Abstract
Allelotype analysis of adult T-cell leukemia (ATL) was undertaken for the first time to identify chromosomal loci relevant to the development of acute/lymphomatous ATL. Loss of heterozygosity (LOH) was screened using 94 highly polymorphic microsatellite markers, distributed among all nonacrocentric, autosomal chromosomes. In each of the 22 cases, DNA obtained from their leukemic cells in acute/lymphomatous phase was compared with their constitutional DNA from mononuclear cells in chronic or remission phase. Allelic losses of at least on one chromosome arm occurred in 91% of the cases (20 individuals). Among 39 chromosome arms, allelic losses were observed on 31 arms at least for one sample. A high frequency of allelic loss (>30%) was seen on chromosome arms 6q (41%) and 17p (48%). The mean fractional allelic loss (FAL) was 0.109. These findings suggest that a novel tumor suppressor gene on chromosome arm 6q, as well as the p53 gene on chromosome arm 17p, probably have an important role in the development of acute/lymphomatous ATL.
© 1998 by The American Society of Hematology.
INITIATION AND PROGRESSION of neoplasia can include inactivation of one or several tumor suppressor genes. The paradigm of alterations of a tumor suppressor gene is a mutation of one allele and loss of the second allele.1 Such losses may involve deletions too small to be detected by conventional cytogenetics. This reduction to homozygosity can be detected by the finding of loss of heterozygosity (LOH) of polymorphic sequences in tumor DNA as compared with constitutional DNA from the same individual. Several tumor suppressor genes, such as the DCC gene, have been cloned from the hot spot of LOH analysis.2-5 Thus, LOH analysis is an indirect method to search for a tumor suppressor gene. Allelotype analysis is an extensive survey of allelic loss throughout the genome, screening multiple loci for regions containing tumor suppressor genes that are likely to be altered.5-15 This technique has recently become extremely powerful with the use of microsatellite markers, which are short tracts of (CA)n repeats with high polymorphism that exists throughout the genome.16 17
Adult T-cell leukemia (ATL) is an aggressive, fatal malignancy of mature CD4+ T lymphocytes.18,19 The human T- cell lymphotropic virus type I (HTLV-I) has been recognized as the etiologic agent of ATL.20-22 However, a long period of clinical latency (mean, 55 years) precedes the developement of ATL23 and only a small percentage of HTLV-I-infected individuals develop this malignancy (one patient/1,000 to 2,000 carriers/year),24,25 indicating that additional genetic events probably are required to develop ATL after viral infection of the target T cells. Statistical analysis suggests that ATL arises following five independent genetic events.26
Studies to date by ourselves and others have implicated deletions or mutations of several tumor suppressor genes in the pathogeneisis of ATL, including p53 in about 40% of acute/lymphomatous ATL,26-28 and the p15INK4B, p16INK4A, and Rb genes in approximately 30%, 35%, and 5% of such cases, respectively.29-32 In addition, loss of expression of p18INK4C was found in ATL cell lines.33 To begin to define further genetic events leading to the evolution of acute/lymphomatous ATL, we performed comprehensive allelotype analysis of ATL using 94 microsatellite markers that covered all autosomal chromosomes.
MATERIALS AND METHODS
Samples.
Patients with acute/lymphomatous ATL were investigated; 21 samples were from Japanese patients and one was from an individual from Panama. The percentage of contaminating normal cells in the acute/lymphomatous phase samples was at most 30% and usually less than 10%. The corresponding control DNAs were obtained from either their peripheral blood after complete remission (n = 17) or during their chronic phase (n = 5). The clinical subtypes of ATL were based on the diagnostic criteria proposed by the Lymphoma Study Group of Japan.34Cytogenetic data were available from five patients. All five cases showed chromosomal abnormalities: patient O, 45X,add(X),-4, -6,del(7)(p15),add(10)(p11), add(11)(q23), add(12)(q11),add(14)(p11),- 21, add(22)(p11),+2mar[6]/46XY[7]; patient P, 45XY,-3; patient Q, 45X,-Y,add(3)(p11q21), add(7)(p11), del(9)(q13q21),add(10)(p13),add(12)(q13), der(15)t(3;15)(p14;p11),-20,+mar[14]/46, idem+Y[10]/46XY[10]; patinet S, 48XY,+2,-3,-6, +add(6)(q32),-10,+add(7)(p13), add(7)(q32),-10, +add(10)(q22), +13,-15,-18,-21,+6,mar; patient U, 47XX,+mar[1]/46XX[3].
Allelic loss analysis.
Primers for polymerase chain reaction (PCR) amplification of microsatellite markers, chosen to represent at least two loci on each chromosomal arm, were obtained from Research Genetics (Huntsville, AL). Ninty-four markers distributed among all 39 autosomal chromosome arms, except for nonacrocentric arms, were analyzed. These markers and their chromosomal regions are detailed in Table1. PCRs were performed as described.35 Briefly, total reaction volumes were 20 μL containing 25 ng DNA, 1.5 mmol/L MgCl2, 10 pmol/L of each of the primers, 2 nmol/L of each of the four deoxyribonucleotide triphosphates (dNTP; Pharmacia, Stockholm, Sweden), 1 U of Taq DNA polymerase (GIBCO-BRL, Gaithersburg, MD), and 2 μCi 32P-labeled deoxycytidine triphosphates (dCTP) (3,000 μCi/mmol; New England Nuclear/Dupont, Boston, MA) with specified buffer provided by the supplier. To ascertain either LOH or duplication of the region, PCR reaction was performed in a multiplex fashion for some of the markers; the reaction mixture contained two primer sets. PCR consisted of 40 seconds at 94°C, 30 seconds at 55°C, and 1 minute at 72°C for 27 to 33 cycles in a Programmable Thermal Controller (MJ Research Inc, Water Town, MA) in order to examine the products in the linear range of signals. PCR products were mixed with a formamide gel-loading solution, heat denatured at 94°C, separated on a denaturing 5% to 8% polyacrylamide gel, and visualized by autoradiography. Allele losses were determined by visual inspection. When visible reduction of radiographic signal was equivocal, a radioanalytic imaging detector (Ambis, Ambis Inc, San Diego, CA) was used to confirm our interpretation. All positive results were repeated for confirmation.
Microsatellite Markers Used for Each Chromosomal Arm for Allelotyping of ATL
| Chromosome Arm . | Markers . |
|---|---|
| 1p | D1S199, D1S244 |
| 1q | D1S196, D1S251 |
| 2p | D2S113, D2S123, D2S146 |
| 2q | D2S122, D2S155 |
| 3p | D3S1266, D3S1285 |
| 3q | D3S1272, D3S1299 |
| 4p | D4S418, D4S419 |
| 4q | D4S408, D4S416 |
| 5p | D5S416, D5S418 |
| 5q | APC, MDF27 |
| 6p | D6S260, D6S265 |
| 6q | D6S284, D6S294, D6S1601, D6S1627 |
| 7p | D7S493, D7S510, D7S517 |
| 7q | D7S487, D7S522, D7S677 |
| 8p | D8S262, D8S283 |
| 8q | D8S272, D8S286 |
| 9p | D9S126, D9S157, D9S165, D9S168, D9S171, INFA |
| 9q | D9S154, D9S176 |
| 10p | D10S191, D10S197 |
| 10q | D10S190, D10S201 |
| 11p | D11S902, D11S907 |
| 11q | D11S924, D11S1391, D11S2179 |
| 12p | D12S89, D12S308, D12S364 |
| 12q | D12S84, D12S346 |
| 13q | D13S156, D13S221 |
| 14q | D14S61, D14S64, D14S72 |
| 15q | D15S126, D15S128, D15S165 |
| 16p | D16S406, D16S418 |
| 16q | D16S402, D16S514 |
| 17p | MDF41, TP53, D17S786 |
| 17q | D17S802, D17S805 |
| 18p | D18S54, D18S452 |
| 18q | DCC, D18S58, D18S67 |
| 19p | D19S221, D19S226, D19S424 |
| 19q | D19S180, D19S214 |
| 20p | D20S98, D20S116 |
| 20q | D20S100, D20S108 |
| 21q | D21S265, D21S270 |
| 22q | D22S280, D22S283 |
| Chromosome Arm . | Markers . |
|---|---|
| 1p | D1S199, D1S244 |
| 1q | D1S196, D1S251 |
| 2p | D2S113, D2S123, D2S146 |
| 2q | D2S122, D2S155 |
| 3p | D3S1266, D3S1285 |
| 3q | D3S1272, D3S1299 |
| 4p | D4S418, D4S419 |
| 4q | D4S408, D4S416 |
| 5p | D5S416, D5S418 |
| 5q | APC, MDF27 |
| 6p | D6S260, D6S265 |
| 6q | D6S284, D6S294, D6S1601, D6S1627 |
| 7p | D7S493, D7S510, D7S517 |
| 7q | D7S487, D7S522, D7S677 |
| 8p | D8S262, D8S283 |
| 8q | D8S272, D8S286 |
| 9p | D9S126, D9S157, D9S165, D9S168, D9S171, INFA |
| 9q | D9S154, D9S176 |
| 10p | D10S191, D10S197 |
| 10q | D10S190, D10S201 |
| 11p | D11S902, D11S907 |
| 11q | D11S924, D11S1391, D11S2179 |
| 12p | D12S89, D12S308, D12S364 |
| 12q | D12S84, D12S346 |
| 13q | D13S156, D13S221 |
| 14q | D14S61, D14S64, D14S72 |
| 15q | D15S126, D15S128, D15S165 |
| 16p | D16S406, D16S418 |
| 16q | D16S402, D16S514 |
| 17p | MDF41, TP53, D17S786 |
| 17q | D17S802, D17S805 |
| 18p | D18S54, D18S452 |
| 18q | DCC, D18S58, D18S67 |
| 19p | D19S221, D19S226, D19S424 |
| 19q | D19S180, D19S214 |
| 20p | D20S98, D20S116 |
| 20q | D20S100, D20S108 |
| 21q | D21S265, D21S270 |
| 22q | D22S280, D22S283 |
RESULTS
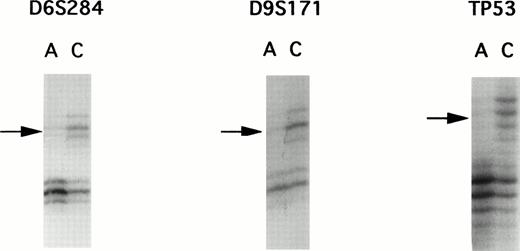
A panel of 94 highly informative microsatellite markers representing every autosomal chromosome were used to screen 22 matched ATL samples. Although PCR allelotyping is capable of showing losses of genetic material in tumors, it is unsuitable for detection of gene amplifications. To rule out the possibility of gene amplification, we performed multiplex PCR to compare the intensity of two loci. Because gene amplification was not found, each of the allelic imbalances were determined to be LOH. Representative examples of autoradiograms demonstrating LOH are shown in Fig 1.
Autoradiographs from LOH analysis with microsatellite markers. PCR products were separated by polyacrylamide gel electrophoresis, showing examples of allelic loss found in patient E. DNA samples isolated from leukemic cells in the acute phase are labelled as A and those DNA samples isolated from corresponding normal peripheral leukocytes after complete remission are designated as C. Microsatellite markers are indicated above the autoradiograms. Arrows, loss of one allele.
Autoradiographs from LOH analysis with microsatellite markers. PCR products were separated by polyacrylamide gel electrophoresis, showing examples of allelic loss found in patient E. DNA samples isolated from leukemic cells in the acute phase are labelled as A and those DNA samples isolated from corresponding normal peripheral leukocytes after complete remission are designated as C. Microsatellite markers are indicated above the autoradiograms. Arrows, loss of one allele.
For each patient, we obtained information for at least 30 (76.9%) chromosomal arms (Table 2). All but two ATLs (patients N and U) showed evidence of a deletion at one or more loci with loss(es) occurring in one to 13 chromosomal arms. Sixteen samples showed losses in more than two different chromosomal arms. Patients E and G had allele loss affecting over 30% of informative chromosomal arms.
Allelic Losses in Chromosomal Arms and FAL
| Samples . | Chromosomal Arms With LOH . | No. of Informative Arms . | FAL . |
|---|---|---|---|
| A | 16p, | 30 | 0.033 |
| B | 20p, 20q, | 31 | 0.065 |
| C | 2p, 7q, 8p, 17q, 18q, 22q, | 35 | 0.171 |
| D | 3q, 4p, 5q, 6q, 9p, 18q, 22q, | 34 | 0.206 |
| E | 2q, 6p, 6q, 9p, 9q, 14q, 17p, 17q, 18q, 22q, | 30 | 0.333 |
| F | 6q, 17p, 18q, | 34 | 0.088 |
| G | 6p, 6q, 8p, 10p, 12p, 12q, 13q, 16p, 17p, 17q, 18q, 19p, 22q, | 35 | 0.371 |
| H | 6p, 6q, 7q, | 32 | 0.094 |
| I | 10q, | 33 | 0.030 |
| J | 17p, | 31 | 0.032 |
| K | 17p, | 34 | 0.029 |
| L | 6q, 4q, 13q, 17p, | 37 | 0.108 |
| M | 6p, 17q, | 33 | 0.061 |
| N | 31 | 0 | |
| O | 2q, 4p, 4q, 12p, 12q, 17p, | 36 | 0.167 |
| P | 6q, 7p, 7q, 12p, | 36 | 0.111 |
| Q | 8q, 17p, | 31 | 0.065 |
| R | 7p, | 37 | 0.027 |
| S | 4p, 6q, 7p, 7q, 8p, 10p, 10q, 18p, | 34 | 0.235 |
| T | 6q, 2p, 20q, | 33 | 0.091 |
| U | 33 | 0 | |
| V | 7q, 9p, 17p, | 33 | 0.091 |
| Samples . | Chromosomal Arms With LOH . | No. of Informative Arms . | FAL . |
|---|---|---|---|
| A | 16p, | 30 | 0.033 |
| B | 20p, 20q, | 31 | 0.065 |
| C | 2p, 7q, 8p, 17q, 18q, 22q, | 35 | 0.171 |
| D | 3q, 4p, 5q, 6q, 9p, 18q, 22q, | 34 | 0.206 |
| E | 2q, 6p, 6q, 9p, 9q, 14q, 17p, 17q, 18q, 22q, | 30 | 0.333 |
| F | 6q, 17p, 18q, | 34 | 0.088 |
| G | 6p, 6q, 8p, 10p, 12p, 12q, 13q, 16p, 17p, 17q, 18q, 19p, 22q, | 35 | 0.371 |
| H | 6p, 6q, 7q, | 32 | 0.094 |
| I | 10q, | 33 | 0.030 |
| J | 17p, | 31 | 0.032 |
| K | 17p, | 34 | 0.029 |
| L | 6q, 4q, 13q, 17p, | 37 | 0.108 |
| M | 6p, 17q, | 33 | 0.061 |
| N | 31 | 0 | |
| O | 2q, 4p, 4q, 12p, 12q, 17p, | 36 | 0.167 |
| P | 6q, 7p, 7q, 12p, | 36 | 0.111 |
| Q | 8q, 17p, | 31 | 0.065 |
| R | 7p, | 37 | 0.027 |
| S | 4p, 6q, 7p, 7q, 8p, 10p, 10q, 18p, | 34 | 0.235 |
| T | 6q, 2p, 20q, | 33 | 0.091 |
| U | 33 | 0 | |
| V | 7q, 9p, 17p, | 33 | 0.091 |
Abbreviation: FAL, fractional allelic loss.
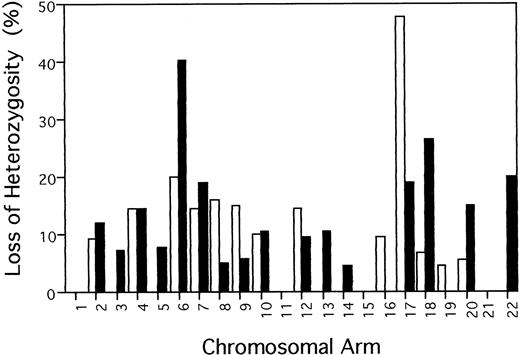
Of the 39 chromosomal arms anaylzed, 29 (74.4%) showed LOH in one or more samples. Figure 2 summarizes the percentage of allelic loss at each chromosomal arm, which was calculated by dividing the number of tumors with LOH at any marker on the chromosmome arm by the total number of informative cases. Frequency of LOH for individual chromosomal arms varied between 0% (1p, 1q, 3p, 5p, 11p, 11q, 15q, 16q, 19q, and 21q) and 47.6% (17p). Over 30% of informative acute/lymphomatous samples showed LOH at chromosomal arms 6q (9/22, 40.9%) and 17p (10/21, 47.6%). Of the 22 samples, five, six, and four cases had LOH on 6q alone, 17p alone, and both 6q and 17p, respectively. In total, 15 samples had LOH on 6q and/or 17p.
Frequency of allelic loss on each chromosome arm in 22 acute/lymphomatous ATLs. Percentage of LOH was calculated for each chromosome arm by dividing the number of tumors with LOH at any marker on the chromosmome arm by the total number of informative cases. (□) p; (▪) q.
Frequency of allelic loss on each chromosome arm in 22 acute/lymphomatous ATLs. Percentage of LOH was calculated for each chromosome arm by dividing the number of tumors with LOH at any marker on the chromosmome arm by the total number of informative cases. (□) p; (▪) q.
DISCUSSION
To our knowledge, this is the first genomic search in ATL using microsatellite markers; the aim was to identify chromosomal regions likely to contain important tumor suppressor genes involved in the progression of acute/lymphomatous ATL. The analysis showed two hot-spots of LOH (>30%), one on 6q and the other one on 17p. Although it is possible that candidate tumor suppressor genes are present in the undeleted segments of DNA, our result suggests that tumor suppressor genes for progression of acute/lymphomatous ATL probably reside on chromosome 6q and 17p.
With regard to tumor suppressor genes, mutations of p53 are among the most common genetic changes in human tumors.36 The greatest frequency of LOH in acute/lymphomatous ATL was observed at the TP53 locus (seven of 15, 46.7%) on chromosome 17p. This result is congruent with previous reports by ourselves and others that the p53 gene was mutated in about 40% of acute/lymphomatous ATL samples.26-28 Although point mutations of the p53 gene in our samples were not examined, we suspect that those with LOH at TP53 had mutations of the gene.
The high frequency of allelic loss affecting 6q is a novel finding in ATL and suggests that this is the site of an as yet uncharacterized tumor suppressor gene(s). The LOH for 6q is in keeping with results in several other types of tumors, including those of the kidney,37 breast,4,38 ovary,39,40liver,41 and prostate,42 as well as melanoma,43-46 lymphoma/lymphoblastic leukemia,47-52 and parathyroid adenoma.53Further studies are required to define the altered gene on chromosome 6q in ATL and determine whether inactivation of the same gene occurrs in all of the cancer types with LOH in this chromosomal region.
The tumor suppressor gene p16INK4A was mapped to chromosome 9p21.54,55 We previously found that this gene was deleted in about 35% of acute/lymphomatous ATL samples.28 29 In this study, we analyzed for LOH at D9S171, which maps to 9p21 about 1 megabase proximal to the p16INK4A gene. Only eight cases were informative at D9S171 and two of them had LOH. The frequency of heterozygosity for this locus is lower than that in the Genome Database (0.79). Such discordance could be due to homogeneity of the Japanese population. In addition, we may have missed some homozygous deletions because homozygous deletions for a polymorphic marker may appear as retention of heterozygosity as a consequence of amplification of DNA from the contaminating normal cells. Therefore, we could have underestimated the frequency of LOH at D9S171.
ATL is frequently characterized by an acquisition of nonrandom chromosomal changes,56 but cytogenetic changes from chronic to acute ATL have been rarely reported. Additionally, small deletions are below the limits of resolution of cytogenetic analysis. Karyotype analyses were available for only five samples in our study. As expected, in four of these five samples, LOH studies identified additional abnormalities not detected in the cytogenetic analysis. Thus, cytogenetic studies have probably missed most of the interstitial deletions. Nevertheless, some of the chromosomal deletions shown by cytogenetic analyses were not detected by our LOH analyses. Most of these samples had noninformative microsatellite loci in these regions. The discrepancy between cytogenetic and LOH studies in several samples can be ascribed to a failure to examine a microsatellite marker in the deleted chromosomal region.
The mean FAL was 0.109 in acute/lymphomatous ATL, which is lower than that in solid tumors.4,7,10,57 58 This suggests that ATL cells may be genetically more stable than those of solid tumor cells.
In summary, the comprehensive allelotyping of ATL has provided novel signposts for future efforts at positional candidate cloning of human tumor suppressor genes involved in progression of ATL. Likewise, these genes will be likely targets for development of other hematologic malignancies, as well as other human tumors.
Supported in part by Grants No. CA26038, CA42710, and CA70675 from the National Institues of Health, Bethesda, MD; the Concern Foundation, Beverly Hills, CA; and the Parker Hughes Fund, Glendale, CA.
Address reprint requests to Yoshihiro Hatta, MD, Division of Hematology/Oncology, Department of Medicine, Cedars-Sinai Research Institute, UCLA School of Medicine, 8700 Beverly Blvd, B-208, Los Angeles, CA 90048.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal