Abstract
Using a highly sensitive fluorescence in situ hybridization method with probes for BCR and ABL1 (D-FISH), we studied 37 paired sets of bone marrow and blood specimens, collected within 24 to 96 hours of each other, from 10 patients before and during treatment for chronic myeloid leukemia (CML). The normal range for 500 interphase nuclei was ≤4 (≤0.8%) nuclei based on 10 bone marrow and 10 blood specimens from normal individuals. The percentage of neoplastic nuclei was usually lower in blood than bone marrow. However, changes in the percentage of neoplastic nuclei in blood and bone marrow tracked closely over the course of therapy and with the results of quantitative cytogenetic studies on bone marrow. This result indicates that D-FISH is useful to test blood from patients with CML to monitor therapy. Moreover, by analysis of 6,000 nuclei with D-FISH, residual disease was identified in bone marrow and blood for patients in complete cytogenetic remission. Consequently, D-FISH analyses of interphase nuclei from blood could substitute for Q-cytogenetic studies on bone marrow. Thus, it may not be necessary to collect bone marrow samples so frequently to monitor therapy in CML.
CONVENTIONAL CYTOGENETIC studies are used in clinical practice to monitor the effectiveness of various forms of treatment for patients with chronic myeloid leukemia (CML), especially interferon therapy. Considerable evidence exists to show a strong correlation between changes in percentage of Ph-positive metaphases after interferon therapy and prognosis.1-3 The best outcome for survival and prolonged chronic phase seems to be enjoyed by patients with CML in whom the percentage of Ph-positive metaphases is reduced to less than 33%.2-5
In clinical practice, physicians usually collect bone marrow aspirates from patients with CML on interferon therapy at 3- to 6-month intervals to obtain cytogenetic data. For technical reasons, such as a packed bone marrow or hypoplasia, it is not always possible to obtain suitable bone marrow specimens for chromosome studies. Moreover, undergoing bone marrow aspiration or biopsy is painful and costly. Although peripheral blood is easier to collect from patients, in our experience, the number of mitotic cells in blood after treatment is usually inadequate to accurately quantify disease.
The advent of fluorescence in situ hybridization (FISH) to detect BCR/ABL fusion in interphase nuclei for patients with CML has become important to quantify disease.6-8 Until recently, the most common FISH procedure for CML used different colored probes for BCR and ABL and detected a single BCR/ABL fusion signal in cells with a Ph chromosome. For purposes of this paper, we refer to this method as S-FISH to imply a single BCR/ABL fusion signal. Some investigators have used S-FISH to show the presence of nuclei with BCR/ABL fusion in blood, but few data are available to show the efficacy of FISH to study peripheral blood to monitor therapy in CML.6-12 S-FISH lacks sensitivity to detect low levels of minimal residual disease and also lacks precision to quantify disease accurately before and after treatment.8 13
New FISH strategies are now available that are highly sensitive to detect BCR/ABL fusion in interphase nuclei.14,15 Recently we investigated the use of one of these new methods called D-FISH to study bone marrow.15 D-FISH detects double or two BCR/ABL fusion signals in cells with a t(9;22)(q34;q11.2) in most patients with CML and the false positive and false negative frequency of D-FISH approaches zero. D-FISH accurately quantifies disease in bone marrow from patients with CML within a few percentage points at diagnosis and at all times after treatment including cytogenetic remission. In addition, D-FISH identifies all known variants of the Ph chromosome translocation and the percentage of abnormal interphase nuclei correlates closely with quantitative cytogenetic studies (Q-cytogenetics) for bone marrow.
The present investigation was designed to test the usefulness of D-FISH to study peripheral blood for purposes of monitoring the effectiveness of interferon α-2b therapy for patients with CML. To do this, paired sets of blood and bone marrow specimens were collected from a series of patients enrolled in the CML National Study Group who were undergoing therapy with either interferon α-2b alone or interferon α-2b and cytosine arabinoside (ara-C). The results of this investigation show that changes in the percentage of neoplastic nuclei in blood over the course of therapy were a good predictor of corresponding changes in bone marrow. In a previous study, the percentage of interphase nuclei with BCR/ABL fusion was strongly correlated with Ph-positive metaphases by Q-cytogenetics.15 D-FISH was also useful to identify residual disease in both bone marrow and peripheral blood specimens for patients in complete cytogenetic remission.
MATERIALS AND METHODS
This investigation used D-FISH to study 37 paired sets of bone marrow and peripheral blood specimens from 10 patients undergoing treatment for CML, 10 normal peripheral blood specimens, 10 normal bone marrow specimens, and 4 serial dilutions with known percentages of Ph-positive nuclei.
Each patient with CML was a participant in the CML National Study Group clinical trial and was randomly receiving treatment with interferon α-2b with or without ara-C. Each patient was known to have cells with a Ph chromosome that produced a typical D-FISH pattern for t(9;22)(q34;q11.2).15 For each patient a paired set of bone marrow and peripheral blood specimens was collected before treatment and at two or more times at approximately 4-month intervals during treatment. Each paired set of peripheral blood and bone marrow specimens was obtained on the same day except for specimens collected before treatment in patients 3 (blood and bone marrow were collected 1 day apart), 5, and 8 (blood and bone marrow were collected 4 days apart).
Uncultured bone marrow and peripheral blood specimens were processed by conventional procedures for cytogenetic and FISH studies. These specimens were stored as fixed pellets at −70°C in methanol:acetic acid (3:1) until FISH studies could be performed. To prepare specimens for D-FISH, specimens were washed twice with fresh fixative and cells were placed on microscope slides and allowed to air-dry in a CDS-5 cytogenetic drying chamber (Thermotron, Holland, MI) adjusted to 50% relative humidity and 25°C.16 Slides were further dried for 1 hour in a 65°C oven and then treated with 2× standard saline citrate solution (SSC; 300 mmol/L sodium chloride, 30 mmol/L sodium citrate) for 1 hour at 37°C. Slides were then dehydrated with a series of 70% to 85% to 100% ethanol at −20°C for 2 minutes each, and air-dried.
Q-cytogenetic studies were performed on each bone marrow specimen by analyzing 25 consecutive G-banded or Q-banded metaphases in which chromosomes 9 and 22 could be observed.17 Hypermetaphase studies using S-FISH with probes for BCR and ABL were performed on many of these specimens.18 The D-FISH procedure was performed according to the method of Dewald et al.15 Chromosomal DNA was denatured in 70% formamide/2× SSC for 2 minutes at 70°C. Slides were dehydrated with an ethanol series (70%, 85%, and 100%) for 2 minutes each and air-dried. The probe was denatured in a water bath at 70°C for 5 minutes. Then 10 μL of BCR/ABL probes were added to each slide, and a 22 × 22–mm coverslip was placed on the slide and sealed with rubber cement. Slides were hybridized for 18 to 20 hours at 37°C in a humidified chamber. After the coverslips were removed, slides were washed for 2 minutes in 0.4× SSC at 70°C, and then in 1× phosphate-buffered detergent for 2 minutes. Chromatin was counterstained in blue with 10 μL of 1% solution of 4′,6′-diamidine-2-phenylindole in Vectashield antifade. Representative cells were captured using a computer-based imaging system (Quips XL Genetics Workstation; Vysis, Inc, Downers Grove, IL).
D-FISH was performed using directly labeled BCR and ABL1 probes (Oncor Inc, Gaithersburg, MD) to show two BCR/ABL fusion signals in cells with a t(9;22)(q34;q11.2), one on the abnormal chromosome 9 and the other on the abnormal chromosome 22. The ABL1 (400 kb) probe set was directly labeled with Rhodamine Green (green signal) and included several DNA sequences that hybridized to 9q34 and spanned the 200-kb breakpoint region of ABL. The BCR (300 kb) probe-set was directly labeled with Texas Red (red signal) and included several DNA sequences that hybridized to 22q11.2 and spanned the common breakpoints in both the major and minor BCR.
The specimens were studied in random order and in a blind fashion by two microscopists (I.B. and W.A.W.) using strict scoring criteria for D-FISH.15 For purposes of this paper, red BCR signals are referred to as R, green ABL signals as G, and BCR/ABL fusion signals as F. For scoring purposes, fusion signals were defined as merging or touching R and G signals. The scoring process was limited to normal nuclei with 2R2G, and abnormal nuclei with 1R1G2F or 2R2G1F (one Ph chromosome), and 1R1G3F or 2R2G2F (two Ph chromosomes). For each specimen, each microscopist scored 250 consecutive qualifying interphase nuclei from different areas of the same slide. At the conclusion of the study, the intermicroscopist agreement was sufficient to pool their results on each specimen in subsequent analyses of the data. Thus, the final statistical analyses were based on 500 nuclei per specimen.
The normal range for D-FISH was calculated for peripheral blood specimens collected from 10 patients without any malignant hematologic disorder and for bone marrow specimens collected from 10 normal bone marrow transplant donors. The four serial dilutions were prepared by mixing cells from a normal individual and a Ph-positive specimen to create a series of specimens determined by repeated blind studies before this investigation to contain specified mean percentages of Ph-positive nuclei.
The D-FISH results for each patient's specimens from both peripheral blood and bone marrow samples were calculated as the proportion of abnormal cells (number of abnormal cells per 500 scored cells). Because the proportion of abnormal cells among the specimens ranged from 0 to 1 (ie, 0% to 100%), a sin-1 transformation was used to stabilize variances and provide a more nearly Gaussian distribution of values. Then, the differences (delta value) between bone marrow and peripheral blood in transformed proportions were computed for each patient's specimens. The proportion of abnormal cells by Q-cytogenetics was also transformed to sin-1.
The delta values for each paired set of bone marrow and blood specimens were then analyzed using a repeated measures regression analysis (PROCEDURE MIXED in SAS).19 To assess the effects of sampling interval in this analysis, the approximate 4-month sampling intervals relative to commencement of therapy were considered nominal predictor variables and the transformed proportion from Q-cytogenetics was included as a covariate. The within-patient correlation of delta values among respective specimen collection times was specified as an autocorrelation structure depending on the actual number of days between sampling times, ie, smaller correlations between sequential values for longer times between sampling episodes. To assess the usefulness of within-subject changes between sampling intervals in blood specimens as indicators of within-subject changes in bone marrow, a similar regression analysis was examined. In this analysis the (within-subject) changes in bone marrow were regressed on corresponding changes in blood, and a test for an equiangular (y = x) regression line computed.
The classification scheme for response to therapy was based on Q-cytogenetics and was similar to the Italian Cooperative Group,2 ie, no response, minimal, minor, major, and complete remission when 100%, 99% to 67%, 66% to 33%, 32% to 1%, and 0% of metaphases are Ph positive, respectively.
RESULTS
Success of different genetic tests.
The goal for D-FISH was to study 500 nuclei for each bone marrow and blood specimen. The goal for Q-cytogenetics was to study 25 metaphases from each bone marrow specimen. The goal for hypermetaphase studies was to study 200 metaphases from bone marrow. D-FISH was successful on 37 of 37 blood specimens and 37 of 37 bone marrow specimens. Q-cytogenetics was successful in 32 of 37 bone marrow specimens. Hypermetaphase was successful in 14 of 24 bone marrow specimens.
Normal range of D-FISH for peripheral blood and bone marrow.
Based on 500 nuclei from each of 10 normal bone marrow specimens, the mean percentage and standard deviation (SD) of nuclei with false BCR/ABL fusion was 0.1% ± 0.1% (range, 0 to 1 per 500 nuclei). Based on 500 nuclei from each of 10 normal peripheral blood specimens, the mean percentage and SD of nuclei with false BCR/ABL fusion was 0.04% ± 0.08% (range, 0 to 1 per 500 nuclei). Based on this data, the upper bound of a one-sided 95% confidence interval for observing 1 of 500 (0.2%) neoplastic cells in either bone marrow or peripheral blood was calculated using the binomial distribution. For both bone marrow and peripheral blood, this calculation implied a cut-off greater than 4/500 (>0.8%) nuclei with BCR/ABL fusion to classify any specimen as abnormal.
Abnormal reference range for D-FISH in untreated CML.
The results of D-FISH for specimens from seven patients (nos. 2 through 7 and 9) that were collected before treatment and that were not mosaic by Q-cytogenetic studies were used to establish an abnormal reference range. We believe these specimens generally represent patients with untreated CML in clinical practice. Among these seven specimens, the mean percentage of abnormal cells was 97.6% ± 1.38% (range, 95.4 to 99.0) for bone marrow, and 86.1% ± 13.59% (range, 61.6 to 98.5) for blood.
Serial dilutions.
The observed percentage of neoplastic cells in each of the four serial dilution specimens was 97.6%, 49.2%, 8.2%, and 1.8%. The expected mean percentage of neoplastic cells in these specimens was 98.2%, 49.1%, 10.7%, and 2.8%, respectively. Thus, the difference between observed and expected values for each of these specimens was 0.6%, 0.1%, 2.5%, and 1.0%, respectively.
Patient specimens.
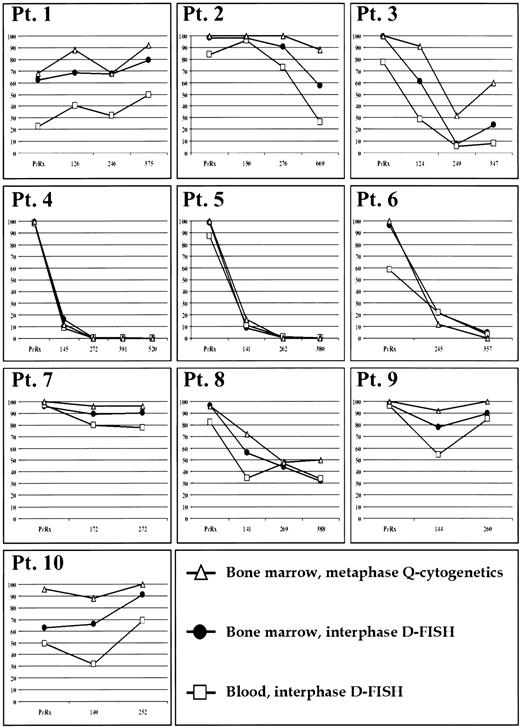
Results for Q-cytogenetic studies for bone marrow and D-FISH for bone marrow and blood for each patient specimen are shown in Fig 1. Based on Q-cytogenetics, three patients (nos. 4, 5, and 6) achieved a complete cytogenetic remission, one patient (no. 3) briefly achieved a major response, and the rest of the patients were classified as minimal, minor, or nonresponders.
Percentage of Ph-positive cells (y-axis) before therapy and during treatment at approximately 4-month sampling intervals (x-axis in days) in bone marrow by Q-cytogenetics and D-FISH, and blood by D-FISH.
Percentage of Ph-positive cells (y-axis) before therapy and during treatment at approximately 4-month sampling intervals (x-axis in days) in bone marrow by Q-cytogenetics and D-FISH, and blood by D-FISH.
Each bone marrow specimen that had any abnormal metaphases by Q-cytogenetics was also abnormal for interphase nuclei by D-FISH in blood and bone marrow. Six specimens from three patients (nos. 4, 5, and 6) had only normal metaphases by Q-cytogenetics. For patient 6, D-FISH results were abnormal at 357 days in both bone marrow (4.8% abnormal nuclei) and blood (3.0% abnormal nuclei). For patient 5 at 262 days, the peripheral blood was marginally abnormal (1.0% abnormal nuclei), but bone marrow was within normal limits (0.6% abnormal nuclei). Each of the remaining four specimens with only normal metaphases by Q-cytogenetics were within normal limits for D-FISH in both bone marrow and blood.
Additional studies were performed to look for minimal residual disease on the paired sets of bone marrow and blood specimens that were normal by Q-cytogenetics and D-FISH. In a blind study, D-FISH was used to score 6,000 nuclei from four of the bone marrow specimens and five of the peripheral blood specimens in this series (Table 1), and three blood and bone marrow specimens from normal individuals. In a prior study, the normal range for D-FISH for 6,000 nuclei was calculated to be <0.079%.15 Based on this cut-off, each of the normal blood and bone marrow specimens was correctly classified as normal. Three of the four patient bone marrow specimens and each of the patient peripheral blood specimens had minimal residual disease. It was not possible to perform further studies on bone marrow no. 5 from patient 4 because this specimen had no leftover cells. The paired-blood specimen for this sampling time was in the abnormal range for D-FISH when 6,000 nuclei were studied and the bone marrow had one Ph-positive metaphase among 169 metaphases that were examined by hypermetaphase FISH studies.
Search for Minimal Residual Disease in Specimens That Were Normal by Q-Cytogenetics for 25 Metaphases and D-FISH for 500 Nuclei
| Pt . | Spec . | 500 Nuclei . | 6,000 Nuclei . | Hypermetaphase Bone Marrow % Abn (Ph Positive/Metaphases Analyzed) . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bone Marrow . | Peripheral Blood . | Bone Marrow . | Peripheral Blood . | |||||||
| % Abn . | Abn Nuclei . | % Abn . | Abn Nuclei . | % Abn . | Abn Nuclei . | % Abn . | Abn Nuclei . | |||
| 4 | 3 | 0.6 | 3 | 0.2 | 1 | 0.22 | 13 | 0.10 | 6 | 0.0 (0/27) |
| 4 | 4 | 0.4 | 2 | 0.2 | 1 | 0.23 | 14 | 0.08 | 5 | 0.0 (0/15) |
| 4 | 5 | 0.0 | 0 | 0.0 | 0 | NA | NA | 0.13 | 8 | 0.6 (1/169) |
| 5 | 3 | 0.6 | 3 | 1.0 | 5 | 1.30 | 78 | 0.95 | 57 | 0.0 (0/136) |
| 5 | 4 | 0.0 | 0 | 0.2 | 1 | 0.05 | 3 | 0.12 | 7 | 0.0 (0/126) |
| Normal cut-off | >0.8 | >4 | >0.8 | >4 | >0.079 | >4 | >0.079 | >4 | ||
| Pt . | Spec . | 500 Nuclei . | 6,000 Nuclei . | Hypermetaphase Bone Marrow % Abn (Ph Positive/Metaphases Analyzed) . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bone Marrow . | Peripheral Blood . | Bone Marrow . | Peripheral Blood . | |||||||
| % Abn . | Abn Nuclei . | % Abn . | Abn Nuclei . | % Abn . | Abn Nuclei . | % Abn . | Abn Nuclei . | |||
| 4 | 3 | 0.6 | 3 | 0.2 | 1 | 0.22 | 13 | 0.10 | 6 | 0.0 (0/27) |
| 4 | 4 | 0.4 | 2 | 0.2 | 1 | 0.23 | 14 | 0.08 | 5 | 0.0 (0/15) |
| 4 | 5 | 0.0 | 0 | 0.0 | 0 | NA | NA | 0.13 | 8 | 0.6 (1/169) |
| 5 | 3 | 0.6 | 3 | 1.0 | 5 | 1.30 | 78 | 0.95 | 57 | 0.0 (0/136) |
| 5 | 4 | 0.0 | 0 | 0.2 | 1 | 0.05 | 3 | 0.12 | 7 | 0.0 (0/126) |
| Normal cut-off | >0.8 | >4 | >0.8 | >4 | >0.079 | >4 | >0.079 | >4 | ||
Abbreviations: Abn, abnormal; NA, not available; spec, specimen.
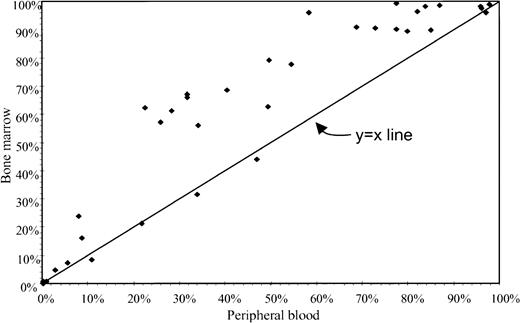
The actual proportions of neoplastic cells from bone marrow specimens were plotted against the corresponding proportions from peripheral blood samples (Fig 2). This plot implied that the proportion of abnormal cells from bone marrow specimens was typically greater (above y = x line) than for peripheral blood.
Percentage of neoplastic cells for paired sets of bone marrow (y-axis) and peripheral blood (x-axis).
Percentage of neoplastic cells for paired sets of bone marrow (y-axis) and peripheral blood (x-axis).
For D-FISH, the mean 4-month intersample differences in percentage of abnormal nuclei between paired sets of bone marrow and peripheral blood were not statistically different (P > .3) (Table 2). These deltas for D-FISH were associated (P < .05) with the transformed proportion of abnormal cells based on Q-cytogenetics of the paired bone marrow specimen. This is important because Q-cytogenetics of bone marrow is widely recognized as the “gold standard” for monitoring response to interferon therapy.1-5
Analysis of Differences in Paired Sets of Bone Marrow and Blood Over Approximate 4-Month Sampling Occasions for Patients Undergoing Treatment for CML
| Sample . | Pts . | Mean Proportions (+SE) Original Scale . | Adjusted Mean Delta (+SE)* (Transformed Scale) . | |
|---|---|---|---|---|
| Bone Marrow . | Blood . | |||
| Dx | 10 | 0.91 (±0.05) | 0.75 (±0.08) | 0.165 (±0.047) |
| 4 mos | 10 | 0.56 (±0.10) | 0.41 (±0.09) | 0.177 (±0.042) |
| 8 mos | 10 | 0.49 (±0.13) | 0.39 (±0.11) | 0.150 (±0.042) |
| 12 mos | 6 | 0.32 (±0.13) | 0.20 (±0.08) | 0.181 (±0.054) |
| Sample . | Pts . | Mean Proportions (+SE) Original Scale . | Adjusted Mean Delta (+SE)* (Transformed Scale) . | |
|---|---|---|---|---|
| Bone Marrow . | Blood . | |||
| Dx | 10 | 0.91 (±0.05) | 0.75 (±0.08) | 0.165 (±0.047) |
| 4 mos | 10 | 0.56 (±0.10) | 0.41 (±0.09) | 0.177 (±0.042) |
| 8 mos | 10 | 0.49 (±0.13) | 0.39 (±0.11) | 0.150 (±0.042) |
| 12 mos | 6 | 0.32 (±0.13) | 0.20 (±0.08) | 0.181 (±0.054) |
Abbreviations: Dx, diagnosis; pts, patients; delta, difference in percentage of abnormal nuclei; SE, standard error.
Adjusted for Q-cytogenetics and within-subject correlations. Not statistically different (P > .3).
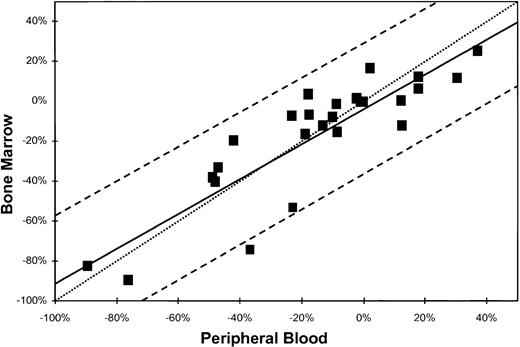
Based on these results, an additional regression analysis was performed to assess the relationship between within-subject changes (sample interval) in the proportion of abnormal cells that would be obtained from bone marrow specimens versus the within-subject changes in peripheral blood samples. This regression analysis is displayed in Fig 3 along with an approximate 95% confidence interval for a new predicted observation (delta bone marrow) given a (new delta) peripheral blood value (prediction interval). In addition, the expected equiangular regression line (y = x) was calculated and included in Fig 3. This analysis indicated a significant (P < .001) linear relationship but was not different than the equiangular (y = x) line (P > .2).
Linear regression analysis (solid line) of the within-subject changes in the proportion of neoplastic cells from bone marrow on the within-subject changes in the proportion of neoplastic cells for peripheral blood. Dashed lines are the 95% prediction interval, and the dotted line is the equiangular (y = x) line.
Linear regression analysis (solid line) of the within-subject changes in the proportion of neoplastic cells from bone marrow on the within-subject changes in the proportion of neoplastic cells for peripheral blood. Dashed lines are the 95% prediction interval, and the dotted line is the equiangular (y = x) line.
DISCUSSION
In the present investigation, the differences between the percentage of neoplastic nuclei in bone marrow and blood were consistent over the follow-up period. This implies that the percentage of neoplastic nuclei in blood during follow-up tracked the corresponding percentage of neoplastic metaphases and nuclei in bone marrow over the course of interferon α-2b therapy. In an earlier study, we showed that the percentage of neoplastic nuclei in the bone marrow strongly correlated with the percentage of Ph-positive metaphases.15 The reduction in percentage of Ph-positive metaphases correlates with a prolonged chronic phase and increased survival in CML, and the results of D-FISH on blood correlates with Q-cytogenetics. This suggests that D-FISH is an efficient and sufficiently accurate method to test periodic peripheral blood specimens from patients with CML to monitor the effectiveness of interferon therapy.
The analysis of 500 nuclei with D-FISH in bone marrow and peripheral blood detects less than 1% disease and is at least as sensitive as Q-cytogenetics. Thus, the use of D-FISH to score 500 interphase nuclei could substitute for Q-cytogenetics for purposes of monitoring response to therapy for CML. Moreover, by analyzing 6,000 nuclei it was possible to identify residual disease in specimens that were normal by Q-cytogenetics and by initial D-FISH studies (Table 2). Thus, using D-FISH has considerable potential to detect very low levels of minimal disease in both blood and bone marrow.
We are aware of only one other report that compares the results of FISH studies of paired sets of bone marrow and peripheral blood to monitor therapy in CML. Mühlmann et al11 recently used S-FISH to study 49 peripheral blood smears and 30 bone marrow specimens from 36 patients in chronic phase CML at different stages of cytogenetic remission. Although S-FISH is significantly less accurate than D-FISH, the results of their study also suggest that FISH studies of blood are useful to monitor the effect of interferon therapy.
In the present investigation, at most times before and after therapy, the percentage of nuclei with BCR/ABL fusion was usually lower in blood than in bone marrow. Nevertheless, it was possible to use a simple linear regression model to predict the changes in percentages of Ph-positive nuclei in bone marrow using D-FISH data from peripheral blood. Because only 10 patients were studied, the 95% prediction interval provided a wide interval estimate of these changes in neoplastic nuclei in bone marrow. The investigation of a larger series of patients could produce narrower interval estimates of neoplastic cells in bone marrow based on studies of blood. This information could be an important outcome of investigations performed by cooperative groups that focus their studies on CML.
The present limitation to predict precisely the actual percentage of neoplastic nuclei in bone marrow based on data from blood should not limit the use of blood to monitor therapy in clinical practice. The results of the present investigation indicate that it is best to assess response to therapy based on changes in percentage of neoplastic nuclei using the same tissue over time. In other words, assess the changes in percentage of neoplastic cells using studies of blood as a predictor of changes in bone marrow samples. This is important because the percentage of abnormal nuclei in blood and bone marrow vary similarly within most patients over their course of therapy (Fig 3).
Three patients (nos. 4, 7, and 9) in our investigation had relatively similar percentages of neoplastic nuclei in blood and bone marrow before therapy; 1 achieved a complete cytogenetic remission (Fig 1). The remaining 7 patients had somewhat greater differences in percentages of neoplastic nuclei in blood and bone marrow and 2 of these patients achieved complete cytogenetic remission. Although only 10 patients were investigated, these results suggest that chances of achieving a complete cytogenetic remission may not be affected by differences in percentage of abnormal nuclei in blood and bone marrow before therapy.
We wondered why some patients had similar percentages of neoplastic cells in their bone marrow and blood, whereas other patients did not. Many investigators have shown that the Ph chromosome occurs in cells of different hematopoietic compartments in different patients with CML.20 Perhaps patients with a similar percentage of neoplastic nuclei in bone marrow and blood have a form of CML that involves stem cells that give rise to both lymphocyte and myelocytes. In contrast, patients with CML that have different percentages of neoplastic nuclei in blood and bone marrow have a form of CML that involves stem cells of only myeloid cell lines.
Among patients with untreated CML, approximately 90% have a Ph chromosome in each of their metaphases and the remaining 10% show mosaicism, ie, a mixture of normal and Ph-positive metaphases.21 Thus, it was not surprising to find three patients (nos. 1, 8, and 10) in this investigation who were mosaic before therapy. The results of D-FISH studies on 43 patients in this study and an earlier investigation suggest that all patients with CML may have both normal and neoplastic cells in their bone marrow before treatment; this is not apparent by Q-cytogenetics.15
Most classification schemes for assessing response to therapy are based on the observed percentage of Ph-positive metaphases. To adjust for mosaicism when using Q-cytogenetics and D-FISH, it may be useful to standardize the percentage of neoplastic cells after therapy to the percentage of neoplastic cells before treatment. One approach is to divide the percentage of neoplastic cells before therapy into the percentage of neoplastic cells after therapy and then multiply by 100.
Cells from each patient in this study that had a classical t(9;22)(q34;q11.2) displayed a D-FISH pattern that matched our strict scoring criteria. However, some patients with CML have Ph chromosomes that produce abnormal D-FISH patterns, but the signal patterns are different than our strict scoring criteria.15 For these patients, it is necessary to develop special scoring criteria for D-FISH by first examining signal patterns in metaphase cells. For patients with atypical D-FISH patterns, the normal and abnormal reference ranges for nuclei with apparent BCR/ABL fusion are different than for patients with CML that have typical D-FISH patterns. Moreover, the quantification of disease is less accurate than it is for patients with typical D-FISH signal patterns. We have studied patients with CML who have cells in their bone marrow and blood with atypical D-FISH patterns and believe that the D-FISH signal patterns do not change in Ph-positive nuclei over time in these patients.15 Thus, it should be possible to use modified D-FISH criteria to study blood for purposes of detecting changes in the percentage of nuclei with BCR/ABL fusion to monitor therapy.
The results of the present investigation and Mühlmann et al11 show great potential for using FISH to study blood for purposes of monitoring the response to therapy in CML. In clinical practice, we believe that cytogenetic studies on bone marrow should continue to be performed at diagnosis to identify patients that are Ph positive and to rule out chromosome abnormalities that could indicate neoplasms other than CML. We also believe that it is important to establish a pretreatment baseline for the percentage of nuclei with BCR/ABL fusion with D-FISH; this could be performed on bone marrow and blood. For patients on therapy, D-FISH could then be performed on peripheral blood at periodic intervals to assess the effectiveness of therapy. Consequently, bone marrow may not need to be collected to monitor therapy as frequently as it is in current practice.
ACKNOWLEDGMENT
We are indebted to the following investigators who supplied bone marrow and samples that were analyzed in this study as part of the CML National Study Group: Luis Baez, MD, VA Medical Center; Stephanie Elkins, MD, University of Mississippi Medical Center; Eric Feldman, MD, New York Medical College; Bruce Lewis, MD, St Paul, MN; Romeo A. Mandanas, MD, University of Oklahoma; Hussain I. Saba, MD, H. Lee Moffitt Cancer Center; and Richard Silver, MD, New York Hospital. We are also grateful to Bernadette Clarke, BSN, at the Chronic Myeloid Leukemia National Study Group Coordinating Center and Larry Heller from Integrated Therapeutics Group Inc for assistance with coordinating this project.
The Chronic Myeloid Leukemia National Study Group provided specimens for this study and was supported in part by Integrated Therapeutics Group Inc. Probes for BCR and ABL1, and salaries for personnel who performed fluorescence in situ hybridization studies were provided by Oncor Inc. Research support was also provided by the United Leukemia Fund Inc and the Cancer Research and Treatment Fund Inc. I.B. was a fellow visiting Mayo Clinic and was supported by Comunidad Autónoma de Madrid, Spain.
Address reprint requests to Gordon W. Dewald, PhD, Cytogenetics Laboratory, Hilton 970, Mayo Clinic, Rochester, MN 55905.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal