Abstract
X-linked hyper IgM syndrome (XHIM) is a primary immunodeficiency disorder caused by mutations of the gene encoding CD40 ligand (CD40L). We correlated mutations of the CD40L gene, CD40L expression, and the clinical manifestations observed in XHIM patients from 30 families. The 28 unique mutations identified included 9 missense, 5 nonsense, 9 splice site mutations, and 5 deletions/insertions. In 4 of 9 splice site mutations, normally spliced and mutated mRNA transcripts were simultaneously expressed. RNase protection assay demonstrated that 5 of 17 mutations tested resulted in decreased levels of transcript. The effect of the mutations on CD40L expression by activated peripheral blood mononuclear cells (PBMC) and T-cell lines or clones was assessed using one polyclonal and four monoclonal antibodies and a CD40-Ig fusion protein. In most patients, the binding of at least one antibody but not of CD40-Ig was observed, suggesting nonfunctional CD40L. However, activated PBMC from three patients and activated T-cell lines from two additional patients, each with different genotype, bound CD40-Ig at low intensity, suggesting functional CD40L. Thus, failure of activated PBMC to bind CD40-Ig is not an absolute diagnostic hallmark of XHIM and molecular analysis of the CD40L gene may be required for the correct diagnosis. Patients with genotypes resulting in diminished expression of wild-type CD40L or mutant CD40L that can still bind CD40-Ig appear to have milder clinical consequences.
X-LINKED HYPER IgM syndrome (XHIM) is a primary immunodeficiency syndrome, characterized by recurrent infections, hypogammaglobulinemia, and normal or elevated serum levels of IgM.1,2 Most patients with XHIM become symptomatic during the first or second year of life when they present with recurrent infections including otitis media, respiratory tract infections, or Pneumocystis carinii pneumonia (PCP).1,2 Transient or chronic neutropenia, present in about half of XHIM patients, may contribute to the development of infections.1,2Cryptosporidium infection causing chronic diarrhea and possibly cholangiopathy or liver cirrhosis,3 malignancies,1,3 and autoimmune disorders1 are other known complications.
The CD40 ligand (CD40L, CD154, gp39, TRAP, orT-BAM) gene, encoding a type II membrane glycoprotein of 261 amino acids mainly expressed by activated CD4+ T cells,4-6 has been identified as the gene responsible for XHIM.7-11 The gene, mapped to Xq26,5,7 consists of five exons and four introns spreading over 12 kb.12,13 CD40L is a member of the tumor necrosis factor (TNF) superfamily4 and its functional unit consists of a trimer.7 The crystal structure of soluble CD40L, representing the extracellular region of CD40L with the entire TNF homology domain, has recently been elucidated,14 and computer-assisted structural analysis has been instrumental in assessing the effect of mutations on CD40L structure and CD40L-CD40 interaction.15 CD40, a counter-receptor of CD40L, is expressed by B cells,16 macrophages, and monocytes,17-19 dendritic cells,20 and vascular endothelial cells.21,22 The interaction of CD40L on activated T cells and CD40 on B cells induces B-cell proliferation and Ig isotype switching, is important for the formation of germinal centers, and prevents B cells from undergoing apoptosis.16
CD40L expressed by activated CD4+ T cells from normal individuals is easily detected by immunostaining using anti-CD40L monoclonal antibodies (MoAbs) or CD40-Ig, a fusion protein consisting of the extracellular region of human CD40 and the Fc portion of Ig. In contrast, activated T cells from XHIM patients fail to express functional CD40L.7-11 The failure of properly activated T cells to bind anti-CD40L MoAbs and CD40-Ig, respectively, is considered a diagnostic hallmark of XHIM, along with a family history and clinical manifestations characteristic of XHIM. The diagnosis of XHIM is confirmed by sequence analysis of the CD40L gene.
A total of 75 unique mutations, affecting the transmembrane and extracellular domains of CD40L, have been collected by the European XHIM registry23 and include missense and nonsense mutations, and mutations affecting splicing of mRNA transcripts, and insertions and deletions. To date, no apparent genotype and phenotype relationship has been reported.
In this study, we have examined 30 unrelated families with XHIM, identified a mutation of the CD40L gene in each family, explored various diagnostic strategies, and attempted to correlate genotype with phenotype.
MATERIALS AND METHODS
Study population.
Thirty unrelated families with 45 affected males were included in this study. They consist of 19 white families (30 affected males), 9 Asian families (13 affected males), and 2 African-American families (2 affected males). Thirty-five patients are alive. All participating patients including those reported previously by us3,7,13,15,24-26 or others11,27-29 are described in Table 1. Ten patients died during the course of this study (between 1991 and 1997); 4 died of malignancies3 including carcinoid (GS, family 16), bile duct carcinoma (MC, family 22), and adenocarcinoma of unknown origin (JJ and CJ, family 30); 2 died of liver failure (CF, family 4 and DS, family 16); 1 (JW, family 6) had a stroke at age 26 due to severe hypertension caused by acute onset nephritis at 10 years of age; 1 (KyS, family 13) died of generalized cryptococcosis27; and 1 patient (KA, family 24) died in a traffic accident. One patient (JoC, family 22) died of PCP at 8 months of age before this study was begun. The current ages of the living patients, shown in Table 1, range from 1 to 28 years (median, 13 years). Serum Ig levels at onset were available from 32 affected males. IgG was consistently low (mean, 92 mg/dL, range, 12 to 267 mg/dL). IgA (<10 mg/dL in 21 patients, 10 to 30 mg/dL in 4 patients, and >30 mg/dL in 7 patients; range <10 to 93 mg/dL) and IgM levels (<50 mg/dL in 4 patients; 50 to 150 mg/dL in 15 patients; >150 mg/dL in 13 patients; range 36 to 1,524 mg/dL) varied considerably.
Mutations of the CD40L Gene and Immunostaining Types in Patients With XHIM
| Family* . | Patient . | Age† . | cDNA Mutation‡ (nucleotide no.) . | Expected Protein Alteration (amino acid no.) . | Genomic DNA Mutation‡ (nucleotide no.) . | Location . | Staining Type§ . | References∥ . |
|---|---|---|---|---|---|---|---|---|
| Missense and nonsense mutations | ||||||||
| 1 (W) | CD | 18 | 405T → A + 407A → G | S128R + E129G | 405T → A + 407A → G | Exon 4 | 3 | 7, 26 |
| 2 (W) | LB | 11 | 461C → A | T147N | 461C → A | Exon 5 | 2 | 15 |
| 3 (W) | JC/SC | 16/7 | 530A → G | Y170C | 530A → G | Exon 5 | 4 | 15 |
| 4 (W) | CF | (29) | 701G → T | G227V | 701G → T | Exon 5 | 3 | 11, 29 |
| 5 (A) | TY/AY | 12/5 | 713T → C | L231S | 713T → C | Exon 5 | ND | 25 |
| 6 (W) | JW | (26) | 724G → C | A235P | 724G → C | Exon 5 | 4 | 7, 26 |
| 7 (W) | TA | 18 | 782C → T | T254M | 782C → T | Exon 5 | 2 | 15, 25,40 |
| 8 (A) | IN | 24 | 782C → T | T254M | 782C → T | Exon 5 | 2 | 25, 40, 59 |
| 9 (W) | YL/JoB/JaB | 21/3/1 | 794T → C | L258S | 794T → C | Exon 5 | 3 | 15 |
| 10 (W) | DA/PA | 28/26 | 794T → C | L258S | 794T → C | Exon 5 | 3 | 15 |
| 11 (W) | DJ | 18 | 52C → T | R11X | 52C → T | Exon 1 | 1 | 60 |
| 12 (A) | NC | 21 | 187G → T | E56X | 187G → T | Exon 2 | 4 | |
| 13 (A) | KyS/KiS | (6)/5 | 440G → A | W140X | 440G → A | Exon 5 | 4 | 8, 13, 25, 27, 28 |
| 14 (W) | MB | 4 | 577C → T | Q186X | 577C → T | Exon 5 | 4 | |
| 15 (W) | WB | 11 | 682C → T | Q221X | 682C → T | Exon 5 | 4 | 29, 38 |
| Splice site mutations | ||||||||
| 16 (W) | DS/GS/DB | (40)/(24)/15 | del 82 bp (nt 96-177) | pt at aa 25 | nt 177 + 1g → t | Intron 1 | 4 | 3, 7, 11, 29 |
| del 43 bp (nt 135-177) | pt at aa 38 | |||||||
| del 10 bp (nt 168-177) | pt at aa 49 | |||||||
| del 14 bp (nt 168-181) | pt at aa 50 | |||||||
| Wild-type mRNA¶ | Wild type CD40L | |||||||
| 17 (A) | YA/TaA | 24/17 | del 132 bp (nt 178-309) (exon 2 skipping) | In-frame del 44 aa | nt 309 + 1g → a | Intron 2 | 2 | 25 |
| ins 19bp (5′ part of intron 2) | pt at aa 117 | |||||||
| 18 (W) | PS | 20 | Same as family 17 | Same as family 17 | nt 309 + 2t → a | Intron 2 | 1 | |
| 19 (W) | AM | 13 | del 58 bp (nt 310-367) (exon 3 skipping) | pt at aa 107 | nt 367G → A | Exon 3 | 1 | |
| Wild-type mRNA¶ | G116S | |||||||
| 20 (B) | MS | 1 | del 58 bp (nt 310-367) (exon 3 skipping) | pt at aa 107 | nt 367 + 2t → c | Intron 3 | 1 | |
| Wild-type mRNA¶ | Wild-type CD40L | |||||||
| 21 (W) | JE | 6 | Same as family 20¶ | Same as family 20 | nt 367 + 5g → a | Intron 3 | 1 | |
| 22 (W) | JoC/SeC/MC/ RoC/RiC | (0.7)/22/ (17)/15/11 | del 63 bp (nt 368-430) (exon 4 skipping) | In-frame del 21 aa | nt 430 + 1g → c | Intron 4 | 3 | 3, 25, 26, 29 |
| ins 23 bp, 73 bp, or 152 bp (5′ part of intron 4) | pt at aa 121 | |||||||
| 23 (A) | RA | 2 | Same as family 22 | Same as family 22 | nt 430 + 1g → a | Intron 4 | 3 | 10, 25 |
| 24 (A) | KA | (19) | del 8bp (nt 431-438) | pt to aa 138 | del nt 431-1g | Intron 4 | ND | 25 |
| Deletions and insertions | ||||||||
| 25 (A) | SU | 9 | del ATAG (nt 178-181) | pt at aa 64 | del ATAG between nt 178-5a and nt 182A | Intron 2exon 3 | ND | 25,39 |
| 26 (A) | JiB/TP | 16/6 | No mRNA | No protein | del more than 10 kb | Upstream of exon 4 | 5 | |
| 27 (W) | MR | 2 | ins G between nt 403A and nt 405T | pt at aa 128 | ins G between nt 403A and nt 405T | Exon 4 | 4 | 29 |
| 28 (W) | JG | 9 | Same as family 27 | Same as family 27 | Same as family 27 | Exon 4 | 4 | 29 |
| 29 (B) | DG | 15 | ins A between nt 445G and nt 451G | pt at aa 163 | ins A between nt 445G and nt 451G | Exon 5 | 4 | |
| 30 (W) | JJ/CJ | (23)/(24) | ins CAGCC between nt 593C and nt 599T | pt at aa 196 | ins CAGCC between nt 593C and nt 599T | Exon 5 | 4 | 24 |
| Family* . | Patient . | Age† . | cDNA Mutation‡ (nucleotide no.) . | Expected Protein Alteration (amino acid no.) . | Genomic DNA Mutation‡ (nucleotide no.) . | Location . | Staining Type§ . | References∥ . |
|---|---|---|---|---|---|---|---|---|
| Missense and nonsense mutations | ||||||||
| 1 (W) | CD | 18 | 405T → A + 407A → G | S128R + E129G | 405T → A + 407A → G | Exon 4 | 3 | 7, 26 |
| 2 (W) | LB | 11 | 461C → A | T147N | 461C → A | Exon 5 | 2 | 15 |
| 3 (W) | JC/SC | 16/7 | 530A → G | Y170C | 530A → G | Exon 5 | 4 | 15 |
| 4 (W) | CF | (29) | 701G → T | G227V | 701G → T | Exon 5 | 3 | 11, 29 |
| 5 (A) | TY/AY | 12/5 | 713T → C | L231S | 713T → C | Exon 5 | ND | 25 |
| 6 (W) | JW | (26) | 724G → C | A235P | 724G → C | Exon 5 | 4 | 7, 26 |
| 7 (W) | TA | 18 | 782C → T | T254M | 782C → T | Exon 5 | 2 | 15, 25,40 |
| 8 (A) | IN | 24 | 782C → T | T254M | 782C → T | Exon 5 | 2 | 25, 40, 59 |
| 9 (W) | YL/JoB/JaB | 21/3/1 | 794T → C | L258S | 794T → C | Exon 5 | 3 | 15 |
| 10 (W) | DA/PA | 28/26 | 794T → C | L258S | 794T → C | Exon 5 | 3 | 15 |
| 11 (W) | DJ | 18 | 52C → T | R11X | 52C → T | Exon 1 | 1 | 60 |
| 12 (A) | NC | 21 | 187G → T | E56X | 187G → T | Exon 2 | 4 | |
| 13 (A) | KyS/KiS | (6)/5 | 440G → A | W140X | 440G → A | Exon 5 | 4 | 8, 13, 25, 27, 28 |
| 14 (W) | MB | 4 | 577C → T | Q186X | 577C → T | Exon 5 | 4 | |
| 15 (W) | WB | 11 | 682C → T | Q221X | 682C → T | Exon 5 | 4 | 29, 38 |
| Splice site mutations | ||||||||
| 16 (W) | DS/GS/DB | (40)/(24)/15 | del 82 bp (nt 96-177) | pt at aa 25 | nt 177 + 1g → t | Intron 1 | 4 | 3, 7, 11, 29 |
| del 43 bp (nt 135-177) | pt at aa 38 | |||||||
| del 10 bp (nt 168-177) | pt at aa 49 | |||||||
| del 14 bp (nt 168-181) | pt at aa 50 | |||||||
| Wild-type mRNA¶ | Wild type CD40L | |||||||
| 17 (A) | YA/TaA | 24/17 | del 132 bp (nt 178-309) (exon 2 skipping) | In-frame del 44 aa | nt 309 + 1g → a | Intron 2 | 2 | 25 |
| ins 19bp (5′ part of intron 2) | pt at aa 117 | |||||||
| 18 (W) | PS | 20 | Same as family 17 | Same as family 17 | nt 309 + 2t → a | Intron 2 | 1 | |
| 19 (W) | AM | 13 | del 58 bp (nt 310-367) (exon 3 skipping) | pt at aa 107 | nt 367G → A | Exon 3 | 1 | |
| Wild-type mRNA¶ | G116S | |||||||
| 20 (B) | MS | 1 | del 58 bp (nt 310-367) (exon 3 skipping) | pt at aa 107 | nt 367 + 2t → c | Intron 3 | 1 | |
| Wild-type mRNA¶ | Wild-type CD40L | |||||||
| 21 (W) | JE | 6 | Same as family 20¶ | Same as family 20 | nt 367 + 5g → a | Intron 3 | 1 | |
| 22 (W) | JoC/SeC/MC/ RoC/RiC | (0.7)/22/ (17)/15/11 | del 63 bp (nt 368-430) (exon 4 skipping) | In-frame del 21 aa | nt 430 + 1g → c | Intron 4 | 3 | 3, 25, 26, 29 |
| ins 23 bp, 73 bp, or 152 bp (5′ part of intron 4) | pt at aa 121 | |||||||
| 23 (A) | RA | 2 | Same as family 22 | Same as family 22 | nt 430 + 1g → a | Intron 4 | 3 | 10, 25 |
| 24 (A) | KA | (19) | del 8bp (nt 431-438) | pt to aa 138 | del nt 431-1g | Intron 4 | ND | 25 |
| Deletions and insertions | ||||||||
| 25 (A) | SU | 9 | del ATAG (nt 178-181) | pt at aa 64 | del ATAG between nt 178-5a and nt 182A | Intron 2exon 3 | ND | 25,39 |
| 26 (A) | JiB/TP | 16/6 | No mRNA | No protein | del more than 10 kb | Upstream of exon 4 | 5 | |
| 27 (W) | MR | 2 | ins G between nt 403A and nt 405T | pt at aa 128 | ins G between nt 403A and nt 405T | Exon 4 | 4 | 29 |
| 28 (W) | JG | 9 | Same as family 27 | Same as family 27 | Same as family 27 | Exon 4 | 4 | 29 |
| 29 (B) | DG | 15 | ins A between nt 445G and nt 451G | pt at aa 163 | ins A between nt 445G and nt 451G | Exon 5 | 4 | |
| 30 (W) | JJ/CJ | (23)/(24) | ins CAGCC between nt 593C and nt 599T | pt at aa 196 | ins CAGCC between nt 593C and nt 599T | Exon 5 | 4 | 24 |
Polymorphism, nt 169T → C (L50L) described by Aruffo et al,7 was found in families 1, 6, 9, 12, 19, and 21.
Abbreviations: nt, nucleotide number; del, deletion; ins, insertion; pt, premature termination; aa, amino acid; ND, not done.
*Race: W, white; B, African-American; A, Asian.
†Age in years as of September 1997; ( ), age at death.
‡Nucleotide number is based on the sequence data described by Hollenbaugh et al.4
§Immunostaining pattern (types 1-5) of activated T-cell lines or CD4+ T-cell clones using one pAb, four MoAbs, and bCD40-Ig. See details in Table 2 and Fig 2.
∥References that report the patient listed with or without detailing the mutation. Reference numbers that are underlined indicate those describing the same mutations found by others in unrelated subjects.
¶Wild-type mRNA transcripts (or normally spliced mRNA transcripts in the case of family 19) were identified by RT-PCR and subsequent cloning.
Cell preparations and culture.
Peripheral blood mononuclear cells (PBMC) were isolated from heparinized venous blood by Ficoll-Hypaque (Pharmacia Biotech, Piscataway, NJ) centrifugation. RPMI1640 supplemented with 10% fetal calf serum (FCS; Hyclone, Logan, UT), 2 mmol/L glutamine, 100 U/mL penicillin, and 100 μg/mL of streptomycin (complete media) was used for cell culture. To establish and maintain cultures of interleukin-2 (IL-2)–dependent T-cell lines and clones, we decreased FCS concentration to 8% and added 2% human AB serum (Biowhittaker, Walkersville, MD) in complete media. T-cell lines were established from PBMC using a standard method30 and CD4+ T-cell clones were established by limiting dilution. CD4+ T-cell lines, prepared from established T-cell lines using a magnetic cell sorting (MACS) system (Miltenyi Biotec Inc, Auburn, CA), were maintained throughout the study period at greater than 95% purity.
Detection of CD40L expression with anti-CD40L antibodies and biotinylated CD40-Ig by flow cytometry.
The expression of CD40L was evaluated by immunostaining as described previously.31 We used four different preparations of anti-human CD40L MoAbs including MoAb 106 and MoAb 1.7 (both mouse IgG1, provided by Dr Tony Siadak, Bristol-Myers Squibb Co, Pharmaceutical Research Institute, Seattle, WA), MoAb 5c8 (mouse IgG2a, provided by Biogen, Cambridge, MA), and MoAb TRAP (mouse IgG1), and a polyclonal antibody (pAb) (rabbit anti-human CD40L antiserum) (the latter two reagents were provided by Dr Richard A. Kroczek, Robert Koch-Institute, Berlin, Germany). In addition, we identified CD40L expression with an Ig fusion protein consisting of its natural counterpart, biotinylated CD40-Ig (bCD40-Ig) (provided by Dr Stephen J Klaus, Department of Microbiology, University of Washington, Seattle). After activation for 8 hours with phorbol 12-myristate 13-acetate (PMA) at 10 ng/mL (GIBCO-BRL, Gaithersburg, MD) and ionomycin at 1 μg/mL (Sigma, St Louis, MO), immunostaining of PBMC was performed using three MoAbs (106, 1.7, and 5c8) and bCD40-Ig. For the immunostaining of T-cell lines and CD4+ T-cell lines or clones, cells were activated for 4 hours with the same stimulants and immunostaining was performed using four MoAbs, one pAb, and bCD40-Ig. The following fluorochrome-conjugated reagents were used: fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG to detect MoAb binding, FITC-conjugated goat anti-rabbit IgG to detect pAb binding, and phycoerythrin-conjugated streptavidin to detect bCD40-Ig binding (all reagents were purchased from Biosource International, Camarillo, CA). The cell suspensions were then analyzed by flow cytometry (FACScan; Becton Dickinson, San Jose, CA); a minimum of 10,000 events were collected and analyzed by Lysis II software (Becton Dickinson). Propidium iodide staining was used to eliminate nonviable cells from the analysis.
Sequence analysis of the CD40L gene in XHIM patients.
Total RNA was isolated from activated PBMC (6 hours incubation with PMA at 10 ng/mL and ionomycin at 1 μg/mL) with TRIzol (GIBCO-BRL). Reverse transcription of mRNA followed by polymerase chain reaction (RT-PCR) was performed as follows: 10 μg of RNA in a total volume of 20 μL was reverse-transcribed into cDNA using oligo-dT primer and SuperScript RNaseH− reverse transcriptase (GIBCO-BRL) and amplification was performed using 1 μL of cDNA in a total volume of 50 μL containing 10 mmol/L Tris-HCl (pH 9.0 at 25°C), 50 mmol/L KCl, 1.5 mmol/L MgCl2, 0.1% Triton X-100, 500 nmol/L of each oligonucleotide primer, 200 μmol/L dNTP, and 1.25 U ofTaq DNA polymerase (Promega, Madison, WI). Two oligonucleotide primers were selected, P1 (5′GCCAGAAGATACCATTTCAAC3′, sense) and P2 (5′CCGCTGTGCTGTATTATGAA3′, antisense), to cover the entire coding region of the CD40L gene. Amplification was performed with 30 cycles of denaturation (93°C, 1 minute), annealing (55°C, 1 minute), and extension (72°C, 2 minutes). The PCR products were size-fractionated by agarose gel electrophoresis, purified, and subjected to direct sequencing using theThermo Sequenase cycle sequencing kit (Amersham, Arlington Heights, IL) according to the manufacturer's instruction. Because theCD40L gene is relatively small, we directly sequenced the entire coding region to determine a mutation. When DNA fragments with different sizes were generated by RT-PCR, each population was cloned into the pCR 2.1 vector (Invitrogen, Carlsbad, CA) and subsequently sequenced; at least 10 to 20 clones were analyzed. After a specific mutation was identified in the CD40L cDNA, we amplified the corresponding exons/introns by PCR using genomic DNA isolated from peripheral blood leukocytes.13 The PCR products were directly sequenced to confirm the mutations, including splice site mutations.
CD40L mutations affecting splice donor sites.
Multiple DNA fragments present in RT-PCR products suggest a splicing abnormality. To further characterize these multiple splicing products by RT-PCR, we designed different pairs of primers for each intron containing splice donor site mutation (summarized in Fig 3A): for intron 1, we selected P1 and P3 (5′CTTTCTCCTGTGTTGCATCTC3′, antisense), which generate a DNA fragment of 262 bp; for intron 2, we selected P4 (5′ATAGAAGATGAAAGGAATCT3′, sense) and P5 (5′CTTTTTGCATTTCAAAGCTGT3′, antisense), resulting in a fragment of 190 bp; for intron 3, we designed P6 (5′GATATAATGTTAAACAAAGAGG3′, sense) and P7 (5′ACTTTAGGCAGAGGCTGGCT3′, antisense), generating a 301-bp fragment; for intron 4, we selected P7 and P8 (5′GTGATCAGAATCCTCAAAATT3′, sense), which generate a fragment of 243 bp. PCR was performed as described above with 30 cycles of denaturation (93°C, 30 seconds), annealing (55°C, 30 seconds), and extension (72°C, 1 minute).
RT-PCR in patients with splice donor site mutations of the CD40L gene. (A) Design and location of primers used in RT-PCR. Shown are the coding regions of CD40L cDNA only. The numbers on top indicate the starting nucleotide number of each exon. The location of primers are shown by arrows [sense (→) and antisense (←)]. Note that sense primers used for amplification of introns 2, 3 , and 4 splice donor site mutations were designed to be localized in an exon that may be skipped. (B) Agarose gel electrophoresis of RT-PCR products. RT-PCR was performed using primer pairs P1 and P3 (lanes 1 through 3), P4 and P5 (lanes 4 through 7), and P6 and P7 (lanes 8 through 11), and P7 and P8 (lanes 12 through 15) (see Materials and Methods). An aliquot of the RT-PCR product was size-fractionated on 2% agarose gel and stained with ethidium bromide. DNA size markers are shown on the left. cDNA used for RT-PCR was derived from a normal control (lanes 1, 4, 8, and 12), DB (family 16) in lane 2, PS (family 18) in lane 5, TaA (family 17) in lane 6, MS (family 20) in lane 9, AM (family 19) in lane 10, SeC (family 22) in lane 13, RA (family 23) in lane 14; lanes 3, 7, 11, and 15 are negative controls (no cDNA in the reaction mixture). Note that patients with splice donor site mutations involving introns 1 (lane 2) and 3 (lanes 9 and 10) but not those involving introns 2 (lanes 5 and 6) and 4 (lanes 13 and 14) have bands which are indistinguishable from those derived from normally spliced mRNA.
RT-PCR in patients with splice donor site mutations of the CD40L gene. (A) Design and location of primers used in RT-PCR. Shown are the coding regions of CD40L cDNA only. The numbers on top indicate the starting nucleotide number of each exon. The location of primers are shown by arrows [sense (→) and antisense (←)]. Note that sense primers used for amplification of introns 2, 3 , and 4 splice donor site mutations were designed to be localized in an exon that may be skipped. (B) Agarose gel electrophoresis of RT-PCR products. RT-PCR was performed using primer pairs P1 and P3 (lanes 1 through 3), P4 and P5 (lanes 4 through 7), and P6 and P7 (lanes 8 through 11), and P7 and P8 (lanes 12 through 15) (see Materials and Methods). An aliquot of the RT-PCR product was size-fractionated on 2% agarose gel and stained with ethidium bromide. DNA size markers are shown on the left. cDNA used for RT-PCR was derived from a normal control (lanes 1, 4, 8, and 12), DB (family 16) in lane 2, PS (family 18) in lane 5, TaA (family 17) in lane 6, MS (family 20) in lane 9, AM (family 19) in lane 10, SeC (family 22) in lane 13, RA (family 23) in lane 14; lanes 3, 7, 11, and 15 are negative controls (no cDNA in the reaction mixture). Note that patients with splice donor site mutations involving introns 1 (lane 2) and 3 (lanes 9 and 10) but not those involving introns 2 (lanes 5 and 6) and 4 (lanes 13 and 14) have bands which are indistinguishable from those derived from normally spliced mRNA.
RNase protection assay.
To detect and quantitate the CD40L mRNA transcripts, we used the RNase protection assay (RPA) originally described by Melton et al.32 Since the mutations of the CD40L gene are highly heterogeneous, three different regions were selected for designing probes (summarized in Fig 4A). Probe A (nucleotides [nt] 163-571), was used in all XHIM patients for which total RNA from activated PBMC was available, except those with splice site mutations affecting introns 2 and 3. Probe B (nt 1-275) was used to specifically quantitate mRNA transcripts in patients with intron 2 splice site mutations (families 17 and 18) causing exon 2 skipping, and probe C (nt 163-367) for quantitating normally spliced mRNA transcripts in patients with mutations affecting intron 3 splicing (families 19 and 20). The template plasmids for the synthesis of these probes, which was prepared by in vitro transcription, were constructed as follows. For probe A, the RT-PCR product generated with primers P9 (5′GCGGGATCCAGAAGGTTGGACAAGATAGAA3′, sense,BamHI recognition sequence underlined) and P2 (antisense) was digested with BamHI and HindIII and the fragment cloned into pTRIamp19 (Ambion, Austin, TX). For probe B, the RT-PCR products generated with P10 (5′CGCGGATCCATTTCAACTTTAACACAGC3′, sense,BamHI recognition sequence underlined) and P11 (5′TCACAGTTCAGTAAGGATAAG3′, antisense) were digested withBamHI, then ligated into the same vector linearized withBamHI and HincII. For probe C, the RT-PCR products generated with primers P9 and P5 were cloned as described for probe B. As an internal control, we selected a probe for the CD3δ chain, using the BamHI-Xba I fragment of CD3δ cDNA33(nt 393-635) cloned into pTRIamp19. The plasmid constructs were individually sequenced to confirm the correct sequence. Cloned plasmids were linearized by EcoRI digestion before synthesizing the radiolabeled probes.
Quantitation of CD40L mRNA transcripts by RNase protection assay. (A) Design of probes used in the RNase protection assay (RPA). The upper open bar shows the domain structure of the CD40L molecule; IC, intracellular tail; TM, transmembrane domain; ECU, extracellular unique region; and TNFH, TNF-homology domain. The numbers above the upper open bars indicate the starting amino acid residue for each domain. The lower open bar shows the contribution of each exon to the CD40L domains. The three closed bars indicate the locations and nucleotide borders of the RPA probes used. (B) Autoradiograms of representative RPAs using probes A, B, and C. In each autoradiogram, arrowheads indicate the protected bands derived from CD40L mRNA transcripts and horizontal bars indicate those derived from CD3δ mRNA transcripts. Total RNA was isolated from activated PBMC of normal controls in lanes 1, 3, and 5, and selected XHIM patients: JG (family 28) in lane 2, PS (family 18) in lane 4, and AM (family 19) in lane 6. Radioactivity of protected CD40L and CD3δ mRNA transcripts was quantitated by the PhosphorImager analysis system and the ratio of CD40L/CD3δ mRNA transcripts calculated as follows: lane 1, 0.883; lane 2, 0.205; lane 3, 1.021; lane 4, total 1.072 (0.692 for 275 bp and 0.380 for 178 bp); lane 5, 0.837; and lane 6, total 0.838 (0.290 for 205 bp and 0.548 for 147 bp). (C) Quantitation of CD40L mRNA transcripts isolated from activated PBMC of XHIM patients. The ratio of CD40L/CD3δ mRNA transcripts was plotted on the ordinate. The number attached to each symbol (representing an individual XHIM patient) corresponds to the patient's family number (see Table 1). In patients with intron 2 splice site mutations (families 17 and 18), mRNA transcripts in which exon 2 was skipped (▿) or 19 nucleotides were inserted (▵) were quantitated separately using probe B; the total amount of transcripts is indicated by an open diamond (◊). In patients with intron 3 splice site mutations (families 19 and 20), normally spliced (▾) and exon 3-skipped (▴) mRNA transcripts were quantitated separately using probe C; the total amount is indicated by a closed diamond (⧫).
Quantitation of CD40L mRNA transcripts by RNase protection assay. (A) Design of probes used in the RNase protection assay (RPA). The upper open bar shows the domain structure of the CD40L molecule; IC, intracellular tail; TM, transmembrane domain; ECU, extracellular unique region; and TNFH, TNF-homology domain. The numbers above the upper open bars indicate the starting amino acid residue for each domain. The lower open bar shows the contribution of each exon to the CD40L domains. The three closed bars indicate the locations and nucleotide borders of the RPA probes used. (B) Autoradiograms of representative RPAs using probes A, B, and C. In each autoradiogram, arrowheads indicate the protected bands derived from CD40L mRNA transcripts and horizontal bars indicate those derived from CD3δ mRNA transcripts. Total RNA was isolated from activated PBMC of normal controls in lanes 1, 3, and 5, and selected XHIM patients: JG (family 28) in lane 2, PS (family 18) in lane 4, and AM (family 19) in lane 6. Radioactivity of protected CD40L and CD3δ mRNA transcripts was quantitated by the PhosphorImager analysis system and the ratio of CD40L/CD3δ mRNA transcripts calculated as follows: lane 1, 0.883; lane 2, 0.205; lane 3, 1.021; lane 4, total 1.072 (0.692 for 275 bp and 0.380 for 178 bp); lane 5, 0.837; and lane 6, total 0.838 (0.290 for 205 bp and 0.548 for 147 bp). (C) Quantitation of CD40L mRNA transcripts isolated from activated PBMC of XHIM patients. The ratio of CD40L/CD3δ mRNA transcripts was plotted on the ordinate. The number attached to each symbol (representing an individual XHIM patient) corresponds to the patient's family number (see Table 1). In patients with intron 2 splice site mutations (families 17 and 18), mRNA transcripts in which exon 2 was skipped (▿) or 19 nucleotides were inserted (▵) were quantitated separately using probe B; the total amount of transcripts is indicated by an open diamond (◊). In patients with intron 3 splice site mutations (families 19 and 20), normally spliced (▾) and exon 3-skipped (▴) mRNA transcripts were quantitated separately using probe C; the total amount is indicated by a closed diamond (⧫).
Total RNA was isolated by ultracentrifugation on CsCl cushion from guanidium isothiocyanate lysate of activated PBMC (3 μg/mL phytohemagglutinin [PHA] and 10 ng/mL PMA for 6 hours). The ethanol precipitated RNA preparations were stored at −20°C until use. RPA was performed using in vitro transcription and an RPA kit (Pharmingen, San Diego, CA), according to the manufacturer's instruction. Protected RNA bands were quantitated by the PhosphorImager (Model 400S; Molecular Dynamics, Sunnyvale, CA) analysis system.34
Immunization with bacteriophage φX174.
To assess the effect of CD40L mutations on in vivo antibody responses to a T-cell–dependent neoantigen, we immunized normal controls and a selected group of XHIM patients with bacteriophage φX174, after Institutional Review Board (IRB) approval (University of Washington) and informed consent were obtained. Bacteriophage φX174 was cloned, harvested, and purified as previously described.35 36 The material was adjusted to a final concentration of 1 × 1011 plaque-forming units (PFU)/mL and, after obtaining informed consent, administered intravenously at a dose of 2 × 109 PFU/kg body weight. A second dose was given 6 weeks later. Blood samples for antibody titers were collected immediately before immunization and at 1, 2, and 4 weeks after immunization. Serum was stored at −20°C until analyzed. Antibody activity was determined by a neutralizing antibody assay and expressed as the first-order rate constant (Kv) of phage inactivation using a standard formula. Neutralizing antibodies resistant to 2-mercaptoethanol were considered to be IgG.
RESULTS
CD40L expression by PBMC.
CD40L expression by ionomycin/PMA activated PBMC using MoAbs and bCD40-Ig was studied in normal controls and in affected members of all XHIM families. Representative immunostainings using MoAb 106 and bCD40-Ig are shown in Fig 1. Activated control PBMC expressed CD40L at high intensity (Fig 1A and B); in contrast, very low levels of CD40L expression were detected in unstimulated control PBMC (Fig 1C). Similarly activated PBMC of most XHIM patients from our study group could not express functional CD40L and failed to bind bCD40-Ig (data not shown). However, activated PBMC of patients from families 11, 19, and 21 showed binding of bCD40-Ig, although at reduced intensity. Mutations identified in these families included a nonsense mutation resulting in a deletion of the cytoplasmic domain of CD40L (family 11) and splice site mutations allowing the generation of normally spliced and mutated transcripts (families 19 and 21). Representative immunostaining of patient AM from family 19 are shown in Fig 1D and E. The pattern of CD40L expression by activated PBMC of this subgroup of patients is similar to that observed in a group of patients with common variable immunodeficiency (CVI) previously reported.37 An example of CD40L expression by a CVI patient with normal CD40L gene based on sequence analysis is shown in Fig 1F and G.
CD40L expression by activated PBMC, T-cell lines, and CD4+ T-cell clones. After stimulation with ionomycin and PMA for 8 hours (PBMC) or 4 hours (T-cell lines and CD4+T-cell clones), immunostaining of cells was performed using MoAb 106 or bCD40-Ig. The immunostaining with MoAb 106 or bCD40-Ig is shown by a bold line and the immunostaining with isotype-matched control reagent is indicated by a thin line. The fluorescence intensity is expressed on the abscissa on a log scale and the number of events is expressed on the ordinate. (A and B) Activated PBMC from a normal control; (C) PBMC from a normal control without activation; (D and E) activated PBMC from patient AM (family 19) with the splice site mutation in exon 3 (nt 367G → A); (F and G) activated PBMC from a CVI patient with confirmed normal CD40L sequence; (H, I, and K) patient TA (family 7) with the missense mutation T254M and includes activated PBMC (H), an activated T-cell line (I), and an activated CD4+ T-cell clone (K).
CD40L expression by activated PBMC, T-cell lines, and CD4+ T-cell clones. After stimulation with ionomycin and PMA for 8 hours (PBMC) or 4 hours (T-cell lines and CD4+T-cell clones), immunostaining of cells was performed using MoAb 106 or bCD40-Ig. The immunostaining with MoAb 106 or bCD40-Ig is shown by a bold line and the immunostaining with isotype-matched control reagent is indicated by a thin line. The fluorescence intensity is expressed on the abscissa on a log scale and the number of events is expressed on the ordinate. (A and B) Activated PBMC from a normal control; (C) PBMC from a normal control without activation; (D and E) activated PBMC from patient AM (family 19) with the splice site mutation in exon 3 (nt 367G → A); (F and G) activated PBMC from a CVI patient with confirmed normal CD40L sequence; (H, I, and K) patient TA (family 7) with the missense mutation T254M and includes activated PBMC (H), an activated T-cell line (I), and an activated CD4+ T-cell clone (K).
CD40L expression by cultured T cells.
To avoid repeated shipments of fresh blood from patients not directly under our care, we analyzed CD40L expression by activated IL-2–dependent T-cell lines, CD4+ T-cell lines, or CD4+ T-cell clones. As shown for patient TA from family 7, activated T-cell lines (Fig 1I) and CD4+ T-cell lines or clones (Fig 1K) showed more pronounced CD40L expression than activated PBMC (Fig 1H). A similar increase in CD40L expression was observed if T-cell lines or clones from normal controls (Fig 2) were studied. Based on these observations, we established T-cell lines and CD4+ T-cell lines/clones from affected members of 27 unrelated XHIM families.
Representative histograms of CD40L expression in patients with XHIM using an anti-CD40L pAb, four MoAbs, and bCD40-Ig. The histograms are similar to those described in Fig 1. The reagents used for immunostaining are indicated on the left and the mutations represented by the cell lines and the assigned staining type are listed at the top. See Table 2 for the definition of the immunostaining type.
Representative histograms of CD40L expression in patients with XHIM using an anti-CD40L pAb, four MoAbs, and bCD40-Ig. The histograms are similar to those described in Fig 1. The reagents used for immunostaining are indicated on the left and the mutations represented by the cell lines and the assigned staining type are listed at the top. See Table 2 for the definition of the immunostaining type.
To assess the quality and quantity of CD40L expression by activated T-cell lines or clones, we performed the immunostaining using one pAb, four different MoAbs, and bCD40-Ig. Based on the staining pattern of these cultured T-cell populations, five different types of immunostaining were recognized (Table 2). Representative histograms and mutations are shown in Fig 2. Type 1 pattern, observed in five families and represented by the splice site mutation nt 367G → A, is characterized by the binding of all reagents, suggesting the expression of functional CD40L molecules. In type 2, exemplified by the missense mutation T147N, all antibodies bind, but CD40-Ig does not, suggesting expression of nonfunctional protein due to its inability to bind CD40. Type 3, represented by the missense mutation G227V, that leads to a more severe alteration of the CD40L molecule, is characterized by the binding of the pAb and one or more MoAbs (in this example, the binding of MoAb 106). In type 4, represented by the nonsense mutation W140X, only the pAb binds. Whereas types 2, 3, and 4 suggest that nonfunctional CD40L is expressed, patients with type 5 pattern lack protein expression, as is the case in the two affected cousins of family 26 who have a large genomic deletion. Mutations of the CD40L gene and the immunostaining patterns found in 30 XHIM families are summarized in Table 1. Type 1 staining is observed in normal individuals whose activated T cells express wild-type CD40L or a normal variant (polymorphism) of CD40L, as exemplified by R181W,15 and in XHIM patients with mutations of lesser consequences such as: (1) the nonsense mutation in a cytoplasmic domain (R11X) resulting in the generation of CD40L that lacks a cytoplasmic domain; (2) leaky splice site mutations (nt 367G → A, nt 367 + 2t → c, and nt 367 + 5g → a) that allow activated T cells to generate reduced amounts of wild type CD40L; and (3) intron 2 splice site mutation (nt 309 + 2t → a) that causes in-frame amino acid deletion and generates a truncated CD40L that can still bind CD40-Ig. Three genotypes (T147N, T254M, and nt 309 + 1g → a) were classified as type 2 and 5 genotypes (1 double missense, 2 missense and 2 splice site mutations) as type 3. Type 4 staining was the most common with 10 (42%) of 24 genotypes tested. Only one mutation was identified that resulted in complete absence of CD40L. When comparing the immunostaining between activated PBMC and cultured T-cell lines/clones, although we did not perform the immunostaining of PBMC with pAb and TRAP, the only difference noticed was the intensity but not the type of immunostaining, with three exceptions: activated PBMC from patient CF of family 4 (immunostaining type 3) failed to bind any of the MoAbs tested, and those from patient PS of family 18 and MS of family 20 (both immunostaining type 1) failed to bind bCD40-Ig. Two mutations affecting intron 2 splice donor site, nt 309 + 1g → a (family 17) and nt 309 + 2t → a (family 18), were classified as type 2 and type 1, respectively, although the effect on the splicing of CD40L mRNA transcripts is identical. In both instances, exon 2 is skipped, generating a truncated CD40L which lacks the exon 2–encoded extracellular stalk, but preserving the entire TNF homology domain of CD40L. This truncated CD40L retains some functional qualities; however, the fact that if expressed in COS cells the mutant binds bCD40-Ig with less intensity compared with wild-type CD40L (data not shown), suggests that the biological function of this mutated CD40L is decreased. Quantitation of mRNA transcripts suggests that the difference observed in immunostaining is a direct consequence of variations in gene expression (see below).
Immunostaining Pattern of CD40L in Activated T-Cell Lines or Clones Established From Patients With XHIM
| Type . | No. of Genotypes . | polyAb . | MoAb 106 . | MoAb 1.7 . | MoAb 5c8 . | MoAb TRAP . | CD40-Ig . |
|---|---|---|---|---|---|---|---|
| 1 | 5 | (+) | (+) | (+) | (+) | (+) | (+) |
| 2 | 3 | (+) or + | (+) or + | (+) or + | (+) or + | (+) or + | − |
| 3 | 5 | (+) or + | (+) | (+) or − | (+) or − | (+) or − | − |
| 4 | 10 | (+) or + | − | − | − | − | − |
| 5 | 1 | − | − | − | − | − | − |
| Type . | No. of Genotypes . | polyAb . | MoAb 106 . | MoAb 1.7 . | MoAb 5c8 . | MoAb TRAP . | CD40-Ig . |
|---|---|---|---|---|---|---|---|
| 1 | 5 | (+) | (+) | (+) | (+) | (+) | (+) |
| 2 | 3 | (+) or + | (+) or + | (+) or + | (+) or + | (+) or + | − |
| 3 | 5 | (+) or + | (+) | (+) or − | (+) or − | (+) or − | − |
| 4 | 10 | (+) or + | − | − | − | − | − |
| 5 | 1 | − | − | − | − | − | − |
Activated T-cell lines or clones were exposed to one pAb, four MoAbs, and bCD40-Ig, and the immunostaining pattern (types 1-5) was determined (see Materials and Methods). The staining intensity of the patients' activated T-cell lines or clones was compared with that of normal T-cell lines or T-cell clones; +, equal to normal cells; (+), significantly reduced, but clearly positive; −, no binding.
Mutations of the CD40L gene.
The 28 unique mutations identified in 30 unrelated families include 9 missense (1 double mutation found in family 1 was counted as 2 mutations, but 1 genotype) and 5 nonsense mutations, 9 splice site mutations, and 5 deletions or insertions in genomic DNA (Table 1). The mutations were distributed throughout the entire CD40L molecule and affected all but the transmembrane domain. Ten of the 28 unique mutations identified here are novel and unreported, and the remaining 18 mutations identified in our patients were published previously,7,13,15,25,26 or reported by others.8,9,11,29 38-40
All nine missense mutations were located in the TNF homology domain, underscoring its functional significance for CD40L-CD40 interaction. Using the classification of Bajorath et al15 based on the effect of the missense mutation on the CD40L monomer structure, 4 missense mutations observed (L231S, A235P, T254M, and L258S) affect the internal core packaging of the CD40L monomer (class I), 2 (Y170C and G227V) interfere with trimer formation (class II), and 3 (S128R, E129G, and T147N) compromise directly or indirectly the CD40 binding site (class III). Four of the 5 nonsense mutations identified are expected to result in the premature termination of translation and the generation of truncated proteins. In contrast, R11X, a nonsense mutation in the cytoplasmic domain, is expected to generate a CD40L molecule that lacks the cytoplasmic domain by using a new initiation site near the end of the cytoplasmic domain. This possibility was further supported by the fact that activated T cells with the mutation R11X (patient DJ, family 11) were able to bind all four MoAbs tested and the bCD40-Ig construct (Tables 1 and 2), and finally confirmed by [35S]-methionine metabolic labeling experiments followed by immunoprecipitation (data not shown).
Mutations of the CD40L gene affecting splicing were found in the splice donor site (5′ splice site) of each intron and in the splice acceptor site (3′ splice site) of intron 4 (Table 1). Three groups of patients with splice donor site mutations affecting introns 2, 3, and 4, respectively, were identified. Although the families within each group had different alterations of the consensus sequence in a given splice donor site, their splicing products were identical: families 17 and 18 had mutations affecting the intron 2 splice donor site; families 19, 20, and 21 had mutations affecting the intron 3 splice donor site; and families 22 and 23 had mutations affecting the intron 4 splice donor site. Using RT-PCR and subsequent cloning, multiple species of mRNA transcripts were identified in activated PBMC from all patients with splice donor site mutations. In patient KA (family 24) who has a mutation affecting a splice acceptor site of intron 4, only one species of mRNA transcripts was detected. In addition to variously misspliced mRNA transcripts, normally spliced (wild-type) mRNA transcripts were identified in patients carrying the mutation nt 177 + 1g → t in intron 1, the mutations nt 367 + 2t → c and nt 367 + 5g → a in intron 3, and the mutation nt 367G → A in exon 3. The normally spliced mRNA transcripts found in patient AM (family 19) with the nt 367G → A mutation generate a glycine to serine substitution at position 116 (G116S); however, this amino acid substitution did not affect CD40-Ig binding becaue COS cells transfected with the cDNA carrying G116S mutation showed normal staining intensity with bCD40-Ig (data not shown). Wild-type mRNA transcripts were not found in activated patients' cells with mutations affecting splice donor sites within introns 2 and 4. These latter mutations resulted in mRNA transcripts that skipped exon 2 (families 17 and 18) or exon 4 (families 22 and 23) and contained insertions derived from intron 2 (families 17 and 18) or intron 4 (families 22 and 23) by using cryptic splice sites. The finding of multiple splicing products in all splice donor site mutations and of normally spliced mRNA transcripts in mutations affecting the splice donor site of introns 1 and 3 were further confirmed by RT-PCR using a pair of primers designed to flank each corresponding exon (Fig 3A). Although the mRNA transcript level in patients with intron 1 splice donor site mutation (family 16) was determined to be extremely low by RNase protection assay (see below), RT-PCR successfully detected both normally spliced and misspliced products as shown in Fig 3B (lane 2), whereas a negative control reaction of RT-PCR yielded nothing (Fig 3B, lane 3).
Three unique insertions (in four families) and two unique deletions (in two families) were identified in genomic DNA (Table 1). In all but one family, a premature termination of translation and generation of a truncated protein are predicted. A deletion of more than 10 kb with a breakpoint upstream of exon 4 of the CD40L gene was found in genomic DNA of two affected cousins of family 26. RT-PCR in affected members of this family failed to yield DNA fragments while DNA fragments derived from glyceraldehyde-3-phosphate dehydrogenase mRNA transcripts were amplified normally. PCR of the genomic DNA successfully generated DNA fragments derived from exons 4 and 5 but not from exons 1, 2, and 3. Southern blotting confirmed a large deletion of more than 10 kb in genomic DNA (data not shown).
Quantitation of the CD40L mRNA transcript by RPA.
CD40L mRNA transcripts were quantitated in activated PBMC from 19 XHIM patients with 17 different mutations using RPA (Fig 4). CD3δ mRNA transcripts were analyzed for quantitative normalization. To determine the normal levels of CD40L mRNA transcripts, we isolated total RNA from activated PBMC of 20 normal controls and estimated the quantity of mRNA transcripts using probe A (Fig 4A and B). The mean (geometric) CD40L/CD3δ mRNA transcripts ratio of controls was 1.064 (95% confidence interval 0.409 to 2.772) (Fig 4C). The CD40L/CD3δ mRNA transcript ratio of unstimulated PBMC, determined in five normal controls, was 0.064 (geometric mean) (Fig 4C), indicating that transcription of theCD40L gene increases approximately 17-fold after in vitro activation with PHA and PMA.
Analyses of missense (6 genotypes) and nonsense (4 genotypes) mutations, one intron 1 splice donor site mutation, and insertions (2 genotypes) were performed using probe A. Most of these mutations did not affect transcription of the CD40L gene and allowed generation of mRNA transcripts comparable to those of normal controls (Fig 4C). However, CD40L/CD3δ mRNA transcript ratio was low in five mutations including two nonsense mutations (0.235 in family 11 and 0.385 in family 14), the intron 1 splice donor site mutation (0.008 in family 16), and two insertions (0.296/0.205 in families 27/28 with identical mutation, and 0.389 in family 30) (Fig 4C). Activated T-cell lines from all patients belonging to these five families with low CD40L/CD3δ mRNA transcript ratio were found to bind pAb, although at very low intensity. These results were reproducible and did not seem to be a background immunostaining since activated T-cell lines from affected members of family 26, who had most of the CD40Lgene deleted, consistently failed to bind pAb.
For patients with intron 2 splice donor site mutations (families 17 and 18), we used probe B to quantitate separately the two species of transcripts, one with exon 2-skipped, the other with a 19 nucleotide insertion (Fig 4A and B). The former mRNA transcripts are expected to generate a truncated protein with an in-frame deletion of 44 amino acids that is capable of CD40-Ig binding, but less efficiently than wild-type CD40L. Although the two different intron 2 splice donor site mutations have identical effects on the splicing of mRNA transcripts, they have a markedly different impact on the level of transcripts: total CD40L/CD3δ mRNA transcript ratio of activated PBMC was 0.464 in the mutation nt 309 + 1g → a (family 17) and 1.072 in the mutation nt 309 + 2t → a (family 18). In both of these mutations, the transcripts with a 19-nt insertion are dominant; however, the exon 2–skipped transcripts were threefold more abundant in nt 309 + 2t → a (PS, family 18) than in nt 309 + 1g → a (TaA, family 17) (0.380 in PS v 0.128 in TaA) (Fig4C). Interestingly, the clinical phenotype of patient PS with the mutation nt 309 + 2t → a is mild whereas patients YA and TaA of family 17 with the mutation nt 309 + 1g → a had classic XHIM. A similar situation was observed in families 19 and 20 with different mutations affecting the intron 3 splice donor site. We used probe C to quantitate separately the normally spliced transcripts and the exon 3–skipped transcripts (Figs 4A and B). Exon 3–skipped transcripts were the dominant mRNA species in both of the two splice site mutations; however, patient AM (family 19) with a mild phenotype and the mutation nt 367G → A had almost twice the amount of normally spliced transcript than patient MS of family 20 (0.290 in AMv 0.156 in MS) (Fig 4C). In all XHIM patients with splice site mutations that allow the generation of normally spliced mRNA transcripts, the levels of normally spliced mRNA transcripts were decreased. Patient DB of family 16 (nt 177 + 1g → t) had less than 1%, patient MS of family 20 (nt 367 + 2t → c) had 15%, and patient AM of family 19 (nt 367G → A) had 27% of the mean level of mRNA transcripts from normal controls (n = 20). Similarly, the levels of the mRNA transcripts encoding the mutants that retain the ability to bind CD40-Ig were decreased: patient DJ of family 11 (R11X) had 22%; the levels of exon 2–skipped transcripts in patient TaA of family 17 (nt 309 + 1g → a) and in patient PS of family 18 (nt 309 + 2t → a) were 12% and 36%, respectively.
Antibody responses to bacteriophage φX174.
To assess in vivo antibody production to a T-cell–dependent antigen, we immunized with bacteriophage φX174 nine XHIM patients from seven families with unique genotypes. After primary immunization, normal controls reach a peak antibody titer (Kv) at 2 weeks and produce phage-specific antibody that is almost entirely of the IgM isotype. After a second immunization 6 weeks later, antibody titers increase briskly, more than 10-fold, reaching a peak at 1 week (Fig 5). During the secondary response, half of the antibody is of the IgG type. All XHIM patients studied to date, independent of phenotype, had quantitatively and qualitatively abnormal antibody responses to bacteriophage φX174 (Fig 5). After primary immunization, five XHIM patients reached peak antibody titers at 1 week and then decreased. However, three patients with a mild phenotype (TA of family 7, DJ of family 11, and PS of family 18) were able to increase titers between weeks 1 and 2, reaching values that remained within the normal range of controls for the first 2 weeks. Titers decreased less rapidly in these three patients than in the others. After a secondary immunization, all XHIM patients failed to amplify antibody responses and the titers remained below 2 SD of normal controls. Compared with the six classic XHIM patients, the three milder cases responded with higher titers, but only one, patient DJ, produced phage specific IgG antibody (10%).
Antibody responses to bacteriophage ◊X174. Bacteriophage ◊X174 was injected twice (↓), 6 weeks apart and antibody titers determined in 12 normal male controls (•, geometric mean; the hatched area, ± 2 SD), and in nine XHIM patients. Neutralizing antibody is expressed as rate of phage inactivation or K value (Kv) on a log scale. The mean percentage of phage-specific IgG antibody in serum collected 2 weeks after secondary immunization is shown on the right. Antibody responses to bacteriophage ◊X174 were quantitatively and qualitatively abnormal in all XHIM patients studied. They characteristically failed to amplify and to switch from IgM to IgG. The three patients with a mild phenotype, PS, DJ, and TA (open symbols), had the highest titers, and one (DJ) was able to produce phage-specific IgG antibody, although at a low concentration (10%).
Antibody responses to bacteriophage ◊X174. Bacteriophage ◊X174 was injected twice (↓), 6 weeks apart and antibody titers determined in 12 normal male controls (•, geometric mean; the hatched area, ± 2 SD), and in nine XHIM patients. Neutralizing antibody is expressed as rate of phage inactivation or K value (Kv) on a log scale. The mean percentage of phage-specific IgG antibody in serum collected 2 weeks after secondary immunization is shown on the right. Antibody responses to bacteriophage ◊X174 were quantitatively and qualitatively abnormal in all XHIM patients studied. They characteristically failed to amplify and to switch from IgM to IgG. The three patients with a mild phenotype, PS, DJ, and TA (open symbols), had the highest titers, and one (DJ) was able to produce phage-specific IgG antibody, although at a low concentration (10%).
Phenotype and genotype.
The clinical characteristics of 45 affected males from 30 unrelated XHIM families were analyzed and the findings summarized in Table 3. More than half of the patients were diagnosed before they reached 1 year of age and only three patients, all with mild disease, were older than 5 years at the time the diagnosis was established. Intravenous Ig (IVIg) therapy was started in more than half of the patients before 1 year of age. PCP, often the presenting symptom during the first year of life, was observed in 38% of patients. Neutropenia was reported in 27 patients (60%) and was described as transient or intermittent in 15, and chronic in 11 patients. Cholangiolitis and liver cirrhosis were diagnosed in 4 patients; 1 was 3 years at diagnosis (MC, family 22), 2 were 23 years (GS, family 16 and CJ, family 30), and 1 was 24 years (DS, family 16). Five patients developed tumors of the gastrointestinal tract, including hepatic/pancreatic carcinoid (GS, family 16 and DG, family 29), bile duct carcinoma (MC, family 22), and adenocarcinoma of unknown origin (JJ and CJ, family 30); DG, the most recent patient diagnosed with a tumor is the only survivor. All but two patients (IN, family 8 and DJ, family 11) are receiving regular IVIg infusions, with good or fair response in 88%.
Clinical Features of Study Population (45 XHIM males from 30 unrelated families)
| Age at Onset* | Age When Ig Therapy Started | ||
| Age (yr) | No. of Patients | Age (yr) | No. of Patients |
| <1 | 24 (59%) | <1 | 24 (53%) |
| 1-3 | 10 (24%) | 1-3 | 11 (24%) |
| 4-5 | 4 (10%) | 4-5 | 2 |
| 8 | 1 | 6-10 | 2 |
| 14 | 1 | 11-15 | 3 |
| 17 | 1 | 16-20 | 1 |
| Never | 2† | ||
| PCP 17 Patients (38%) | Neutropenia 27 Patients (60%) | ||
| Age (yr) | No. of Patients | No. of Patients | |
| <1 | 14 | Transient | 12 |
| 1-4 | 3‡ | Intermittent | 3 |
| Chronic | 11 | ||
| Associated with B19 infection | 12-153 | ||
| Cholangiolitis and cirrhosis (due to Cryptosporidiuminfection)4 patients (9%) | |||
| Malignancy | 5 Patients (11%) | ||
| Response to IVIg Therapy2-155 | |||
| No. of patients | |||
| Good | 20 (48%) | ||
| Fair | 17 (41%) | ||
| Marginal | 5 (12%) | ||
| Age at Onset* | Age When Ig Therapy Started | ||
| Age (yr) | No. of Patients | Age (yr) | No. of Patients |
| <1 | 24 (59%) | <1 | 24 (53%) |
| 1-3 | 10 (24%) | 1-3 | 11 (24%) |
| 4-5 | 4 (10%) | 4-5 | 2 |
| 8 | 1 | 6-10 | 2 |
| 14 | 1 | 11-15 | 3 |
| 17 | 1 | 16-20 | 1 |
| Never | 2† | ||
| PCP 17 Patients (38%) | Neutropenia 27 Patients (60%) | ||
| Age (yr) | No. of Patients | No. of Patients | |
| <1 | 14 | Transient | 12 |
| 1-4 | 3‡ | Intermittent | 3 |
| Chronic | 11 | ||
| Associated with B19 infection | 12-153 | ||
| Cholangiolitis and cirrhosis (due to Cryptosporidiuminfection)4 patients (9%) | |||
| Malignancy | 5 Patients (11%) | ||
| Response to IVIg Therapy2-155 | |||
| No. of patients | |||
| Good | 20 (48%) | ||
| Fair | 17 (41%) | ||
| Marginal | 5 (12%) | ||
*Four unrelated patients (SC [family 3], AY [family 5], JaB [family 9], and RoC [family 22], each with an older affected brother) are excluded since they were diagnosed before developing symptoms.
JoC (family 22) died of PCP at 8 months of age. IN (family 8), now 24 years of age, is doing well without IVIg infusion.
SC (family 3), AY (family 5), and JaB (family 9) developed PCP after trimethoprim-sulfamethoxazole prophylaxis had been stopped.
Neutropenia developed during parvovirus B19 infection (DJ, family 11).
Three patients cannot be evaluated and were excluded; MS (family 20) underwent bone marrow transplantation at an early age; IN (family 8) was never treated with IVIg; and JoC (family 22) died of PCP at 8 months of age. Clinical course after initiation of IVIg therapy was evaluated according to the following criteria: good, frequency and severity of infection similar to those expected in normal children; fair, infections are not severe but more often than in normal children and less frequent than before IVIg; and marginal, frequency and severity of common infections seem improved, but the clinical symptoms may at times be severe.
Five unrelated patients, all teenagers or young adults, had phenotypes that clearly distinguished them from patients with classic XHIM (Table 4). Two had identical missense mutations affecting exon 5 (T254M), one had a nonsense mutation (R11X), and two had splice site mutations affecting intron 2 (nt 309 + 2t → a) and exon 3 (nt 367G → A), respectively. All were older at onset, never had PCP, and neutropenia was observed in only two cases, one intermittently and one during parvovirus B19 infection. Parvovirus B19-induced red blood cell aplasia was the presenting illness in three of the five patients with a mild phenotype, but was not observed in patients with classic XHIM. In the latter group of patients, Ig therapy was started at a mean age of 1.5 years. In the five patients with mild disease, IVIg therapy was started much later. None of the patients presenting with parvovirus B19-induced anemia had received IVIg prophylaxis at the time they developed symptoms, and at present only three of our patients with a milder phenotype receive regular IVIg infusions. It is conceivable that patients with classic XHIM avoid infection with parvovirus B19 because they are recognized earlier and started on IVIg at a younger age than those XHIM patients with a milder phenotype. Of those with mild disease, none had any problems with recurrent infections regardless of treatment with IVIg. Activated T-cell lines from patients with a mild phenotype were able to bind all four anti-CD40L MoAbs tested (immunostaining type 2) and three could bind CD40-Ig (immunostaining type 1); in contrast, activated T-cell lines from most patients (80%) with a classic XHIM phenotype were classified as immunostaining type 3, 4, or 5 and did not bind CD40-Ig.
Clinical Features of Five Selected Patients Compared With Classic XHIM
| . | Classic Phenotype (n = 40) . | Mild Phenotype (family no.) . | ||||
|---|---|---|---|---|---|---|
| T254M (7) . | T254M (8) . | R11X (11) . | nt309 + 2t → a (18) . | nt367G → A (19) . | ||
| Age (yr) at onset | Mean 1.1 (range, 0.17-5)3-150 | 14 | 5 | 8 | 17 | 5 |
| PCP | 43% | − | − | − | − | − |
| Neutropenia | 68% | − | Intermittent | +3-151 | − | − |
| Cryptosporidium | 10% | − | − | − | − | − |
| Red blood cell aplasia due to B19 infection | 0% | + | − | + | + | − |
| Age (yr) when Ig therapy started | Mean 1.5 (range, 0.5-12) | 14 | Never | 123-152 | 18 | 5 |
| Response to Ig therapy | Good in 42%3-153 | Good | NA | Good | Good | Good |
| Immunostaining type | ≥3, 16/20 (80%)3-155 | 2 | 2 | 1 | 1 | 1 |
| . | Classic Phenotype (n = 40) . | Mild Phenotype (family no.) . | ||||
|---|---|---|---|---|---|---|
| T254M (7) . | T254M (8) . | R11X (11) . | nt309 + 2t → a (18) . | nt367G → A (19) . | ||
| Age (yr) at onset | Mean 1.1 (range, 0.17-5)3-150 | 14 | 5 | 8 | 17 | 5 |
| PCP | 43% | − | − | − | − | − |
| Neutropenia | 68% | − | Intermittent | +3-151 | − | − |
| Cryptosporidium | 10% | − | − | − | − | − |
| Red blood cell aplasia due to B19 infection | 0% | + | − | + | + | − |
| Age (yr) when Ig therapy started | Mean 1.5 (range, 0.5-12) | 14 | Never | 123-152 | 18 | 5 |
| Response to Ig therapy | Good in 42%3-153 | Good | NA | Good | Good | Good |
| Immunostaining type | ≥3, 16/20 (80%)3-155 | 2 | 2 | 1 | 1 | 1 |
Abbreviations: NA, not applicable; Ig, immunoglobulin.
Four patients were excluded (n = 36, see Table 3).
Only during parvovirus B19 infection.
IVIg is administered sporadically every 6 to 18 months to treat B19-induced anemia.
Two patients were excluded (n = 38, see Table 3).
16 of 20 unique genotypes showed an immunostaining type of 3-5; 4 unique genotypes had a type 1 or 2.
DISCUSSION
The discovery of the molecular defect causing the X-linked form of the hyper IgM syndrome has allowed the precise definition of XHIM. Affected males have mutations in the CD40L gene resulting in the inability of T cells to express functional CD40L molecules, and as a consequence, lack B-cell signaling via the CD40 receptor. Mutations responsible for XHIM are distributed throughout the CD40L gene and are highly heterogeneous.7-13,25-29,38-40 A recent report from the XHIM Registry of the European Society for Immunodeficiencies, based on mutation data from 53 families,23 indicated that the most common CD40L mutations were missense mutations, present in 39.5% of XHIM patients, followed by nonsense mutations (18.6%), deletions (19.8%), insertions (10.5%), and splice site mutations (9.3%). Here, we report the individual mutations and their effects on CD40L expression in 30 unrelated families. Of the 28 unique mutations identified, 10 have not been previously described. Although missense mutations of the CD40L gene were the most common with 33% (10 of 30 families), splice site mutations were nearly as frequent (9 of 30 families). Mutations affecting the splice donor site were found in each intron of the CD40L gene. Four of these splice donor site mutations (nt 177 + 1g → t, nt 367G → A, nt 367 + 2t → c, and nt 367 + 5g → a) were leaky and produced both normally and abnormally spliced CD40L mRNA transcripts. The consensus sequence at the 5′ end of introns that is recognized efficiently by the spliceosome is (A/C)AG(−1)g(+1)taagt in which the cleavage occurs between the G nucleotides at positions −1 and +141: the gt dinucleotides located at the 5′ end of each intron is considered invariant and the most preserved sequence. One possible exception is the substitution of the invariant gt dinucleotides with gc dinucleotides as long as the other nucleotides match the consensus.41 In vitro studies of the rabbit β-globin gene showed that the gt → gc mutation at the splice donor site merely reduced the efficiency of splicing.42 Such a situation was observed in patient MS (family 20) with the mutation nt 367 + 2t → c that generated wild-type transcripts, at a reduced amount, as well as transcripts that skipped exon 3. Because the 5′ cleavage site is not determined solely by the invariant gt dinucleotides but rather by the 5′ splice region as a whole,42 it is likely to find normally spliced transcripts whenever multiple RT-PCR products are identified. This is further underscored by the finding that three different mutations affecting the intron 3 splice donor site (mutations at position −1, +2, and +5) allow activated T cells to generate normally spliced CD40L transcripts in addition to misspliced transcripts. An RT-PCR strategy designed to amplify the entire CD40L coding region has the advantage of identifying multiple splicing products, which may be missed if single-strand conformational polymorphism43 or dideoxy fingerprinting44screening, preceded by amplification of several overlapping segments, is used.7,11 29 This RT-PCR strategy used here may explain the high frequency of multiple splicing products, including wild-type CD40L mRNA transcripts, we have observed in patients with splice site mutations.
Using a previously described three-dimensional model of the CD40L extracellular region, we have explored the effect of amino acid substitutions resulting from missense mutations on the structure and function of CD40L.15 Missense mutations affecting the core packaging of the CD40L monomer (class I), found in four of nine unique missense mutations, were the most common, followed by three missense mutations known to interfere directly or indirectly with CD40 binding (class III). Class II missense mutations, thought to affect trimer formation, were found twice. Although most missense mutations resulted in mutants that were recognized by pAb and at least one MoAb (usually MoAb 106) (immunostaining type 3) or by pAb only (immunostaining type 4), mutant T254M (class I) and T147N (class III) were recognized by all four MoAbs tested. Although the immunostaining intensity of cells with the T254M mutation was much lower than that observed in normal controls (Fig 1H, I, and K), the finding that all four MoAbs could bind to this mutant suggests that this amino acid substitution affects CD40L monomer structure less profoundly. The T147N mutant, on the other hand, was able to bind all MoAbs with almost normal intensity (Fig 2). Five residues located along the interface between CD40L monomers (K143, Y145, Y146, R203, Q220) contribute substantially to the interaction with CD40.45 The proximity of codon 147 to the CD40 binding site is the most likely explanation for the interference of T147N with CD40 binding, despite the fact that the resulting amino acid change is conservative and the mutant retains binding activity to four MoAbs. Thus, the classification of missense mutations combined with the immunostaining pattern provides insight into the structural and functional abnormalities of mutated CD40L.
To explore a possible correlation between phenotype and immunostaining type, we selected XHIM patients who presented with a mild phenotype based on clinical characteristics, including age at onset of symptoms, frequency of infections requiring antibiotic therapy, the presence of complications (neutropenia, PCP, and cholangiolitis), and response to IVIg therapy. Five XHIM patients with mild phenotype could be clearly delineated (Table 4): all had late onset of symptoms and never PCP; in three patients the presenting illness was chronic anemia due to red blood cell aplasia caused by parvovirus B19 infection; they had few complications and a good response to IVIg therapy. The mutations identified in these patients were heterogeneous and allowed the generation either of mutant CD40L that can bind CD40-Ig (R11X, nt 309 +2t → a, and nt 367G → A), or of a class I missense mutation (T254M, present in two unrelated patients) resulting in an extracellular domain that can bind all four anti-CD40L MoAbs tested. In contrast, two additional patients (MS of family 20, nt 367 + 2t → c, and JE of family 21, nt 367 + 5g → a) with splice site mutations that allow the generation of wild-type CD40L mRNA transcripts, at reduced amounts, developed PCP during the first 6 months of life, similar to a patient, described by Ameratunga et al,46 who developed PCP in early infancy and was found to have a splice site mutation that resulted in the expression of very low amounts of wild-type CD40L mRNA transcripts. After recovery, patient JE, now 6 years old, has remained asymptomatic on IVIg therapy, and patient MS's subsequent clinical course cannot be evaluated because of successful bone marrow transplantation at the age of 9 months. Although PCP is most often associated with an impaired host defense system and is considered a frequent complication of primary or secondary T-cell deficiencies, Pneumocystis carinii has been identified as an important cause of pneumonia in normal infants. In a prospective study, Stagno et al47 found P carinii antigenemia in 10 of 67 infants, none of which had received immunosuppressive drugs. Possible reasons for this susceptibility include the decreased ability of lymphocytes from young infants to produce interferon-γ and IL-4, and to express CD40L.31 The majority of our XHIM patients who had developed PCP during the first year of life did not develop PCP again, even after PCP prophylaxis was discontinued. We conclude that the patients whose genotypes result in immunostaining types 1 and 2 often present with a mild XHIM phenotype, although they may still be at risk of developing PCP because other factors apart from defective CD40L contribute to the development of PCP.
To confirm the diagnosis of XHIM, a mutated, nonfunctional CD40L protein has to be demonstrated. The most widely accepted techniques include (1) immunostaining of activated T cells with anti-CD40L antibodies; (2) binding of a CD40-Ig construct designed to specifically interact with CD40L; and (3) mutation analysis of the CD40Lgene. Using polyclonal and monoclonal anti-CD40L antibodies, we were able to detect mutated CD40L on the surface of activated T cells of all XHIM patients except those from one family with a large deletion, and to distinguish five immunostaining types. Thus, our study expands the observation of Callard et al,48 who, using a CD40-Ig construct and MoAbs 5c8 and TRAP, have described two different staining patterns for CD40L expression in a group of XHIM patients. In addition, we have identified five mutations (R11X, nt 309 + 2t → a, nt 367G → A, nt 367 + 2t → c, and nt 367 + 5g → a) that allow the binding of a CD40-Ig construct by activated patients' T-cell lines. In three of them (R11X, nt 367G → A, and nt 367 + 5g → a), the binding of CD40-Ig was noted in activated PBMC, the cell preparation most commonly used for the diagnostic immunostaining in patients with XHIM. Nevertheless, patients whose activated lymphocytes bind CD40-Ig must express functional CD40L. The fact that these patients are symptomatic indicates that their activated T cells are not capable of engaging CD40 on B cells effectively. This may be due to a sub-threshold expression of CD40L, a depressed affinity or avidity for CD40 or the formation of “heterotrimers,” consisting of wild-type and mutated CD40L monomers, as is expected in splice site mutations that result in multiple splicing products. All these possibilities will result in deficient cross-linking of CD40. It is important to note that in the genotypes resulting in the immunostaining type 1, 2, or 3, found in 13 of 24 genotypes (54%) from our study group, CD40L expression by activated PBMC may be detected by anti-CD40L MoAbs or CD40-Ig. Indeed, at least one MoAb, usually MoAb 106, has bound to activated PBMC in 12 genotypes and bCD40-Ig has bound to activated PBMC in three genotypes. Thus, 12 genotypes (50%) would have been missed if only one of the four MoAbs available to us had been used for immunostaining of activated PBMC. If immunostaining with MoAbs, together with CD40-Ig binding by activated PBMC, had been used without sequence analysis, 3 of 24 genotypes tested (12.5%) would have been missed. As previously described37 and shown in Fig 1F and G, decreased binding of anti-CD40L MoAbs (and pAb) and CD40-Ig is observed in a subgroup of patients with CVI who have a staining pattern similar to the immunostaining type 1 of XHIM patients. To discriminate XHIM patients from CVI patients (both with an immunostaining type 1), molecular analysis of the CD40L gene is required. Nevertheless, it is reasonable to study CD40L expression by activated T cells with one or two MoAb(s) and with the CD40-Ig construct because this technique will identify approximately 90% of XHIM patients.
Although activated T cells from XHIM patients with certain mutations of the CD40L gene can express either wild-type or mutant CD40L that can bind CD40-Ig, they do so at low density. Carrier females, on the other hand, have been identified with extremely skewed CD40L expression, favoring the X chromosome carrying the abnormal gene, and whose immunologic status nevertheless has been completely normal.26 In these carrier females, a small number of T cells in which the normal X chromosome is active is expected to express CD40L at normal density, suggesting that a relatively small number of T cells with a normal density of CD40L molecules is sufficient for the normal function of the immune system. There is most likely a threshold density of CD40L that is required to ligate CD40 on B lymphocytes efficiently to transduce signals that will ultimately result in increased Ig production and class switching. The reciprocal dialogue between T and B cells through CD40L/CD40 and B7-1 or B7-2/CD28 is important for T cells to be fully activated and exert their effector functions.49-51 Without a sufficient density of functional CD40L expressed by activated T cells, this reciprocal dialogue between T and B cells may not occur or may be inadequate. As a result, T cells may fail to produce IL-4 and IL-10, which are important cytokines for B cells to undergo Ig isotype switching and to produce high-affinity antibody.52,53 On the other hand, CD40 is not only expressed by B cells but also by a variety of cells including monocytes/macrophages,16-19 dendritic cells,20vascular endothelial cells,21,22 and transformed cells.54 These observations suggest that CD40L/CD40 interaction can influence many aspects of T-cell–mediated inflammatory responses, such as cell extravasation, production of inflammatory cytokines, apoptosis of transformed cells or rescue of macrophages from apoptosis, as well as activation of macrophage effector functions.19,54,55 Signaling via CD40 activates monocytes to produce nitric oxide 56,57 involved in the protection from intracellular pathogens.58 Differences in CD40L requirement are suggested by the observation that monocytes can be activated to produce IL-1β, TNF-α, IL-6, and IL-8 in the presence of a soluble CD40L-CD8 fusion protein alone, without the need for other costimulatory molecules.19 Therefore, it is possible that the low density of CD40L expressed on activated T cells in patients with immunostaining type 1 (eg, R11X and several splice site mutations) are able to activate some CD40-expressing cells, eg, monocytes, but not B lymphocytes, because B cells require contact-independent help provided by activated T cells in addition to CD40 ligation. This could explain the fact that PCP and persistent Cryptosporidiuminfection, characteristic for classic XHIM patients, are rarely observed in mild cases whereas persistent parvovirus B19 infection, dependent on specific IgG production for elimination, is a frequent complication. However, other genetic or environmental factors may explain the milder phenotype observed in this subgroup of XHIM patients.
ACKNOWLEDGMENT
We thank the following physicians for referring patients: A.J. Apter, N.K. Day, A. Dorenbaum, R. Good, H. Hasle, H. Hill, A. Huttenlocher, R. Kagan, R. Kobayashi, B. Mazer, R. Roberts, D. Rosen, F.T. Saulsbury, M.J. Schumacher, J. Slater, E.R. Stiehm, K. Sullivan, K. Terada, J. Winkelstein, and D. Williams-Herman. We thank Drs Christopher Wilson and Brian Smart for their critical reading of the manuscript.
Supported in part by the University of Washington Clinical Research Center (RR-37), and by grants from the National Institutes of Health (HD17427 and AI40102), the March of Dimes Birth Defects Foundation (6-FY96-0330), and the Immune Deficiency Foundation.
Address reprint requests to Hans D. Ochs, MD, Department of Pediatrics, University of Washington, School of Medicine, Box 356320, Seattle, WA 98195-6320; e-mail: allgau@u.washington.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

![Fig. 3. RT-PCR in patients with splice donor site mutations of the CD40L gene. (A) Design and location of primers used in RT-PCR. Shown are the coding regions of CD40L cDNA only. The numbers on top indicate the starting nucleotide number of each exon. The location of primers are shown by arrows [sense (→) and antisense (←)]. Note that sense primers used for amplification of introns 2, 3 , and 4 splice donor site mutations were designed to be localized in an exon that may be skipped. (B) Agarose gel electrophoresis of RT-PCR products. RT-PCR was performed using primer pairs P1 and P3 (lanes 1 through 3), P4 and P5 (lanes 4 through 7), and P6 and P7 (lanes 8 through 11), and P7 and P8 (lanes 12 through 15) (see Materials and Methods). An aliquot of the RT-PCR product was size-fractionated on 2% agarose gel and stained with ethidium bromide. DNA size markers are shown on the left. cDNA used for RT-PCR was derived from a normal control (lanes 1, 4, 8, and 12), DB (family 16) in lane 2, PS (family 18) in lane 5, TaA (family 17) in lane 6, MS (family 20) in lane 9, AM (family 19) in lane 10, SeC (family 22) in lane 13, RA (family 23) in lane 14; lanes 3, 7, 11, and 15 are negative controls (no cDNA in the reaction mixture). Note that patients with splice donor site mutations involving introns 1 (lane 2) and 3 (lanes 9 and 10) but not those involving introns 2 (lanes 5 and 6) and 4 (lanes 13 and 14) have bands which are indistinguishable from those derived from normally spliced mRNA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/7/10.1182_blood.v92.7.2421/3/m_blod41924003ax.jpeg?Expires=1765938240&Signature=PLXRgUrf8CgNthJO8OvKtylGLet6SIpiNM9zVJg8FeE6OwfWz9SNuHcU9PbUKvVEpNGmmpbS2ILXg1tR~8nDX7p07FKqHmOI06xgFisEUWw~UYXH5r2Clq5NguhEeEsaEvG6ywajYm6tu6noTYx888kyYFwGxaZHMD2pwwAjSIPIcc9OmekYBg4xk0u~CBnF2bFj4QJ1I5Zu2rAdWs5b9VnzwDJxaynWv8PsN2dpC1lW9XytRyIN469RW6ePkRR84FyRovvV-0~KLV309CIG6aYZOiAZHIj4HtIHYxxcGkGLog8rnIqkvY-36Z0Ssdof9C-pAAVxIJQZbxQ-9Vd8rw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. RT-PCR in patients with splice donor site mutations of the CD40L gene. (A) Design and location of primers used in RT-PCR. Shown are the coding regions of CD40L cDNA only. The numbers on top indicate the starting nucleotide number of each exon. The location of primers are shown by arrows [sense (→) and antisense (←)]. Note that sense primers used for amplification of introns 2, 3 , and 4 splice donor site mutations were designed to be localized in an exon that may be skipped. (B) Agarose gel electrophoresis of RT-PCR products. RT-PCR was performed using primer pairs P1 and P3 (lanes 1 through 3), P4 and P5 (lanes 4 through 7), and P6 and P7 (lanes 8 through 11), and P7 and P8 (lanes 12 through 15) (see Materials and Methods). An aliquot of the RT-PCR product was size-fractionated on 2% agarose gel and stained with ethidium bromide. DNA size markers are shown on the left. cDNA used for RT-PCR was derived from a normal control (lanes 1, 4, 8, and 12), DB (family 16) in lane 2, PS (family 18) in lane 5, TaA (family 17) in lane 6, MS (family 20) in lane 9, AM (family 19) in lane 10, SeC (family 22) in lane 13, RA (family 23) in lane 14; lanes 3, 7, 11, and 15 are negative controls (no cDNA in the reaction mixture). Note that patients with splice donor site mutations involving introns 1 (lane 2) and 3 (lanes 9 and 10) but not those involving introns 2 (lanes 5 and 6) and 4 (lanes 13 and 14) have bands which are indistinguishable from those derived from normally spliced mRNA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/92/7/10.1182_blood.v92.7.2421/3/m_blod41924003bw.jpeg?Expires=1765938240&Signature=ngyuEpHzG67isCrdmsYw9IGwav6jAnX6kdJz9xiS72U9mXM-QzQlCarxaS2pyXCYjW7po-1nG7HAbpe5B5cqYuLScvCPWmz6zVKIiRYNekGeNxm8YaLEiYjzqaMigu6yvYS1I9vd3XSTP1PFIPjCGwOoJrOwMemw-d3iaNv0Ngeh~5j~o-VAmlVbC7Z81W2cLbw~oNY6uwSQHnwrFJlz3efQI03qqbxCN8s1BwyGucy2wimnHti9sYeS~x99oDCXV7qxpeXj~3XVp8ypCNyasg6-WhSAE3IehH5oAorvmLLDfI0So30HRLL7p8i6~f-9NDm9MC5LEhb4Coa5QyHgAA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

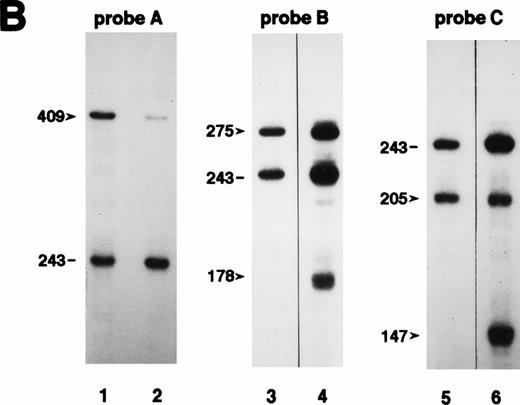
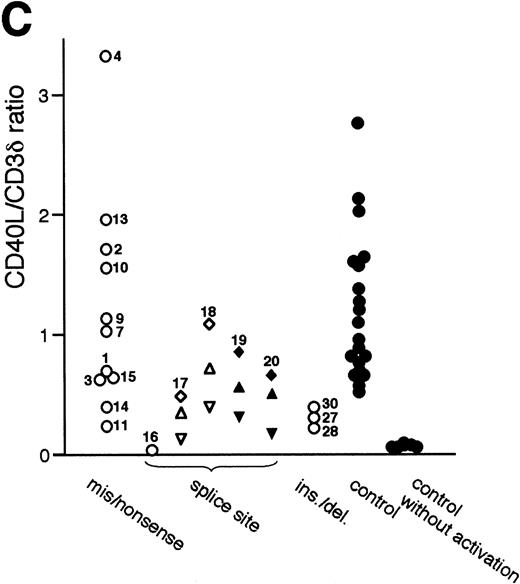
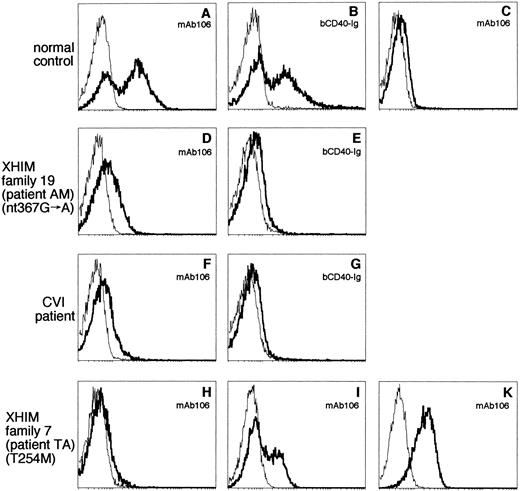
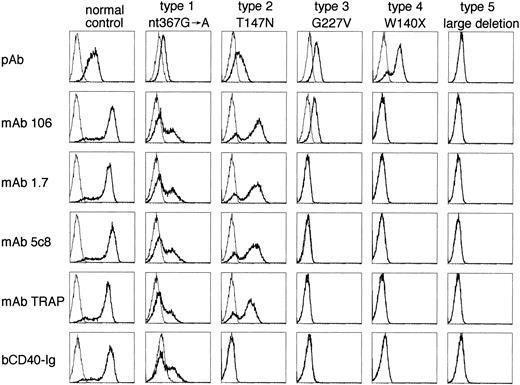
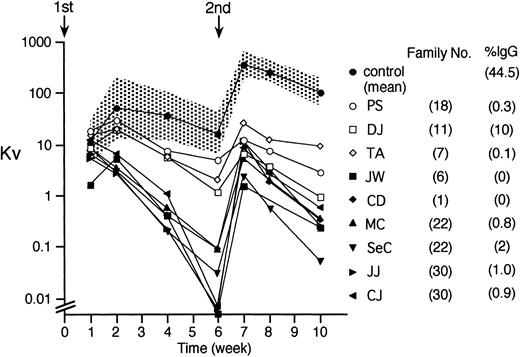
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal