Abstract
Partial tandem duplication within the MLL gene has recently been described as a novel genetic alteration in acute myeloid leukemia (AML). It has been associated with trisomy of chromosome 11, but was also identified in AML patients with normal karyotypes. The current study was performed to investigate whether MLL duplications are restricted to AML, and hence whether they may also occur in normal hematopoietic cells. MLL-duplication transcripts were analyzed by nested reverse-transcriptase polymerase chain reaction (RT-PCR) in peripheral blood in two groups of 45 and 20 patients, respectively, as well as in two bone marrow samples from healthy volunteers. Duplications were detected in two independent nested RT-PCR experiments in the peripheral blood samples of 38 of 45 (84%) and 20 of 20 (100%) of the two groups and in both bone marrow samples. On this basis, MLL duplications seem to occur frequently in a subset of cells in normal hematopoiesis. The type of partially duplicated MLL transcripts varied substantially. Three transcripts were identical to those known from AML. In addition, four new transcripts were characterized. Three of these four were in frame and potentially translatable. MLL duplications were also detected by seminested genomic PCR with intron 9– and intron 1–specific primers in 20 of 20 peripheral blood samples studied, indicating that the duplications are genomically fixed at the DNA level and are not an RT-PCR artifact. In summary, MLL duplications are regularly generated by homologous ALU recombination in a small number of hematopoietic cells of most or even all healthy donors. These data suggest that MLL duplications are not implicated in the malignant transformation in AML, or alternatively, that only a few cells will acquire additional oncogenic mutations necessary to establish the malignant phenotype of AML.
© 1998 by The American Society of Hematology.
CHROMOSOMAL rearrangements are thought to play a crucial role in the development of hematologic malignancies. These disorders arise from a single transformed cell by a multistep process that involves sequentially occurring mutations of genes, which may function as oncogenes or tumor-suppressor genes.1-3Specific chromosomal alterations have been found in association with certain morphologic subtypes of acute myeloid leukemia (AML), as t(8;21) in a subset of M2, and less often in M1,4-6t(15;17) in M3,7 and inv(16)8 in M4eo. Rearrangements of the MLL gene by reciprocal translocations involving chromosome band 11q23 are well described in infants and adults with AML and acute lymphocytic leukemia (ALL), and involve more than 30 chromosomal regions as translocation partners.9,10 About half of the partner genes have been cloned to date. Rearrangements are strikingly diverse, involving partner genes, with various predicted protein functions like transcription factors (AF4,11AF9,12 AF10,13,14 AF17,15ENL10,16), RNA polymerase II elongation factor (ELL17), cell-cycle regulation factor (CBP18), forkhead protein family members (AFX19), or SH3 domain family member EEN,20 or seem to be target genes for RAS such as AF6.21,22 In contrast to the diversity of the partner genes, all translocations occur within a well-defined breakpoint cluster region restricted to an 8.3-kb genomic area of the large 100-kb MLL gene23 encompassing exons 8 to 14 (nomenclature by Nilson et al, 199624). In addition, partial tandem duplications of the NH2 region of this part of the gene have been associated with trisomy of chromosome 11 in AML,25 and recently, have also been reported in AML patients with normal karyotypes.26-29 The chimeric transcripts code for proteins with potential oncogenic activity. The mechanism by which the partial tandem duplication contributes to leukemogenesis is currently unknown, as is the mechanism of all rearrangements involving MLL. It is still unclear whether MLL-fusion genes act as oncogenes or whether the fusion proteins have dominant negative effects. Introduction of an MLL-AF9 transgene resulted in expansion of the myeloid compartment, and after a median interval of 8 months, the development of AML.30 Retroviral transduction of murine progenitor cells with an MLL-ENL transgene leads to immortalization and establishment of myeloid leukemias after several passages in mice hosts.31 These data suggest a gain of function mechanism for MLL-fusion genes and demonstrate that additional events are probably necessary for the establishment of the leukemogenic phenotype. Other data are more consistent with the possibility that loss of function of the MLL gene is important in leukemogenesis.32 Thus, the MLL gene may be a tumor-suppressor gene. However, as MLL-deficient embryonic stem cells are inable to differentiate normally, it is highly suggestive that MLL is involved in the regulation of early stages of hematopoietic development, either directly or indirectly. The MLL translocations in ALL are most commonly associated with a pre–pre-B phenotype,33-35 whereas AMLs with MLL translocations are predominantly seen in the monocytic subtypes.36 The MLL duplication has been found in most AML subtypes37,38 and does not seem to be restricted to specific cell lineages. If a mutation does not fatally affect cells, it might be observed throughout the normal life of an individual. In the current study, we addressed the question whether MLL tandem duplications can be detected via a nested reverse-transcriptase polymerase chain reaction (RT-PCR) in blood samples of healthy donors. We were able to show that MLL duplications occur frequently in a small number of cells in healthy individuals. In addition, our data strongly suggest that MLL duplication transcripts are not splicing artifacts, but most likely result from homologous Alu recombination of the MLL gene, similar to that described for AML.25 26
MATERIALS AND METHODS
Probands.
Normal individuals gave informed consent and the procedure was approved by the Ethics Committee of the University of Göttingen as of June 14, 1996. Equal numbers of males and females were analyzed. The mean age was 45.6 (range, 7 to 78). A description of the AML patients used as controls in this study will be given elsewhere (Schnittger et al, in preparation).
Nonhuman specimens.
For dilution experiments to determine the level of sensitivity of detection of MLL-duplication transcripts, cDNA was prepared from peripheral blood mononuclear cells of Maccaca mulatta, rat kidney, as well as mouse (strain SJL) liver and spleen. Tissue was mechanically dispersed to single-cell suspension for RNA extraction and processed as described later.
Nucleic acid isolation.
DNA was extracted with a salting-out procedure39 from peripheral blood cells after Ficoll separation of mononucleated cells. Total RNA was isolated from peripheral blood or bone marrow after Ficoll separation with RNeasy (Quiagen, Hilden, Germany) following the manufacturer’s instructions. PolyA+ mRNA was separated from total RNA using Oligotexll particels (Quiagen).
RT-PCR.
One microgram total RNA was reverse-transcribed with 200 U Superscript (GIBCO-BRL, Eggenstein, Germany) in a 40-μL reaction using random primers. cDNA equivalent to a quantity of 25 ng reverse-transcribed RNA was amplified for 35 cycles (1 minute at 94°C, 1 minute at 63°C, and 1 minute at 72°C), in 50 μL with 10 pmol of each primer (3.C1 and MLLint), 10 mmol dNTPs, and 1.25 U Taq polymerase (GIBCO-BRL) in the buffer shipped by the supplier. Nested PCR was performed with an aliquot of 1 μL of primary PCR reaction with primers 6.1 and E3AS (Fig1). Nucleotide sequences of the primers were as follows: 3.1C, 5′AGGAGAGAGTTTACCTGCTC3′ (bp 821 to 840); MLLint, 5′CTTCCAGGAAGTCAAGCAAGCAGGT3′ (bp 3869 to 3892); E3AS, 5′ACACAGATGGATCTGAGAGG3′ (bp 567 to 586); and 6.1, 5′GTCCAGAGCAGAGCAAACAG3′ (bp 4013 to 4032). The positions of RT-PCR primers within the MLL gene are given in Fig 1, with sequence numbering according to Tkachuk et al.10
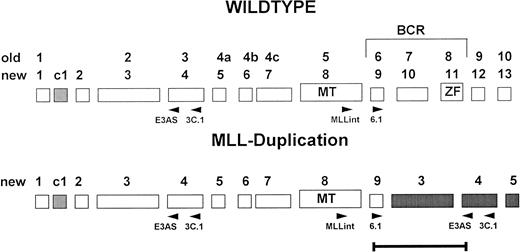
Exon structure of the 5′ region of the MLL gene. Old and new nomenclature (Nilson et al,24) of the exons is indicated. Positions of the primers used for RT-PCR are given below the schematic exons.
Exon structure of the 5′ region of the MLL gene. Old and new nomenclature (Nilson et al,24) of the exons is indicated. Positions of the primers used for RT-PCR are given below the schematic exons.
For each RNA sample, an ABL-specific RT-PCR was performed to control the integrity of RNA using primers abl5′ GGCCAGTAGCATCTGACTTTG and abl3′ ATGGTACCAGGAGTGTTTCTCC. cDNA of an AML with MLL tandem duplication was used as a positive control and water instead of cDNA was included as a blank sample in each experiment.
Peripheral blood from 45 healthy donors was analyzed in a first screening and an additional 20 samples (isolated by caesium chlorid gradient centrifugation and kindly provided by Dr Reis from the Institute of Human Genetics) in a second screening with completely new reagents and performed in a different laboratory building. In this laboratory, no PCR experiments with MLL as a target gene had ever been performed before this series of experiments. In addition, two bone marrow samples from patients with no hematologic malignancies were analyzed.
Genomic PCR.
Seminested PCRs were performed with the same reaction conditions as for RT-PCR with 100 ng of genomic DNA using primers MLLi6 (5′GCTGAGATAGAAGGATTGTCTTG3′ [bp 998 to 1020]40) or MLLi8′ (5′GTCCCAATAATTCCTTTATGGC3′ [bp 5963-5985]40), and MLLi1p (5′AGAGTCAGGTGGCTAACTG3′)41 as external primer in the first 35 cycles. One microliter was further amplified with nested primer MLLi1n 5′CTCCTCTTCAAAGACATCTG3′ (bp 571 to 590; Genbank U66259) for 35 cycles. Both amplifications were performed at 55°C.
Sensitivity.
One microgram of RNA was isolated from approximately 100,000 cells and was reverse-transcribed in one experiment. As we used 1/40 μL of the RT reaction for PCR, we analyzed an equivalent of 25,000 cells per sample. The amount of DNA used for genomic seminested PCR corresponded to approximately 10,000 mononucleated cells.
Dilution experiments with cDNA from a duplication-positive AML and a normal donor sample were performed in 10−1 steps up to 10−8. To equalize the total amount of cDNA in the reaction mixture, cDNA prepared from mouse spleen was added. Mouse spleen RNA was found to be negative for MLL-duplication transcripts.
Sequencing.
RT-PCR products were separated on 2% agarose gels, cut from the gels, and isolated with Quiaex II (Quiagen) following the manufacturer’s instructions. Approximately 100 ng of purified RT-PCR products was directly sequenced with 3.3 pmol of primer 6.1 for forward andE3AS for reverse reactions using the Dye Terminator Cycle Sequencing Kit (Perkin Elmer, Weiterstadt, Germany). After initial denaturation at 95°C for 5 minutes, 25 cycles at 94°C for 15 seconds and 60°C for 4 minutes were performed. Sequence analysis was performed on 6% polyacrylamid gels on an ABI 373 sequencer.
RESULTS
MLL duplication transcripts are detected by nested RT-PCR in peripheral blood of 84% to 100% of normal donors.
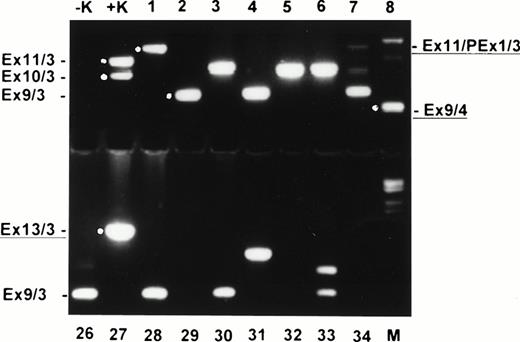
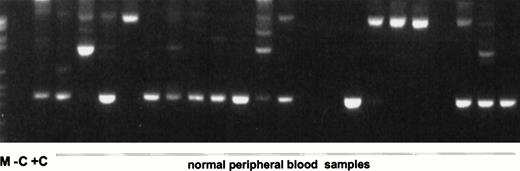
In all 45 normal blood samples, no MLL duplication transcripts were detectable after 35 cycles of the primary PCR (data not shown). After an additional 35 cycles with nested primers, various amplified transcripts were detected in 38 of 45 samples (84.4%) (Fig2). In a second series of experiments in a physically distant laboratory building with completely new reagents, RNA MLL-duplication transcripts were amplified in 20 of 20 samples, probably due to better quality RNA (Fig 3). The sensitivity of the second experiment was 1 log more sensitive than the first series. In the first series, MLL-duplication transcripts were detected solely in undiluted cDNA, whereas in the second series, dilutions of proband material into mouse cDNA at a concentration of 10−1 still showed the duplication transcripts (data not shown).
Ethidium bromide–stained agarose gel with nested RT-PCR products of normal donor blood (samples 1 to 8 and 26 to 34). −K, blank control; +K, AML with MLL duplication; M, molecular weight standard (Boehringer no. VI). Exon fusions are indicated at the left and right. New characterized fusions are underlined.
Ethidium bromide–stained agarose gel with nested RT-PCR products of normal donor blood (samples 1 to 8 and 26 to 34). −K, blank control; +K, AML with MLL duplication; M, molecular weight standard (Boehringer no. VI). Exon fusions are indicated at the left and right. New characterized fusions are underlined.
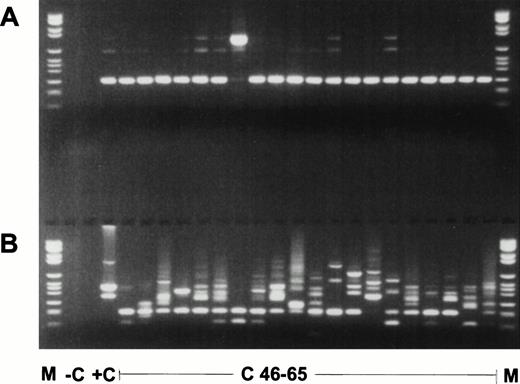
Nested RT-PCR products of the second experiment. (A) ABL PCR; (B) nested PCR for MLL duplication AML with MLL duplication. M, molecular weight standard (Boehringer no. VI); −C, blank control; +C, AML with MLL-duplication; C46-65, pheripheral blood of healthy controls no. 46 to 65.
Nested RT-PCR products of the second experiment. (A) ABL PCR; (B) nested PCR for MLL duplication AML with MLL duplication. M, molecular weight standard (Boehringer no. VI); −C, blank control; +C, AML with MLL-duplication; C46-65, pheripheral blood of healthy controls no. 46 to 65.
Dilution experiments to determine the sensitivity of nested RT-PCR for the detection of MLL-duplication transcripts.
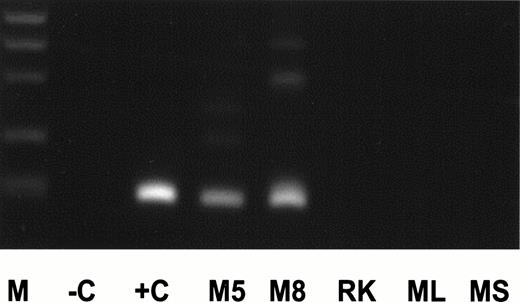
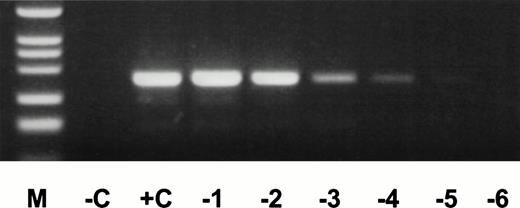
For dilution experiments, tissues of several nonhuman species were tested. Peripheral blood of two individual Maccaca mulattashowed MLL-duplication transcripts with the identical length of an MLL exon 9/exon 3 fusion transcript as in an AML (+C, Fig4). The 200-bp amplification product was sequenced and showed an exon 9/exon 3 fusion. The nucleotide sequence was exactly identical to the human 200-bp fragment. In contrast, no MLL-duplication transcripts were detected in rat kidney, mouse spleen, and liver specimens. Therefore, mouse liver cDNA was used for dilution experiments. cDNA of an AML with an exon 9/exon 3 fusion transcript was diluted into mouse cDNA at concentrations ranging from 100to 10−8. The MLL duplication was detected up to a concentration of 10−5 (Fig5). Thus, the frequency of MLL-duplication transcripts in peripheral blood of healthy individuals is estimated at one in ≤5,000 circulating cells, as 25,000 cells were included per reaction and each sample showed an average of five amplification products.
cDNA of AML positive for an exon 9/exon 3 duplication transcript (+C), peripheral blood of Maccaca mulatta animals no. 5 and 8 (M5, M8), rat kidney (RK), and mouse liver and spleen (ML, MS) was amplified for MLL-duplication transcript. Description of other lanes as in Fig 2.
cDNA of AML positive for an exon 9/exon 3 duplication transcript (+C), peripheral blood of Maccaca mulatta animals no. 5 and 8 (M5, M8), rat kidney (RK), and mouse liver and spleen (ML, MS) was amplified for MLL-duplication transcript. Description of other lanes as in Fig 2.
cDNA of AML positive for an exon 10/exon 3 duplication transcript was diluted into mouse liver cDNA at concentrations ranging from 100 to 10−5 (0 to −5) and MLL-duplication transcripts were amplified by nested PCR. For each experiment, cDNA of ∼−25,000 cells was used. Description of other lanes as in Fig 2.
cDNA of AML positive for an exon 10/exon 3 duplication transcript was diluted into mouse liver cDNA at concentrations ranging from 100 to 10−5 (0 to −5) and MLL-duplication transcripts were amplified by nested PCR. For each experiment, cDNA of ∼−25,000 cells was used. Description of other lanes as in Fig 2.
Sequence analysis of MLL-duplication transcripts.
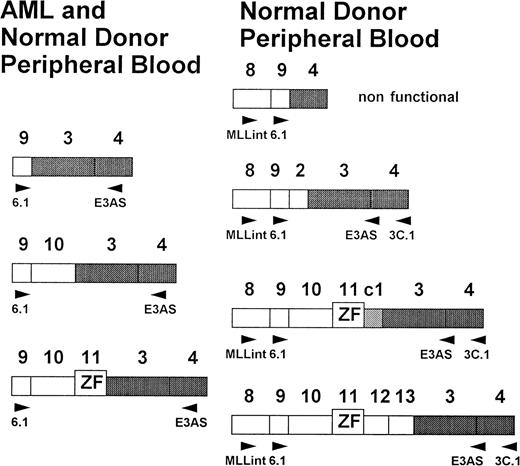
Sequencing of RT-PCR products showed fusions of exons 9, 10, and 11, with exon 3 identical to those found in AML patients.25-27 38 In addition, several new fusion products were identified. Four of these fusions were characterized by sequencing and showed MLL exon 9/exon 4 and exon 9/exon 3, as well as exon 13/exon 3 fusions. One fusion transcript included a cryptic pseudoexon from intron 1 fused to exon 11 (Figs 2 and 6). All transcripts, with the exception of the exon 9/exon 4 fusion, are in frame and potentially translatable. In addition, several yet uncharacterized transcripts were identified (Figs 2 and 3). In the second experiment with high-quality RNA, more than one transcript was detected in all samples (Fig 3). Most samples showed four to six different transcripts. The exon 9/exon 3 fusion transcript, which has been associated with AML, was found to be the most common form (24 of 65 cases). Others, like the large transcript with fusion of exon 11 to a cryptic pseudoexon from intron 1 followed by exon 3 could only be found in single cases. To rule out an RNA-processing artifact in precursor RNA, we isolated mRNA from the total RNA of two normal bone marrow samples and peripheral blood of eight additional normal donors. Amplification products were obtained indicating that at least exon 9/exon 3 and exon 10/exon 3 fusion transcripts, as well as transcripts not observed in AML, were processed to mRNA and thus are potentially functional.
Schematic presentation of the exon fusions (new nomenclature after Nilson et al24). AML specific fusions at the left and new characterized fusion at the right. All fusions can be found in peripheral blood of healthy donors.
Schematic presentation of the exon fusions (new nomenclature after Nilson et al24). AML specific fusions at the left and new characterized fusion at the right. All fusions can be found in peripheral blood of healthy donors.
Genomic MLL duplication in peripheral blood of healthy individuals.
To evaluate whether MLL duplications occur at the genomic level, genomic seminested PCRs for the MLL duplication using intronic primers complementary to introns 1, 9, and 11 were performed. Specific amplification products (Fig 7) identical to those found in AML with MLL duplication were obtained. Sequencing of these products showed Alu sequences (data not shown). This suggests that the duplications are present at the DNA level and are probably Alu-mediated recombinations. Thus, MLL duplications in healthy donors do not seem to be a mechanism at the transcription level or an artifact by RT-PCR. In addition, Southern blots of BamHI andEcoRI with a 0.74-kb MLL cDNA probe encompassing exons 8 to 10 and 12 to 15 (Oncor, Heidelberg, Germany)23only showed germline bands in normal controls, whereas AML samples positive for the duplications all showed additional fragments indicative of MLL rearrangements (data not shown). This is consistent with a higher amount of cells with the duplications in AML versus healthy donors. Our data strongly suggest that MLL duplications occur in a subset of cells in normal hematopoiesis at the genomic level and that MLL-duplication transcripts are detectable in total RNA, as well as in processed mRNA.
Genomic amplification of the duplications using intron 9 and intron 1 primers. M, molecular weight standard; −C, blank control; +C, AML with MLL duplication.
Genomic amplification of the duplications using intron 9 and intron 1 primers. M, molecular weight standard; −C, blank control; +C, AML with MLL duplication.
DISCUSSION
The present study provides convincing evidence that partial tandem duplications within the MLL gene are not restricted to malignant cells in AML, but may also occur in a subset of normal hematopoietic cells. Using a highly sensitive nested RT-PCR approach, duplications within the MLL gene were identified in 38 of 45 and 20 of 20 peripheral blood and two bone marrow samples of healthy donors in two separate sets of analysis. This alteration seems to be a relatively frequent event, as it can be detected in an RNA equivalent of approximately one in 25,000 cells. We found various forms of transcripts, of which all but one are potentially translatable. Genomic PCR data indicate that the MLL duplication transcripts result from MLL duplications at the genomic level and are not a transcriptional or RT-PCR artifact. Furthermore, contamination was formally ruled out by a second series of experiments performed in a physically distant institute with completely new reagents and RNA.
The presence of tumor-associated fusion genes in normal donor blood has been described for the BCL2-IgH transcript specific for germinal center lymphoma with t(14;18) and the BCR-ABL fusion gene, the hallmark of t(9;22)-positive chronic myeloid leukemia. BCL2-IgH transcripts have been shown to be present at a low level, but are detectable with nested RT-PCR in 50% of normal donors.42-44 These findings point to a low-level transcription rate or alternatively to a small subpopulation of cells carrying the translocation t(14;18). Similarly, at least one t(9;22)-specific BCR-ABL fusion transcript in 108 cells has been found in a 30% of healthy donors.45,46 For both translocations, the incidence of fusion transcripts increases with age. It has been postulated that these translocations are first hits in tumor development and that additional genetic alterations are necessary to establish the malignant phenotype. The incidence of MLL duplications in healthy donors seems to be much higher than that of BCL2-IgH or BCR-ABL, as MLL duplications are detectable in nearly all samples and with a less sensitive PCR technique as that described for BCL2-IgH and BCR-ABL. In addition, more than one partially duplicated transcript is usually detected, making up to an average of about 5 different duplications per ≤25,000 cells. Thus, the estimated mean number of duplication-positive cells in healthy blood donors is 1 in ≤5,000 cells, strongly suggesting that the duplication is continuously generated during normal hematopoiesis. Furthermore, new, not previously described transcripts were characterized by sequencing, and genomic fusions were identified by PCR. It should be stressed that in addition to new duplication transcripts, the most frequent duplication transcript is the MLL exon 9/exon 3 fusion, which is also found in AML. The genomic data on the basis of PCR showed identical fragments in healthy donors as in AML. Sequencing of the genomic PCR fragments showed Alu sequences at or near the breakpoints. However, the exact fusion sites were not identified, because genomic fusions resulted into reconstitution of intact ALU sequences. These data are consistent with the hypothesis of homologous Alu recombination within the MLL gene occurring at a relatively high level in most or even all normal individuals. The biologic significance of MLL duplications in healthy individuals is currently unclear. It may be speculated that these transcripts have a special function in a specific subtype of cells. Alternatively, the MLL duplications may represent preleukemic alterations such as the BCL2-IgH and BCR-ABL fusions, and further alterations are required for the induction of the leukemic phenotype. Whether the predominance of only three duplication transcripts in AML is a result of selection for the duplication transcript with higher transforming potential is currently unknown. Another hypothesis may be that highly recombinogenic structures within the MLL gene, probably the frequently occurring ALU repeats, may lead to frequent alterations within the gene. Homologous ALU recombination, leading to partial gene duplication or deletion, has been shown to be implicated in the genesis of a number of nonmalignant diseases.47 A unique feature of partial gene duplication is that the duplicate copy is usually organized in tandem with the original copy in a head to tail orientation.46 Unequal crossing over has generally been accepted as the underlying mechanism. Some of these rearrangements were found to occur within Alu elements,48,49 as was described for duplications within the MLL gene in AML.25 26 In any case, the MLL gene seems to be a target for homologous recombination of Alu repeats in normal individuals and may serve as a model to study Alu recombination in genetic disorders.
Furthermore, our data demonstrate that nested PCR for MLL duplication is not suitable for the detection of minimal residual disease, as has been recently suggested by Satake et al.50 MLL duplication is obviously not a new molecular target for monitoring minimal residual disease in patients with AML and normal karyotype.
NOTE ADDED IN PROOF
ACKNOWLEDGMENT
We greatly acknowledge Dr J. Reis (Institute of Human Genetics, Göttingen) for providing RNA samples. Professor Dönecke (Institute of Biochemistry, Göttingen) is acknowledged for providing laboratory space and equipment for the second series of experiments.
Supported by a program grant of the Deutsche Forschungsgemeinschaft to F.G. and B.W. (SFB500, project A1).
Address reprint requests to Susanne Schnittger, PhD, Department of Hematology/Oncology, University of Göttingen, Robert Koch Str. 40, 37075 Göttingen, Germany.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal