and the LCL target, a B8-positive BL line (OKU) gave negative results in this antigen processing assay. Thus, the line was recognized by the B8-restricted CTL clone when preloaded with the peptide epitope but was unable to process and present EBNA 3A protein endogenously expressed from a vaccinia vector (Fig 3A). In the same way, we showed that HDLM2 could also process and present a defined B44-restricted epitope in EBNA 3C (residues 161-171; data not shown). Similar results were obtained with the three other H-RS cells in the panel which showed easily detectable levels of surface HLA class I expression. Thus, KMH2 (HLA A11,24; B51,62) and L540 (HLA A3,11; B51) were able to present a defined A11-restricted epitope in EBNA 3B (residues 416-424) (Fig 3B), and L1236 (HLA A2; B51) could present a defined A2-restricted epitope in LMP2 (residues 426-434) (Fig 3C).
CTL recognition of BL, HD, and LCL lines expressing EBV protein antigens from recombinant vaccinia vectors. (A) A B8-restricted EBNA 3A-specific CTL clone tested against OKU BL, HDLM2, and a B8-matched LCL target. (B) An A11-restricted EBNA 3B-specific CTL clone tested against KMH2 and L540. (C) An A2-restricted LMP 2-specific CTL clone tested against L1236. All targets had either been preincubated with the cognate peptide epitope or preinfected with a recombinant vaccinia vector expressing the target EBV protein. As controls, targets were preincubated with an equivalent dilution of DMSO solvent (−peptide) or with the vaccinia vector alone (control vacc). E:T ratios ranged from 5:1 to 10:1. All results are expressed as % specific lysis + 1 SD and are representative of several repeated assays.
CTL recognition of BL, HD, and LCL lines expressing EBV protein antigens from recombinant vaccinia vectors. (A) A B8-restricted EBNA 3A-specific CTL clone tested against OKU BL, HDLM2, and a B8-matched LCL target. (B) An A11-restricted EBNA 3B-specific CTL clone tested against KMH2 and L540. (C) An A2-restricted LMP 2-specific CTL clone tested against L1236. All targets had either been preincubated with the cognate peptide epitope or preinfected with a recombinant vaccinia vector expressing the target EBV protein. As controls, targets were preincubated with an equivalent dilution of DMSO solvent (−peptide) or with the vaccinia vector alone (control vacc). E:T ratios ranged from 5:1 to 10:1. All results are expressed as % specific lysis + 1 SD and are representative of several repeated assays.
The analysis was then extended to L428, a HD line which in its trace levels of surface HLA class I expression may serve as an in vitro counterpart of those HD cases where the tumor cells are classified as HLA class I-negative by immunohistochemical staining. This line (HLA class I genotype A3; B35) was assayed using CTL clones recognizing a defined B35-restricted epitope in EBNA 3A (amino acids 458-466). As shown in Fig 4A, L428 was able to present the exogenously loaded epitope peptide but not the EBNA 3A protein endogenously expressed from a vaccinia vector. In this respect L428 resembled the processing defective B35-positive BL line (DH) and was clearly distinct from a processing competent LCL. To investigate the L428 phenotype further, we attempted to increase surface HLA class I levels. This could not be achieved by IFN-γ treatment (data not shown) and therefore we infected the cells with recombinant vaccinia vectors expressing either HLA B35 or HLA A2. However, FACS analysis revealed that vaccinia-mediated expression of these HLA alleles resulted in only a small increase in the levels of surface class I molecules in L428 cells compared with that produced by the same vectors in LCL cells (Fig 4B; data shown for HLA A2 expression only). Furthermore, when L428 was coinfected with vaccinias expressing both HLA B35 and EBNA 3A, there was still no detectable CTL recognition (Fig4C) even though these coinfected targets were clearly sensitized to lysis if pre-exposed to the epitope peptide before testing in the cytotoxicity assay. Therefore, we wondered whether there might be an additional lesion in the HLA class I processing pathway in L428 cells at the level of the TAP transporter function, even though TAP protein expression was detectable. This possibility was tested using CTL recognition of an HLA A2-restricted epitope in the LMP 2 protein (amino acids 426-434) which is very unusual in being processed by a TAP-independent route.42 In this case, L428 cells (and a control LCL from an A2-negative donor) were coinfected with vaccinia vectors expressing HLA A2 and LMP 2 and tested for lysis by A2-restricted CTLs specific for the above epitope. Despite clear lysis of the coinfected LCL target there was no recognition of coinfected L428 cells (Fig 4D) unless these cells were additionally exposed to the synthetic epitope peptide before testing in the assay.
L428 is unable to present endogenously synthesized antigens to HLA class I–restricted CTLs. (A) A B35-restricted EBNA 3A-specific CTL clone tested against DH BL, L428, and a B35-matched LCL target. All targets had either been preincubated with the cognate peptide epitope (EBNA 3A residues 458-466) or preinfected with a recombinant vaccinia vector expressing EBNA 3A. As controls, targets were preincubated with an equivalent dilution of DMSO solvent (−peptide) or with the vaccinia vector alone (control vacc). (B) Expression of HLA A2 from a recombinant vaccinia vector was tested in L428 and an HLA A2-negative LCL. Viable cells were analyzed by FACScan for surface expression of HLA A2 using the MoAb BB7.2. Solid line, BB7.2 staining; dotted line, staining with second step antibody alone. (C) A B35-restricted EBNA 3A-specific CTL clone tested against a B35-negative LCL and L428. Targets were infected with vaccinia recombinants expressing HLA B35 and/or EBNA 3A and in one case targets were also preincubated with the cognate peptide epitope (EBNA 3A residues 458-466). (D) An A2-restricted LMP 2-specific CTL clone tested against an A2-negative LCL and L428. Targets were infected with vaccinia recombinants expressing HLA A2 and/or LMP 2 and in one case targets were also preincubated with the cognate peptide epitope (LMP 2 residues 426-434). E:T ratios = 5:1 (▪) and 1:1 (▨). Results of cytotoxicity assays are expressed as % specific lysis + 1 SD and are representative of several repeated assays.
L428 is unable to present endogenously synthesized antigens to HLA class I–restricted CTLs. (A) A B35-restricted EBNA 3A-specific CTL clone tested against DH BL, L428, and a B35-matched LCL target. All targets had either been preincubated with the cognate peptide epitope (EBNA 3A residues 458-466) or preinfected with a recombinant vaccinia vector expressing EBNA 3A. As controls, targets were preincubated with an equivalent dilution of DMSO solvent (−peptide) or with the vaccinia vector alone (control vacc). (B) Expression of HLA A2 from a recombinant vaccinia vector was tested in L428 and an HLA A2-negative LCL. Viable cells were analyzed by FACScan for surface expression of HLA A2 using the MoAb BB7.2. Solid line, BB7.2 staining; dotted line, staining with second step antibody alone. (C) A B35-restricted EBNA 3A-specific CTL clone tested against a B35-negative LCL and L428. Targets were infected with vaccinia recombinants expressing HLA B35 and/or EBNA 3A and in one case targets were also preincubated with the cognate peptide epitope (EBNA 3A residues 458-466). (D) An A2-restricted LMP 2-specific CTL clone tested against an A2-negative LCL and L428. Targets were infected with vaccinia recombinants expressing HLA A2 and/or LMP 2 and in one case targets were also preincubated with the cognate peptide epitope (LMP 2 residues 426-434). E:T ratios = 5:1 (▪) and 1:1 (▨). Results of cytotoxicity assays are expressed as % specific lysis + 1 SD and are representative of several repeated assays.
The effects of IL-10 on CTL recognition of HD cell lines.
A previous report by Matsuda et al24 described the ability of IL-10 to block antigen-specific CTL-mediated lysis of human melanoma cells and allo-specific CTL-mediated lysis of LCLs by downregulating the surface expression of HLA class I molecules. Because IL-10 is expressed by tumor cells in some cases of HD, we sought to determine whether this cytokine has a similar effect on the ability of H-RS cells to be recognized and lysed by CTL. Using recombinant human IL-10 we first confirmed that this preparation was biologically active by demonstrating its ability to inhibit IFN-γ production by phytohemagglutinin-treated PBMCs (Fig 5A). We then studied the effect of pretreating the H-RS cell lines with IL-10, following the procedure described by Matsuda et al.24 Thus, H-RS cell lines were washed and cultured in AIM-V serum-free medium (Life Technologies) containing 200 U/mL IL-10 for 3 days. During the last five hours of IL-10 pretreatment, H-RS lines were infected with recombinant vaccinia vectors expressing EBV proteins and were then tested for recognition by EBV-specific CTL clones in a chromium release assay. As shown in Fig 5B, HDLM2 expressing EBNA 3A was clearly recognized by a B8-restricted EBNA 3A-specific CTL clone, whereas HDLM2 infected with a control vaccinia vector was not. However, IL-10 pretreatment had no obvious effect on the levels of lysis observed. In the same way, L1236 expressing LMP 2 from a vaccinia vector was efficiently lysed by an A2-restricted LMP 2-specific CTL clone even after IL-10 pretreatment (Fig 5C). Furthermore, the same result was obtained with L540, KMH2, and an A11-positive LCL when tested with an A11-restricted CTL clone specific for EBNA 3B (Fig 5D).
CTL recognition of H-RS lines pretreated with recombinant human IL-10. (A) The biological activity of the human IL-10 was shown by its ability to inhibit IFN-γ production by PHA-treated PBMCs. However, IL-10 pretreatment had no inhibitory effect on CTL-mediated lysis of H-RS lines or an LCL (B-D). (B) A B8-restricted EBNA 3A-specific CTL clone tested against HDLM2 infected with a vaccinia vector expressing EBNA 3A (vE3A) or infected with the vector alone (vTK−). (C) An A2-restricted LMP 2-specific CTL clone tested against L1236 expressing LMP 2 from a vaccinia vector (vLMP2) or the vector alone (vTK−). (D) An A11-restricted EBNA 3B-specific CTL clone tested against L540, KMH2, and an A11-matched LCL expressing EBNA 3B from a vaccinia vector (vE3B) or the vector alone (vTK−). E:T = 5:1 (▪) and 1:1 (▨). All results are expressed as % specific lysis + 1 SD and are representative of several repeated assays.
CTL recognition of H-RS lines pretreated with recombinant human IL-10. (A) The biological activity of the human IL-10 was shown by its ability to inhibit IFN-γ production by PHA-treated PBMCs. However, IL-10 pretreatment had no inhibitory effect on CTL-mediated lysis of H-RS lines or an LCL (B-D). (B) A B8-restricted EBNA 3A-specific CTL clone tested against HDLM2 infected with a vaccinia vector expressing EBNA 3A (vE3A) or infected with the vector alone (vTK−). (C) An A2-restricted LMP 2-specific CTL clone tested against L1236 expressing LMP 2 from a vaccinia vector (vLMP2) or the vector alone (vTK−). (D) An A11-restricted EBNA 3B-specific CTL clone tested against L540, KMH2, and an A11-matched LCL expressing EBNA 3B from a vaccinia vector (vE3B) or the vector alone (vTK−). E:T = 5:1 (▪) and 1:1 (▨). All results are expressed as % specific lysis + 1 SD and are representative of several repeated assays.
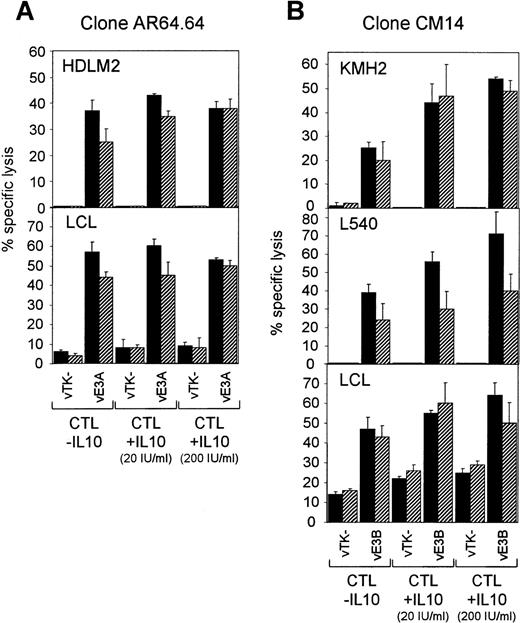
In parallel experiments, we assessed whether pretreatment of the CTL effectors with IL-10 could inhibit their ability to lyse the H-RS cell lines. CTL clones were pretreated with IL-10 at 20 U/mL and 200 U/mL in AIM-V serum-free medium for 3 days and then tested for their ability to lyse H-RS lines expressing the relevant EBV protein from a recombinant vaccinia virus. Testing a B8-restricted EBNA 3A-specific CTL clone against HDLM2 and a B8-positive LCL, we observed no significant effect of IL-10 pretreatment on the levels of CTL-mediated lysis (Fig6A). In addition, we obtained the same result testing an A2-restricted LMP 2-specific CTL clone against both L1236 and an A2-positive LCL (data not shown). IL-10 pretreatment of an A11-restricted CTL clone specific for a defined epitope in EBNA 3B (residues 416-424) again failed to inhibit lysis of KMH2, L540, and an A11-positive LCL expressing EBNA 3B from a vaccinia vector (Fig 6B). Indeed, in this case IL-10 pretreatment appeared to increase the levels of CTL-mediated target cell lysis in a dose-dependent manner. This result was obtained with two different CTL clones specific for this particular epitope and was observed in four of five replicate experiments.
Recognition of H-RS lines and LCLs by CTL clones pretreated with recombinant human IL-10. (A) HDLM2 and a B8-positive LCL target were infected with a vaccinia vector expressing EBNA 3A (vE3A) or with the vector alone (vTK−) and then tested for recognition by a B8-restricted EBNA 3A-specific CTL clone AR64.64 pretreated with IL-10 at the concentrations shown. E:T = 5:1 (▪) and 2:1 (▨). (B) KMH2, L540, and an A11-positive LCL target were infected with a vaccinia vector expressing EBNA 3B (vE3B) or with the vector alone (vTK−) and then tested for recognition by an A11-restricted EBNA 3B-specific CTL clone CM14 pretreated with IL-10 at the concentrations shown. E:T = 4:1 (▪) and 1:1 (▨). All results are expressed as % specific lysis + 1 SD and are representative of several repeated assays.
Recognition of H-RS lines and LCLs by CTL clones pretreated with recombinant human IL-10. (A) HDLM2 and a B8-positive LCL target were infected with a vaccinia vector expressing EBNA 3A (vE3A) or with the vector alone (vTK−) and then tested for recognition by a B8-restricted EBNA 3A-specific CTL clone AR64.64 pretreated with IL-10 at the concentrations shown. E:T = 5:1 (▪) and 2:1 (▨). (B) KMH2, L540, and an A11-positive LCL target were infected with a vaccinia vector expressing EBNA 3B (vE3B) or with the vector alone (vTK−) and then tested for recognition by an A11-restricted EBNA 3B-specific CTL clone CM14 pretreated with IL-10 at the concentrations shown. E:T = 4:1 (▪) and 1:1 (▨). All results are expressed as % specific lysis + 1 SD and are representative of several repeated assays.
DISCUSSION
A prerequisite for any attempt to target EBV genome-positive HD with virus-specific CTL preparations is that the H-RS cells must be capable of processing and presenting antigens to HLA class I–restricted T cells. In this context, two previous immunohistochemical studies of HLA class I expression by H-RS cells18,19 have produced conflicting results. In the present work we found that the H-RS cell population was uniformly HLA class I positive in at least 75% of cases of EBV genome-positive HD, whereas the figure was much lower in EBV genome-negative tumors; more than half of the EBV genome-negative tumors had no detectable HLA class I expression in tumor cells. Because LMP 1 is known to activate HLA class I (and TAP) gene expression in B cells,41 it is tempting to speculate that a similar influence is occurring in the context of HD. However, there clearly are a minority of EBV/LMP1-positive HD cases that lack HLA class I expression. The present findings are in agreement with Oudejans et al19 who reported that 23 of 25 EBV-positive HD cases express medium to high levels of HLA class I (based on staining with the HC10 MoAb) compared with only 5 of 38 EBV-negative cases. The contrast between these findings and the results of Poppema and Visser18 may be more apparent than real because the latter authors did not assess the EBV status of HD biopsies. Thus it is conceivable that the large number of HLA class I–negative cases that they identified (22 of 28) may have reflected an unusually large proportion of EBV-negative biopsies studied. Our analysis of TAP expression in H-RS cells found that almost all HD cases, regardless of EBV status, were positive for TAP 1 (again as reported by Oudejans et al19) and for TAP 2 (Table 1and Fig 1). Thus, unlike some carcinomas, the lack of HLA class I molecules on the surface of some H-RS cells is not due to the downregulation of either TAP 1 or TAP 2, but rather, must involve an alternative mechanism.
Although our results indicated that the majority of EBV-positive H-RS cells express HLA class I molecules, this did not prove that the class I processing pathway is intact because some processing-defective BL cells also express relatively high levels of HLA class I molecules on the cell surface.41,43 To begin to analyze this question experimentally, we used a panel of five cell lines derived from HD biopsies. These lines have been extensively analyzed and carry many phenotypic markers characteristic of H-RS cells including CD15, CD30, and CD71.29,44 L1236 is a HD line of particular interest because it has also been shown to carry the same immunoglobulin gene rearrangement sequences as H-RS cells in the bone marrow of the original HD patient.29 Analyzing H-RS cell lines for HLA class I and TAP expression showed that all expressed high levels of TAP 1 and TAP 2 and that, with the exception of L428, all expressed high levels of surface HLA class I molecules (Fig 2). Using EBV-specific HLA class I–restricted CTL clones, we demonstrated that all 4 HD-derived lines with high HLA class I expression were able to process and present endogenously synthesized EBV proteins (Fig 3). Moreover, in the case of L1236 we were able to show that one of the EBV proteins known to be expressed in some H-RS cells, namely LMP 2, can be processed and presented through the HLA class I pathway. Note that, in this particular case, although TAP expression in L1236 is not downregulated, processing of the LMP 2-derived target epitope might also have occurred via a TAP-independent route.42
The only HD line which failed to present endogenously synthesized antigen to a class I–restricted CTL was L428 (Fig 4A). This cell line expresses only trace levels of surface HLA class I molecules but high levels of TAP 1 and 2 (Fig 2B) and may therefore be representative of HLA class I–negative H-RS cells present in some HD cases (Table 1). In this case there seemed to be a fundamental lesion in the antigen processing pathway because it was not possible to achieve CTL recognition even when both the necessary HLA class I heavy chain and the necessary EBV target antigen were expressed within the cells from vaccinia vectors. This was the case whether the target in question was a conventional TAP-dependent epitope in EBNA 3A (Fig 4C) or an unconventional TAP-independent epitope in LMP 2 (Fig 4D). These negative results were not due to an inability to infect L428 cells with vaccinia viruses because immunofluoresence screening on fixed cell populations showed that two independent indicator proteins (β-galactosidase and LMP 2) were efficiently expressed in L428 cells when delivered using vaccinia vectors (S.P.L., data not shown). A particularly interesting result was the very low level of surface HLA class I molecules that appeared at the L428 cell surface despite efficient infection with vectors expressing HLA A2 and B35 heavy chains. This suggests a profound block to mature HLA class I assembly in L428 cells and is consistent with the functional data from antigen presenting assays. More work is required to determine the nature of this defect and its relevance to the situation in HLA class I–negative H-RS cells in vivo.
The balance of findings from immunohistochemical staining of H-RS cells in EBV genome-positive HD and from the functional assays with HLA class I-positive H-RS lines would suggest that in most cases there is no fundamental block to recognition and lysis of such cells by EBV-specific effectors. However, it remains possible that IL-10 (which is expressed by H-RS cells in vivo in the majority of EBV genome-positive cases) might offer the tumor a means of local immune evasion. Thus it has been reported that IL-10 pretreatment of target cells protects them from lysis by tumor- and allo-specific CTLs in a dose-dependent manner.24 However, we followed this protocol precisely and were unable to show any change in the sensitivity of the H-RS cell lines or of a reference LCL target to CTL lysis. Furthermore, and again in contrast to this earlier report, IL-10 pretreatment had no effect on the surface expression of HLA class I molecules by either type of line (SPL, data not shown). Indeed, at least one of the H-RS cell lines (L1236) constitutively expresses IL-1045 and yet is clearly susceptible to CTL-mediated lysis (Fig 3C). IL-10 is also known to suppress the induction of cellular immune responses by inhibiting IFN-γ and IL-2 production by the Th1 subset of T-helper cells20; this could be explained by an effect on antigen presentation because IL-10 is known to downregulate major histocompatibility complex class II expression and cytokine synthesis by macrophages21 and may also inhibit the recruitment and maturation of dendritic cells.46 Whether this cytokine might also impair CTL recognition at the effector phase is perhaps a more relevant question in the present context, because CTL therapy for HD would involve the adoptive transfer of effector cells that had already been activated in vitro. We examined this question in our in vitro system and found that IL-10 did not inhibit H-RS target cell lysis by preactivated CTL clones; in fact, in some cases IL-10 pretreatment of effector cells increased the levels of killing observed. Such results are consistent with an earlier finding that EBV-specific CTLs reactivated in the presence of viral IL-10, a homologue of the human cytokine and encoded by an EBV lytic cycle gene, mediated increased levels of LCL lysis when tested 14 days later.47
In summary, several of the requirements for successful T-cell–based tumor therapy seem to be satisfied in at least the majority of cases of EBV genome-positive HD. First, at least one of the three viral antigens expressed in tumor cells, namely LMP 2, is a known target for CTL responses on several common HLA class I alleles, including HLA A*0201, A*2402, and B*4001, that together cover greater than 50% of the white population.48 Transcripts for LMP 2 are detectable in most cases of EBV-positive HD and, even using MoAbs of limited sensitivity, it is often possible to detect the protein in H-RS cell by immunohistochemical staining. Second, circumstantial evidence from HLA class I staining of biopsy material and from functional assays on cell lines suggests that the class I processing pathway is usually intact. Third, there is no indication from in vitro work that IL-10, endogenously produced by H-RS cells, could protect the tumor from virus-specific CTL recognition. Therefore, the challenge is to optimize ways of generating and expanding LMP-specific CTL preparations in vitro and of targeting them to the tumor site in vivo.
ACKNOWLEDGMENT
The authors thank Dr M. Rowe, Dr J. Trowsdale, and Dr H. Ploegh for generously providing reagents for immunohistochemistry; and Dr J. Wolf, Dr V. Diehl, and Dr H. Kamesaki for supplying us with the H-RS cell lines. We also thank Dr M. Kurilla and Dr J. Yewdell for providing the vaccinia recombinants, Dr B. Moss for vRB21 and pRB21, and Dr E. Scotet for HLA B*3501 cDNA.
Note that, as described previously, many EBV-specific CTL clones reactivated in vitro with the autologous LCL mediate little or no lysis of the LCL in a 5-hour chromium release assay.49 Using such clones throughout the present study ensured that background levels of lysis of the LCL alone were low even though this target constitutively expresses EBV proteins.
Supported by the Medical Research Council and by the Cancer Research Campaign, UK. S.P.L. is supported by a Medical Research Council Career Development Award.
Address reprint requests to Steven P. Lee, PhD, Institute for Cancer Studies, University of Birmingham, Vincent Dr, Edgbaston, Birmingham B15 2TA, UK; e-mail: s.p.lee@bham.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal