Abstract
Rhnull disease includes the amorph and regulator types that are thought to result from homozygous mutations at theRH30 and RH50 loci, respectively. Here we report an unusual regulator Rhnull where two G→A nucleotide (nt) transitions occurred in trans, targeting different regions of the two copies of Rh50 gene. The nt 836 G→A mutation was a missense change located in exon 6; it converted Gly into Glu at position 279, a central amino acid of the transmembrane segment 9 (TM9). While cDNA analysis showed expression of the 836A(Glu279) allele only, genomic studies showed the presence of both 836A(Glu279) and 836G(Gly279) alleles. A detailed analysis of gene organization led to the identification in the Rh50(836G) allele of a defective donor splice site, caused by a G→A mutation in the invariant GT element of intron 1. This is the first known example of such mutations that has apparently abolished the functional splicing of a pre-mRNA encoding a multipass integral membrane protein. With a silent phenotypic copy intrans, the negatively charged Glu279 residue may disrupt TM9 and impair the interaction of the missense protein with Rh30 polypeptides. To evaluate the significance of the mutation, we took a comparative genomic approach and identified Rh50 homologues in different species. We found that Gly279 is a conserved residue and its adjacent amino acid sequence is identical fromCaenorhabditis elegans to human. These findings provide new insight into the diversity of Rhnull disease and suggest that the C-terminal region of Rh50 may also participate in protein-protein interactions involving Rh complex formation.
© 1998 by The American Society of Hematology.
RH30 POLYPEPTIDES and Rh50 glycoprotein are members of the same family expressed mainly in erythroid cell lineages.1-5 They share a significant sequence homology, as well as a similar 12-transmembrane (TM) topology, but their genetic loci are localized to chromosomes 1p34-36 and 6p11-21.1, respectively.6-8 The RH30 locus comprises two tightly linked, highly homologous genes referred to as RHD andRHCE: the former expresses D antigen and the latter CcEe antigens in the ce, cE, Ce, or CE allelic combinations.9-13The RH50 locus appears to harbor a single copy gene8 14 and its product, the Rh50 glycoprotein, does not carry Rh antigens, but may serve as a coexpressor by forming a membrane complex with Rh30s through protein-protein interactions.
The rare occurrence of Rhnull disease, which manifests a varying degree of chronic hemolytic anemia and stomatocytosis,15,16 highlights a critical role of Rh proteins in maintaining the integrity of the erythrocyte membrane. Classic genetic studies established Rhnull as a recessive disorder that occurs by two distinct mechanisms.15 The amorph type arises by mutations at the RH30 locus itself that silence the gene function, whereas the regulator type results from mutations at a separate suppressor locus. This hypothesis has been confirmd by the identification of various molecular defects in the Rh30 and Rh50 genes in unrelated Rhnullpatients.14 17-19
Recently we characterized the organization of RH50, showing that its coding sequence is distributed in 10 exons.14 This information has facilitated our search for the underlying suppressor mutations in regulator Rhnull probands. Here we describe an unusual example where the phenotypic expression of Rh50 gene is inactivated by two different mutations in trans configuration. One mutation is a G→A transition at +1 position of intron 1, which inactivates the cis-acting GT element of the donor splice site. We show that this mutation has apparently abolished RNA splicing of the mutated Rh50 allele. The other mutation is also a G→A transition, but it causes a Gly279→Glu missense change in the TM9 segment, as recently described by Hyland et al18 in family studies. By searching Rh50 homologues, we show that the Gly279→Glu missense mutation has targeted a protein domain conserved from Caenorhabditis elegansto humans.
MATERIALS AND METHODS
Blood samples, Rh phenotyping, and Western blotting.
Control blood samples were drawn from normal human blood donors. The Rhnull blood sample was from a female proband (YT) of Australian origin. Serologic test confirmed the null status of Rh antigens (D-C-E-c-e- and Rh17-). Typing of Rh-related antigens showed that the proband’s red blood cells lacked the 2D10, LW, and U antigens, but weakly expressed the CD47, S, and s antigens. Western blot was performed using the monoclonal antibodies 2D10 against Rh50 and LOR-15C9 against RhD, as described.20 21
Southern blotting.
Genomic DNA was isolated from leukocytes22 and digested with BamHI and Sph I enzymes. The blots were hybridized with 32P-cDNA probes labeled by the random primer extension method.23 The Rh50 probe was a full-length cDNA isolated from a placenta cDNA library constructed with the Marathon Amplification Kit (Clontech, Palo Alto, CA). This cDNA contains all of the coding sequence of 409 amino acids and spans nucleotides (nt) -6 to 1256.14 Rh30 cDNA probes are region-specific and span, respectively, the 5′ (exon 1-3, nt 1-480), middle (exon 4-7, nt 515-1073) and 3′ portions (exon 8-10 plus 3′-untranslated region [UTR], nt 1074-1456).24
Reverse transcriptase-polymerase chain reaction (RT-PCR).
Total RNA was isolated from hemolysates25 and extracted with Trizol reagent (GIBCO, Gaithersburg, MD). Rh50 and Rh30 transcripts were analyzed by gene-specific RT-PCR and direct sequencing.14,24 Primers for cDNA synthesis and PCR are shown in Table 1. The Rh50 or Rh30 mRNA was converted into cDNA with a 3′-UTR primer and avian myeloblastosis virus (AMV) RT (Promega, Madison, WI). The cDNA was amplified by two pairs of specific primers spanning the entire coding region of Rh50 or Rh30. Conditions for cDNA synthesis and PCR were as detailed previously.14 24 The cDNA was directly sequenced, subcloned, and subjected to single strand conformation polymorphism (SSCP) analysis.
Synthetic Oligonucleotides for Analysis of Rh50 Glycoprotein Gene
| Category* . | Sequence . | nt Position-151 . | Location . |
|---|---|---|---|
| I. cDNA PCR | |||
| 3′-UTa | 5′-AATGGGAAAGGAAGCTGGAGAGCA-3′ | 1321-1298 | 3′-UTR |
| Ex-1s | 5′-AGTGTGCCTCTGTCCTTTGCCACA-3′ | −27-−4 | Exon 1 |
| Ex-4s | 5′-GAAGAGTCCGCATACTACTCAGAC-3′ | 601-624 | Exon 4 |
| Ex-5a | 5′-CTGTTTGTCTCCAGGTTCAGCAAT-3′ | 708-605 | Exon 5 |
| Ex-10a | 5′-CCATGTCCATGGAACTGATTGTCA-3′ | 1256-1233 | Exon 10 |
| II. Genomic PCR | |||
| 5′P2 | 5′-TCCAGCTTTGCTTGACTGATTGCC-3′ | −679-−656 | 5′ region |
| 5′P3 | 5′-CACAGGCTGTGAGATGTTGATAAG-3′ | −36-−54 | 5′ region |
| 1Ps | 5′-AGAACGACAATTAGATTACAGAGC-3′ | −202-−179 | 5′ region |
| In-1a | 5′-CAAAGAGCTCAAGGGTTTATAGTA-3′ | 72-95 | Intron 1 |
| In-1s | 5′-TCCTAGTGTGTTATACAAAGGTGG-3′ | 111-88 | Intron 1 |
| In-2a | 5′-AACACTCTGGCTTCATGACCTGGT-3′ | 117-130 | Intron 2 |
| In-2s | 5′-TGGTGGGAGATGGGATTGCACTTT-3′ | 57-80 | Intron 2 |
| In-3a | 5′-CAAGATTTGCTCCCATATACCGTA-3′ | 72-95 | Intron 3 |
| In-3s | 5′-ATTGACTTTCCTCCCATAAATGAG-3′ | 46-69 | Intron 3 |
| In-4a | 5′-CTGGCTTGGATGTTCTTTTTGAGC-3′ | 81-104 | Intron 4 |
| In-4s | 5′-TGACAGAGGGACACTCCATCTCAA-3′ | 101-124 | Intron 4 |
| In-5a | 5′-CAGCAGTGATGCAGAAATTACCTA-3′ | 67-90 | Intron 5 |
| In-5s | 5′-TTTCAACTGAAGAATGTGATGTGG-3′ | 85-107 | Intron 5 |
| In-6a | 5′-CCAAAAAGAAGGCAAATGGTCTCC-3′ | 111-134 | Intron 6 |
| Ex-6a | 5′-AGTCAGGAACTTGTATCCAAGCAC-3′ | 945-922 | Exon 6 |
| In-6s | 5′-GCCCAGCTATAGCTGTGTTTCAGT-3′ | 57-80 | Intron 6 |
| In-7a | 5′-CTAATGATCTTCTCTCAGGCGCGT-3′ | 129-152 | Intron 7 |
| In-7s | 5′-ATGTTACACTCTGGCCTGAAGATG-3′ | 21-44 | Intron 7 |
| In-8a | 5′-ACTGCCAGAAGTTTAGAACATTTT-3′ | 91-114 | Intron 8 |
| In-8s | 5′-TGTTCTAAACTTCTGGCAGTAAAG-3′ | 111-134 | Intron 8 |
| In-9a | 5′-CCTACCATGCCAGGCTAACTCAAT-3′ | 101-124 | Intron 9 |
| In-9s | 5′-TCCCAGTTCTTTCCAAGCTTGAAT-3′ | 95-118 | Intron 9 |
| 3′-UTa | 5′-AATGGGAAAGGAAGCTGGAGAGCA-3′ | 1321-1298 | 3′ region |
| Category* . | Sequence . | nt Position-151 . | Location . |
|---|---|---|---|
| I. cDNA PCR | |||
| 3′-UTa | 5′-AATGGGAAAGGAAGCTGGAGAGCA-3′ | 1321-1298 | 3′-UTR |
| Ex-1s | 5′-AGTGTGCCTCTGTCCTTTGCCACA-3′ | −27-−4 | Exon 1 |
| Ex-4s | 5′-GAAGAGTCCGCATACTACTCAGAC-3′ | 601-624 | Exon 4 |
| Ex-5a | 5′-CTGTTTGTCTCCAGGTTCAGCAAT-3′ | 708-605 | Exon 5 |
| Ex-10a | 5′-CCATGTCCATGGAACTGATTGTCA-3′ | 1256-1233 | Exon 10 |
| II. Genomic PCR | |||
| 5′P2 | 5′-TCCAGCTTTGCTTGACTGATTGCC-3′ | −679-−656 | 5′ region |
| 5′P3 | 5′-CACAGGCTGTGAGATGTTGATAAG-3′ | −36-−54 | 5′ region |
| 1Ps | 5′-AGAACGACAATTAGATTACAGAGC-3′ | −202-−179 | 5′ region |
| In-1a | 5′-CAAAGAGCTCAAGGGTTTATAGTA-3′ | 72-95 | Intron 1 |
| In-1s | 5′-TCCTAGTGTGTTATACAAAGGTGG-3′ | 111-88 | Intron 1 |
| In-2a | 5′-AACACTCTGGCTTCATGACCTGGT-3′ | 117-130 | Intron 2 |
| In-2s | 5′-TGGTGGGAGATGGGATTGCACTTT-3′ | 57-80 | Intron 2 |
| In-3a | 5′-CAAGATTTGCTCCCATATACCGTA-3′ | 72-95 | Intron 3 |
| In-3s | 5′-ATTGACTTTCCTCCCATAAATGAG-3′ | 46-69 | Intron 3 |
| In-4a | 5′-CTGGCTTGGATGTTCTTTTTGAGC-3′ | 81-104 | Intron 4 |
| In-4s | 5′-TGACAGAGGGACACTCCATCTCAA-3′ | 101-124 | Intron 4 |
| In-5a | 5′-CAGCAGTGATGCAGAAATTACCTA-3′ | 67-90 | Intron 5 |
| In-5s | 5′-TTTCAACTGAAGAATGTGATGTGG-3′ | 85-107 | Intron 5 |
| In-6a | 5′-CCAAAAAGAAGGCAAATGGTCTCC-3′ | 111-134 | Intron 6 |
| Ex-6a | 5′-AGTCAGGAACTTGTATCCAAGCAC-3′ | 945-922 | Exon 6 |
| In-6s | 5′-GCCCAGCTATAGCTGTGTTTCAGT-3′ | 57-80 | Intron 6 |
| In-7a | 5′-CTAATGATCTTCTCTCAGGCGCGT-3′ | 129-152 | Intron 7 |
| In-7s | 5′-ATGTTACACTCTGGCCTGAAGATG-3′ | 21-44 | Intron 7 |
| In-8a | 5′-ACTGCCAGAAGTTTAGAACATTTT-3′ | 91-114 | Intron 8 |
| In-8s | 5′-TGTTCTAAACTTCTGGCAGTAAAG-3′ | 111-134 | Intron 8 |
| In-9a | 5′-CCTACCATGCCAGGCTAACTCAAT-3′ | 101-124 | Intron 9 |
| In-9s | 5′-TCCCAGTTCTTTCCAAGCTTGAAT-3′ | 95-118 | Intron 9 |
| 3′-UTa | 5′-AATGGGAAAGGAAGCTGGAGAGCA-3′ | 1321-1298 | 3′ region |
*Sense (s) and antisense (a) primers to 3′-UT, exon (Ex), 5′-region (P) and intron (In) are denoted.
Nucleotide positions of exon primers are accounted from the first base of initiation codon ATG, whereas intron primers are designated by their distance from the exons they encompass.
Genomic amplification and mutation screening.
The Rh50 and Rh30 gene sequences were amplified using total genomic DNA as template. Based on our published report,14 primers were designed (Table 1) to amplify the Rh50 gene in 12 segments, which together cover the 5′ promoter, 3′-UTR, 10 exons, and flanking splice junctions. Except for 5′P2, 5′P3, 1Ps, Ex-6a, and 3′-UTa, the others primers all reside in noncoding introns. The genomic products were analyzed by restriction digestion, SSCP, subcloning, and sequencing. Exonic sequences of RhD and RhCE were amplified and analyzed by diagnostic restriction enzymes, as detailed previously.24
Subcloning and DNA sequencing.
After an initial identification of the two alterations in Rh50 gene, both cDNA and genomic fragments were subcloned to verify the genotype of the Rhnull proband. The amplified products were ligated into the pCR2.1 TA vector according to the supplier’s specifications (Invitrogen, San Diego, CA). After transformation, positive clones with the insert were screened by color selection with X-Gal and further identified by PCR with the original amplimers. Multiple clones were picked and miniprep plasmids were sequenced on both strands on a Model 373A sequencer (Applied Biosystems, Foster City, CA).
Identification of Rh50 gene homologues.
The discrete domains of human Rh50 were used as queries to search various databases including dbEST (expressed sequence tags) (National Biotechnology Information Center, National Institutes of Health).26 Based on sequence homology, degenerate or exact oligonucleotide primers were designed and used to amplify cDNA or genomic libraries. For analysis of mouse Rh50, the cDNA library was generated by using total RNA isolated from the mouse erythroleukemic (MEL) cell line. The PCR products were sequenced, giving further information on primer design and cDNA PCR. For identification of the Rh50 homologue in C elegans, total RNA was isolated from mixed stage worms using the standard method.27 The cDNA was synthesized and amplified with two exact primers whose sequences were retrieved from GenBank (accession,Z74026).28 For C elegans Rh-1 homologue, the following primers were used: 5′-CCAGAATGTGGCGGTTCTACATCG-3′ (nt -5 to 19, sense, initiation codon underlined), and 5′-TTCTCAATCTCTCCTGTATCCCCC-3′ (nt 1410-1433, 3′-UTR, antisense). For C elegans Rh-2, the two primers were: 5′-CTCTCTTTCAGTAATTCAACC GAAAC-3′ (nt -31 to -6, 5′-UTR, sense) and 5′-CATGAACGACTAACGAGCAATAAAAC-3′ (3′-UTR, antisense). Full-length cDNA sequences were assembled and regions relevant to the Rhnull mutation were aligned for comparison using the DNASIS program (Hitachi, South San Francisco, CA).
RESULTS
Gross structure of Rh50 and Rh30 genes.
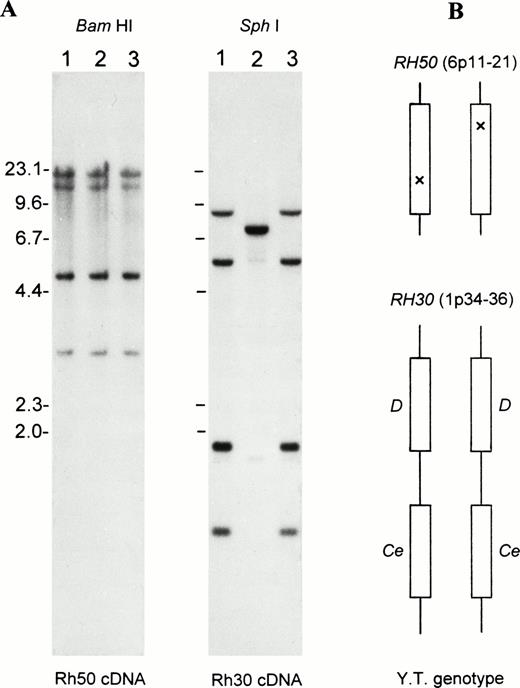
Figure 1 shows blot analysis ofRH50 and RH30 loci. No gross alteration of RH50was noted in the proband (Fig 1A, left). There was also no apparent reduction in band intensity, suggesting the presence of two intact copies of the Rh50 gene. With regard to RH30, the proband showed a banding pattern conforming to the DCe/DCegenotype29 (Fig 1A, right). This genotype was verified by transcript sequencing of RhD and RhCe, whose sequence was identical to that of D+C+c-E-e+ normal subjects. These results excluded the involvement of a mutated RH30 locus in Rhnull(YT), suggesting that some subtle alteration(s) had silenced the phenotypic expression of two copies of the Rh50 gene (Fig 1B).
Gross structure of RH50 and RH30 in Rhnull as determined by Southern blots. (A) Southern blot analysis of the genes encoding the Rh50 and Rh30 proteins. Lane 1, RhD-positive; lane 2, RhD-negative; and lane 3, Rhnull(YT). The BamHI and Sph I blots were probed with the full-length Rh50 cDNA and Rh30 cDNA (exon 4-7), respectively. Size markers (in kb) of Hind III-cleaved lambda phage DNA are shown at the left margin. (B) Schematic representation of the proband’s genotype for RH50 and RH30, whose chromosomal location is indicated. Note that the proband is homozygous for the two tightly linked and actively expressed Rh30 genes, RHD and RHCe. However, the two Rh50 alleles carry different, subtle changes as suppressor mutations (indicated by crosses).
Gross structure of RH50 and RH30 in Rhnull as determined by Southern blots. (A) Southern blot analysis of the genes encoding the Rh50 and Rh30 proteins. Lane 1, RhD-positive; lane 2, RhD-negative; and lane 3, Rhnull(YT). The BamHI and Sph I blots were probed with the full-length Rh50 cDNA and Rh30 cDNA (exon 4-7), respectively. Size markers (in kb) of Hind III-cleaved lambda phage DNA are shown at the left margin. (B) Schematic representation of the proband’s genotype for RH50 and RH30, whose chromosomal location is indicated. Note that the proband is homozygous for the two tightly linked and actively expressed Rh30 genes, RHD and RHCe. However, the two Rh50 alleles carry different, subtle changes as suppressor mutations (indicated by crosses).
Identification of a missense mutation in Rh50 cDNA and Western blot analysis.
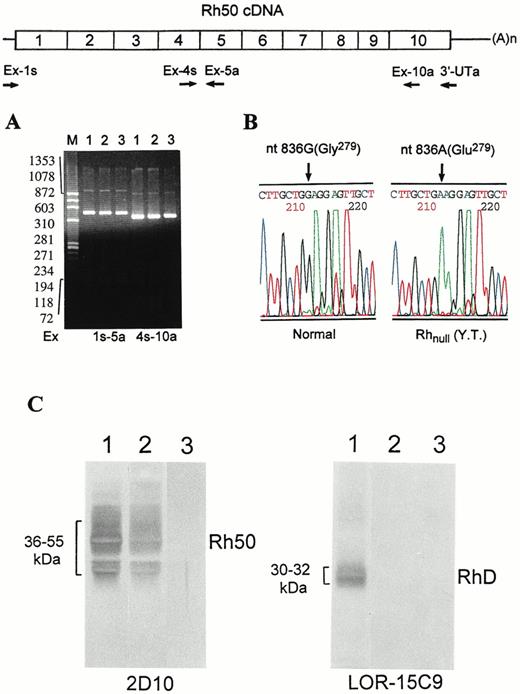
To define the underlying defect, we determined the structure of Rh50 transcripts. Although the Rh50 cDNAs from Rhnull (YT) showed no apparent difference in size (Fig2A), it contained a point mutation, a G→A transition at nt 836 of exon 6 (Fig 2B). This mutation resulted in a Gly279→Glu missense change. Significantly, a single 836A peak is seen in the sequencing profile (Fig 2B), indicating the 836A allele as the only Rh50 transcript expressed in Rhnull cells. To confirm this expression, we subcloned the cDNA, picked 40 independent clones, and performed PCR-SSCP analysis. All of the clones were mutant forms containing the 836A residue (not shown), suggesting that the 836A(Glu279) change is a suppressor mutation. To examine the expression of Rh30 and Rh50, immunoblot analysis of red blood cell membrane proteins was performed. Compared with controls, no bands with either LOR-15C9 or 2D10 were seen in the proband (Fig 2C), suggesting that no significant amount of Rh30 or Rh50 was present in plasma membranes prepared from Rhnull cells.
Identification of nt 836 G→A (Gly279→Glu) missense mutation in the Rh50 transcript. The strategy for synthesis and amplification of Rh50 cDNA is shown. The Rh50 mRNA was reverse-transcribed with the 3′-UTa primer and then amplified with two pairs of upstream primers. (A) The resultant cDNA products, designated by the primers used, were electrophoresed on 1.8% agarose gel. Size markers ofHaeIII-cleaved◊X174 DNA are shown at left. Lanes are designated as in Fig 1. No difference in size is seen between normal subjects and Rhnull(YT). (B) DNA sequencing profiles for the nt 836 G→A missense change. The G residue is seen in normal subjects, whereas the A residue is seen in Rhnull(YT) only. (C) Immunoblot analysis of RhD and Rh50 in red blood cell membranes. Lanes are the same as in (A). The antibodies used are indicated. Note that no band is seen in Rhnull (YT), even though a normal RhD and a missense Rh50 transcript were expressed. Note also that RhD-negative control did not react with LOR-15C9.21
Identification of nt 836 G→A (Gly279→Glu) missense mutation in the Rh50 transcript. The strategy for synthesis and amplification of Rh50 cDNA is shown. The Rh50 mRNA was reverse-transcribed with the 3′-UTa primer and then amplified with two pairs of upstream primers. (A) The resultant cDNA products, designated by the primers used, were electrophoresed on 1.8% agarose gel. Size markers ofHaeIII-cleaved◊X174 DNA are shown at left. Lanes are designated as in Fig 1. No difference in size is seen between normal subjects and Rhnull(YT). (B) DNA sequencing profiles for the nt 836 G→A missense change. The G residue is seen in normal subjects, whereas the A residue is seen in Rhnull(YT) only. (C) Immunoblot analysis of RhD and Rh50 in red blood cell membranes. Lanes are the same as in (A). The antibodies used are indicated. Note that no band is seen in Rhnull (YT), even though a normal RhD and a missense Rh50 transcript were expressed. Note also that RhD-negative control did not react with LOR-15C9.21
Heterozygosity of nt 836A mutation.
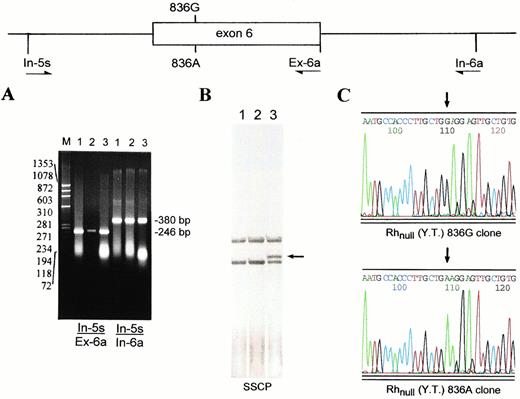
As shown by Southern analysis (Fig 1A), the proband carried two copies of Rh50 gene. The finding that only the 836A transcript was expressed (Fig 2B) suggested that the proband was either a homozygote of the mutation or a double heterozygote with an additional mutation in the other copy of Rh50 gene. To distinguish these possibilities, we determined the genomic sequence encompassing exon 6 (Fig 3A). The exon 6-containing fragment from Rhnull(YT) harbored both G and A at nt 836 (not shown). This allelic coexistence was further indicated by SSCP analysis showing two single-stranded forms (Fig 3B). Sequencing of the subcloned inserts verified the heterozygosity for 836G and 836A alleles in Rhnull (YT) (Fig 3C).
Heterozygosity for the 836G and 836A Rh50 alleles in Rhnull(YT). The genomic region encompassing exon 6 of the Rh50 gene was amplified by two pairs of primers In-5s/Ex-6a and In-5s/In-6a (Table 1). (A) 1.8% agarose gel electrophoresis of the amplified In-5s/Ex-6a (246 bp) and In-5s/In-6a (380 bp) fragments. Lanes are designated as in Fig 1. (B) SSCP analysis of the In-5s/Ex-6a fragments followed by silver staining. The shifted band seen in Rhnull(YT) (lane 3, arrow-indicated) is the single-stranded form containing 836A. (C) Sequencing profiles of the subcloned In-5s/In-6a inserts derived from Rhnull(YT). The presence of both GGA and GAA codons (denoted by two arrows) confirmed the proband to be heterozygous for the missense mutation.
Heterozygosity for the 836G and 836A Rh50 alleles in Rhnull(YT). The genomic region encompassing exon 6 of the Rh50 gene was amplified by two pairs of primers In-5s/Ex-6a and In-5s/In-6a (Table 1). (A) 1.8% agarose gel electrophoresis of the amplified In-5s/Ex-6a (246 bp) and In-5s/In-6a (380 bp) fragments. Lanes are designated as in Fig 1. (B) SSCP analysis of the In-5s/Ex-6a fragments followed by silver staining. The shifted band seen in Rhnull(YT) (lane 3, arrow-indicated) is the single-stranded form containing 836A. (C) Sequencing profiles of the subcloned In-5s/In-6a inserts derived from Rhnull(YT). The presence of both GGA and GAA codons (denoted by two arrows) confirmed the proband to be heterozygous for the missense mutation.
Exon/intron structure and splice donor mutation.
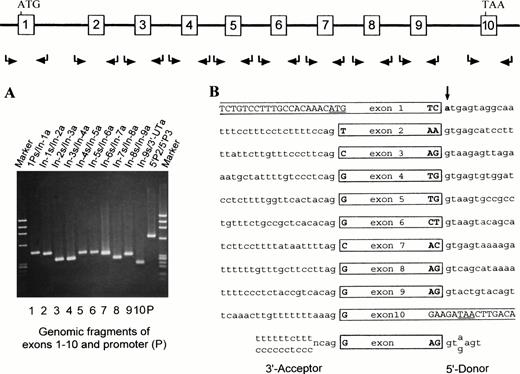
To search for the unknown mutation in the Rh50(836G) allele, genomic fragments for the 5′ and 3′ regions, all exons, and flanking exon/intron junctions were amplified and sequenced (Fig 4A). This screening identified a single G→A transition at +1 position of the donor splice site in intron 1 (Fig 4B). This point mutation was also present in a heterozygous state, as the strength of A and G signals was equal (not shown). Because the G→A transition introduced a novelBspHI site (TC/GTGA to TC/ATGA), a diagnostic assay was done. As shown, the amplified fragments from Rhnull gave rise to two smaller bands for the mutant allele and one undigested band as seen in normal subjects (Fig 5A, see page 1780). Subcloning and sequencing of the proband’s genomic products verified the presence of both wild-type and mutant donor splice sites (Fig 5B, see page 1780). Thus, although the Rh50(836G) allele had a wild-type coding sequence, the splice donor mutation had blocked the functional splicing of its pre-mRNA.
Exon/intron structure and donor splice site mutation of the silent Rh50 (836G) allele. Mutation screening of the silent Rh50(836G) allele in Rhnull (YT) was performed by genomic PCR. The amplified products were each designated by the two primers used (Table 1). (A) 1.8% agarose gel electophoresis of the 11 genomic products. Note that segment 5′P2/5′P3 is upstream of and overlapping with segment 1Ps/In-1a. (B) The exon/intron structure of the silent Rh50 (836G) allele. Nucleotide sequences of the acceptor and donor splice sites are shown and compared with the consensus ones at bottom. Intronic nucleotides are denoted by lower-case letters. The g→a mutation at +1 position of intron 1 that destroys the invariant gt-element of the donor splice site is bolded and marked by an arrow.
Exon/intron structure and donor splice site mutation of the silent Rh50 (836G) allele. Mutation screening of the silent Rh50(836G) allele in Rhnull (YT) was performed by genomic PCR. The amplified products were each designated by the two primers used (Table 1). (A) 1.8% agarose gel electophoresis of the 11 genomic products. Note that segment 5′P2/5′P3 is upstream of and overlapping with segment 1Ps/In-1a. (B) The exon/intron structure of the silent Rh50 (836G) allele. Nucleotide sequences of the acceptor and donor splice sites are shown and compared with the consensus ones at bottom. Intronic nucleotides are denoted by lower-case letters. The g→a mutation at +1 position of intron 1 that destroys the invariant gt-element of the donor splice site is bolded and marked by an arrow.
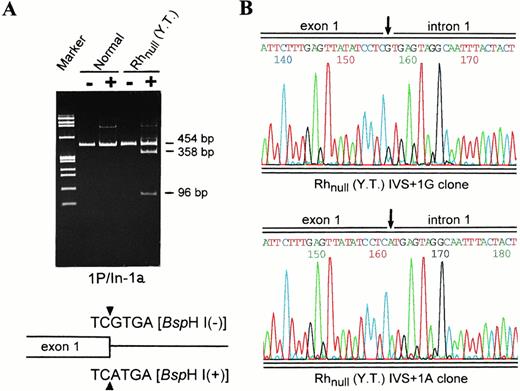
Heterozygosity for the splicing donor mutation as shown by Bsp HI analysis and subcloning. (A) Restriction digestion of genomic segments 1P/In-1a by Bsp HI. Lanes 1 and 2, normal; lanes 3 and 4, Rhnull (YT). (−), uncut; and (+),Bsp HI cut. Note that no digestion is seen in controls. For Rhnull (YT), the size of the uncleaved fragment from the Rh50(836A) allele and the two smaller fragments expected of the Rh50(836G) allele is indicated. Shown schematically at the bottom are the wild-type or mutant exon 1/intron 1 boundary (triangles) and recognition sequence of Bsp HI. (B) Sequencing profiles of the subcloned fragments 1P/In-1a from Rhnull (YT). Two types of the inserts that carry either the G or A residue at +1 position of intron 1 (IVS) were identified, confirming the proband to be heterozygous for the splice donor mutation.
Heterozygosity for the splicing donor mutation as shown by Bsp HI analysis and subcloning. (A) Restriction digestion of genomic segments 1P/In-1a by Bsp HI. Lanes 1 and 2, normal; lanes 3 and 4, Rhnull (YT). (−), uncut; and (+),Bsp HI cut. Note that no digestion is seen in controls. For Rhnull (YT), the size of the uncleaved fragment from the Rh50(836A) allele and the two smaller fragments expected of the Rh50(836G) allele is indicated. Shown schematically at the bottom are the wild-type or mutant exon 1/intron 1 boundary (triangles) and recognition sequence of Bsp HI. (B) Sequencing profiles of the subcloned fragments 1P/In-1a from Rhnull (YT). Two types of the inserts that carry either the G or A residue at +1 position of intron 1 (IVS) were identified, confirming the proband to be heterozygous for the splice donor mutation.
Localization of Gly279→Glu mutation to a conserved protein domain.
Owing to the difficulty in expressing the Rh complex, we took a comparative genomic approach to assess the significance of the Gly279→Glu mutation and to infer its contribution to the Rhnull phenotype. We reasoned that if Gly279 is critical to the function of Rh50, it would be conserved in Rh50 homologues from different species. Homologues were identified in C. elegans and mice and their full-length cDNA sequences were deposited in GenBank. Figure6 shows the alignment of amino acid sequences from the three species in regions relevant to the 836G→A (Gly279→Glu) mutation. Gly279 is indeed a conserved residue from C elegans to human and its adjacent amino acid sequence is identical in a contigous 7-residue stretch among the three species. Notably, the region also is highly conserved in the Rh30 proteins9-13 and in the marine sponge Rh homologue.30
Conservation of Gly279 and TM9 in Rh50 homologues from three species. Comparison of amino acid sequences (equivalent to residues 270-315 in humans) encoded by exon 6 of Rh50 homologues. The Gly279→Glu (G279E) mutation found in Rhnull (YT) is indicated by an arrow. h,Homo sapiens; m, Mus musculus; c,Celegans; and g, Geodia cydonium. Identical amino acids are indicated by colons. Note that exon 6 is identical in size between the human and mouse Rh50, but splits into two exons inC elegans Rh-1. Predicted TM9 and TM10 are boxed. The sequences for human Rh30s, RhCE (upper), and RhD (lower) are also aligned. Note the two consecutive glycine residues (G279G280) and their flanking amino acids are conserved in the Rh family of proteins. In the TM9 of Rh50 homologues, each pair of the three substitutions (A275S, V283I, and T285S) is similar in nature, rendering a 100% similarity between C elegans and Homo sapiens.
Conservation of Gly279 and TM9 in Rh50 homologues from three species. Comparison of amino acid sequences (equivalent to residues 270-315 in humans) encoded by exon 6 of Rh50 homologues. The Gly279→Glu (G279E) mutation found in Rhnull (YT) is indicated by an arrow. h,Homo sapiens; m, Mus musculus; c,Celegans; and g, Geodia cydonium. Identical amino acids are indicated by colons. Note that exon 6 is identical in size between the human and mouse Rh50, but splits into two exons inC elegans Rh-1. Predicted TM9 and TM10 are boxed. The sequences for human Rh30s, RhCE (upper), and RhD (lower) are also aligned. Note the two consecutive glycine residues (G279G280) and their flanking amino acids are conserved in the Rh family of proteins. In the TM9 of Rh50 homologues, each pair of the three substitutions (A275S, V283I, and T285S) is similar in nature, rendering a 100% similarity between C elegans and Homo sapiens.
DISCUSSION
Family studies show that the Rhnull phenotype is often displayed by homozygotes of a given mutation transmitted on consanguineous background.15 However, the regulator form of Rhnull may also result from heterozygosity of compounded mutations in the Rh50 gene. Rhnull (TB) appeared as a case like this, although the exact defect in one of two copies ofRH50 had not been defined.17 Here we characterized the first double heterozygote of regulator Rhnull, showing that RH50 harbored one splice donor mutation and one missense mutation. These two mutations must occur in trans or otherwise the expression of a wild-type allele should be observed. This conclusion is supported by the pattern of family inheritance that is not known to be consanguineous.18 It is also evident that both mutations have contributed to the null phenotype, as neither Rh antigens nor Rh30 and Rh50 proteins were detectable in the red blood cell membrane. Our findings suggest a molecular model in which the mutations each affect RH50 at a different level of gene expression (Fig 7).
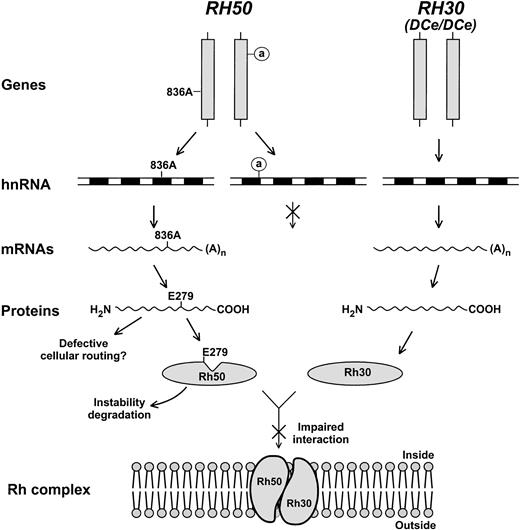
Model for regulator Rhnull caused by two mutations in trans configuration. Of the two mutant copies of RH50 (vertical bars), one contains the 836A mutation and the other carries a defective splice donor in intron 1 (circled a). The RH30 locus is also illustrated whose genomic structure and transcript expression are apparently normal. Diagrammed are the two different levels at which the expression of RH50 is affected (crossed arrows denote blocked steps). The inactive donor is assumed not to alter transcription, but prevents pre-mRNA splicing. This, in turn, leads to degradation of the accumulated hnRNA precursors and no production of mature mRNAs. As a defect likely affecting posttranslational events, E279 (Glu279) may disrupt TM9 of Rh50, alter its conformation (indicated by a notch), and impair the interaction with Rh30. As such, the Rh complex could not be assembled in the cell membrane. Changes of the Rh50 mutant in stability or intracellular routing might also contribute to the failure of Rh complex formation.
Model for regulator Rhnull caused by two mutations in trans configuration. Of the two mutant copies of RH50 (vertical bars), one contains the 836A mutation and the other carries a defective splice donor in intron 1 (circled a). The RH30 locus is also illustrated whose genomic structure and transcript expression are apparently normal. Diagrammed are the two different levels at which the expression of RH50 is affected (crossed arrows denote blocked steps). The inactive donor is assumed not to alter transcription, but prevents pre-mRNA splicing. This, in turn, leads to degradation of the accumulated hnRNA precursors and no production of mature mRNAs. As a defect likely affecting posttranslational events, E279 (Glu279) may disrupt TM9 of Rh50, alter its conformation (indicated by a notch), and impair the interaction with Rh30. As such, the Rh complex could not be assembled in the cell membrane. Changes of the Rh50 mutant in stability or intracellular routing might also contribute to the failure of Rh complex formation.
The intronic G→A transition resides in the invariant GT element of splice donor that directs the processing of exon 1 and, therefore, defines a posttranscriptional splicing defect. Several lines of evidence indicate that the mutation may totally prevent the Rh50(836G) allele from functional splicing in Rhnull cells. (1) No aberrant (whether truncated or elongated) cDNA forms containing 836G were detected. (2) Direct sequencing showed that only the 836A(Glu279) allele was expressed. (3) Forty independent cDNA clones were shown by SSCP analysis to be derived from the 836A allele, but not the 836G allele. As the half-life of hnRNAs is fairly short, the failure to outsplice intron 1 caused by the inactive donor site may lead to a rapid degradation of the mutant pre-mRNA (Fig 7). This is in contrast to the G→A mutation in the splice donor of intron 7 that results in exon skipping and production of a shortened mRNA in another regulator Rhnull patient.14
In a survey of human disease genes related to RNA splicing,31 most mutations occur in internal donor GT and acceptor AG invariant elements.32 Such mutations generally cause a total skipping of the affected exon and may in some cases lead to aberrant splicing by activating cryptic splice sites. Mutations rarely target the splice donor for the extreme 5′ exon. To our knowledge, the splice donor mutation described here is the first example involving a multipass membrane protein gene. The two other examples of such splicing defects are found in human thalassemic β-globin genes that totally abolish the production of mature mRNAs.33,34 This comparison suggests that the splice donor mutations of intron 1 have a more profound effect on pre-mRNA processing than those of internal introns. In fact, the mechanism governing the splicing of 5′ exons in a multiexon gene is not fully understood; the present mutation may offer an interesting model to address how the extreme 5′ exon is scanned and whether its splicing couples with transcription.35 36
As shown, the 836A missense allele was expressed as a normally spliced transcript. Given its location, the point mutation may not impede protein initiation or translation. Thus, its loss of function is more likely related to the Gly279→ Glu change causing some defects in posttranslational events (Fig 7). Gly279 is predicted to lie at center of TM9 of the Rh50 protein,8 and its replacement with a negatively charged, relatively bulky Glu would break up the continuity of hydrophobicity of TM9. This perturbation alone may be sufficient to cause a conformation change, disrupting the assembly of Rh30 and Rh50 as a complex in the cell membrane. Second, Glu279 may render the missense protein defective in cellular routing, as those seen in cystic fibrosis transmembrane conductance regulator (CFTR) and aquaporin-2 mutants.37,38Third, the Glu279-containing mutant itself and Rh30 proteins may become vulnerable to proteolysis if they fail to form a stable, correctly folded complex. Although these hypotheses remain to be examined, the apparent disruption of the Rh complex by the missense change provides evidence for the presence of additional interacting sites in the C-terminal half of Rh50.39
The identification of Rh homologues in the mouse and C elegans shows a high degree of sequence conservation in the TM domains (Fig 6). This finding substantiates our viewpoint that the Gly279→Glu missense change occurs as a loss-of-function mutation. Relevant to this study is the recent characterization of an Rh homologue in the marine sponge G. cydonium.30 These observations together envisage an ancient occurrence of the Rh protein family, reinforcing the hypothesis that the Rh complex and its homologues possess an essential function yet to be identified.1-5 This function is unrelated to antigen presentation, but is critical to the architecture and physiology of plasma membranes, including the red blood cell membrane. Coincidentally, recent database search has related human Rh50 and Rh30 to a large family of genes encoding ammonia transporters.40 It is hoped that a full appreciation of the structure/function relations of the Rh family will emerge from a detailed molecular catalogue of Rhnullmutations along with studies of Rh homologues in other organisms.
ACKNOWLEDGMENT
We are grateful to Marion Reid for the Rhnull blood sample and blood typing, Zhibing Wang for MEL cell total RNA, Anthony Radice for nematode total RNA, and Tian Ye for assistance in library construction. We thank Antonie Blancher for monoclonal antibody LOR-15C9 and Albert von dem Borne for monoclonal antibody 2D10. Thanks are also due to Olga Blumenfeld for comments on the manuscript and Tellervo Huima-Byron and Yelena Oskov for production of photoprints.
Supported in part by Grant No. HL54459 from the National Institutes of Health.
Address reprint requests to Cheng-Han Huang, MD, PhD, Laboratory of Biochemistry and Molecular Genetics, Lindsley F. Kimball Research Institute, New York Blood Center, New York, NY 10021; e-mail:chuang@nybc.org.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal