Abstract
Previous studies have suggested that the B-cell repertoire after stem cell transplantation resembles the developing repertoire in the fetus. Fetal and adult repertoires differ strikingly at the molecular level in Ig heavy chain third complementarity determining region (H CDR3) size distribution and Ig gene utilization. Previously, the posttransplant repertoire has not been studied fully in this regard. In this study, we analyzed H CDR3s posttransplant using CDR3 fingerprinting, single-strand conformation polymorphism (SSCP), and random sequencing. Eleven adult patients who received either autologous (n = 6) or allogeneic adult sibling (n = 5) hematopoietic stem cell transplants were studied. IgM H CDR3 repertoires demonstrated limited clonal diversity within the first 6 to 10 weeks posttransplant. By 3 to 4 months, the IgM H CDR3 repertoires were as diverse as those in healthy adults. Reconstitution of the IgM diversity correlated with the expansion of the multimember VH3 family. By contrast, the contribution of the single-member VH6 family was limited in most patients up to 6 to 9 months. No evidence was seen for greater contribution of VH6 posttransplant. IgG repertoires remained clonally restricted at all times. In all patients, H CDR3 sizes fell within adult limits. Direct nucleotide sequencing of H CDR3s showed adult-type N-nucleotide insertions and Ig gene utilization. These results indicate that the emerging repertoire posttransplant does not resemble the developing fetal repertoire at the molecular level.
© 1998 by The American Society of Hematology.
HIGH-DOSE CHEMO-RADIOTHERAPY with hematopoietic stem cell support has become the standard treatment for several otherwise incurable malignant and nonmalignant diseases. However, the success of stem cell transplantation can be hampered by delayed B-cell immune reconstitution and associated infectious complications. Several studies have suggested that this delay is due in part to the time period required for recapitulation of B-cell ontogeny.1-11 To address this issue at the molecular level, we have analyzed the emerging B-cell repertoire posttransplant using fingerprinting and single-strand conformation polymorphism (SSCP) of Ig heavy chain third complementarity determining regions (H CDR3s) in combination with direct sequencing. This allowed for a global view of the repertoire so that the nature and diversity of the emergent repertoire could be objectively examined.
Ig heavy chain (Ig H) diversity is generated by somatic recombination of variable (VH), diversity (DH), and joining (JH) gene segments.12 Further diversification is accomplished by the addition of nongermline encoded N-nucleotides to the VH-DH and DH-JHjunctions by the enzyme terminal deoxynucleotidyl transferase (TdT). Endonuclease activity and the addition of P-nucleotides provide additional junctional diversity. These mechanisms together generate a hypervariable segment of Ig H termed the third complementarity determining region (H CDR3). H CDR3s provide essential residues for direct interaction with antigens.
Diversification of H CDR3s is differentially regulated during ontogeny resulting in characteristic differences between fetal and adult repertoires. Several studies have shown that the single-member family VH6 is preferentially used in the fetal repertoire,13-19 comprising up to 10% of Ig H transcripts in the second trimester. In contrast, 0% to 5% of transcripts in adults use VH6.19-22 DHQ52, the shortest DH element, is used in up to 40% of the fetal transcripts14,15,17,18,23-25 and is rarely seen in the adult repertoire.20-22,25-27 JH3 and JH4 genes, among the shortest in nucleotide length, are used frequently in the fetal repertoire.14,15,17,18,23Because of limited TdT expression in utero,28 fetal Ig H transcripts often lack or contain fewer numbers of N-nucleotides.14,15,17,18,23 25 Together, these mechanisms result in fetal H CDR3s that are smaller in size than that seen in adults.
It has been argued that the newly acquired repertoire after bone marrow transplantation (BMT) resembles a fetal one, suggesting that the full ontogenetic program must be repeated for successful reconstitution of the B-cell repertoire.6,7,9,11 Previous studies have generally focused on morphologic, phenotypic, and functional characteristics of the posttransplant B-cell repertoire6-8and VH gene utilization.9,11,29 However, other important molecular characteristics that typify fetal and adult repertoires were not examined in detail. In fact, published studies of H CDR3s after high-dose therapy and stem cell transplantation are limited to VH6 containing rearrangements in 2 patients.30
The objectives of this study were to enhance our understanding of B-cell generation posttransplant at the molecular level and to further characterize the developing repertoire. In this study, the Ig H CDR3 size distribution and diversification posttransplant show that the emerging repertoire is clonally restricted. However, the repertoire does not resemble a fetal one, because it displays adult-size H CDR3s, adult-type Ig gene utilization and no evidence of bias towards VH6. IgG repertoire diversification lags significantly behind IgM. This suggests impaired memory reconstitution and provides an explanation for the immunodeficiency after stem cell transplantation.
MATERIALS AND METHODS
Stem cell recipients and control subjects.
Eleven adult patients who underwent hematopoietic stem cell transplantation between June and December 1996 in the BMT Units of the University and the Audie L. Murphy Veterans Affairs Hospitals in San Antonio under Institutional Review Board approved protocols were studied. The patient characteristics are summarized in Table 1. Four patients received allogeneic BMT from HLA-identical siblings. These patients received graft-versus-host disease (GVHD) prophylaxis consisting of cyclosporin and a short-course of methotrexate. One patient with acute myeloid leukemia (AML) who had relapsed after allogeneic BMT received a leukocyte infusion from the same donor (HLA-identical sibling) after cytoreduction. Intravenous immune globulin (IVIG) was administered to allogeneic BMT recipients until day +100 posttransplant. Only 2 patients (patients no. 1 and 5) developed grade II GVHD involving the skin and the gut at 13 and 9 weeks posttransplant, respectively. GVHD was treated with cyclosporin and steroids. The remaining 6 patients received autologous stem cell transplantation (2 peripheral blood stem cell transplantation [PBSCT], 2 BMT, 1 BM plus PBSCT, and 1 CD34 selected PBSCT). None of the autologous stem cell recipients received IVIG posttransplant. Granulocyte colony-stimulating factor (G-CSF) was administered to all patients until the absolute neutrophil count was greater than 5,000/μL in 2 consecutive days. Control subjects were the healthy donors for the allogeneic transplant recipients.
Patient Characteristics
| Patient No. . | Age/Sex . | Diagnosis . | Source of Stem Cells . | Conditioning Regimen . | MNCs (108/kg) . | CD34+ (106/kg) . |
|---|---|---|---|---|---|---|
| 1 | 54/M | AA | Matched sibling BM | Cy/TBI | 2.1 | 2.9 |
| 2 | 44/M | CML | Matched sibling BM | Bu/Cy | 1.4 | 2.4 |
| 3 | 50/M | NHL | Matched sibling BM | CBV | 1.9 | 2.1 |
| 4 | 51/F | AML | Matched sibling BM | Bu/Cy | 1.9 | 2.3 |
| 5 | 30/M | AML | Matched sibling PB | Flud/Ara-C | 10.5 | 6 |
| 6 | 32/M | NHL | Autologous BM + PB | CBV | 8.7 | 1 |
| 7 | 53/M | HD | Autologous BM | CBV | 2.7 | 1 |
| 8 | 47/M | NHL | Autologous BM | CBV | 2.2 | ND |
| 9 | 36/F | Breast Ca | Autologous PB | Cy/Carbo/TT | 9.1 | 2.9 |
| 10 | 66/F | Breast Ca | Autologous PB | Cy/Carbo/TT | 18 | 1.3 |
| 11 | 51/F | MM | Autologous PB CD34+ | Bu/Cy | 0.3 | 3.4 |
| Patient No. . | Age/Sex . | Diagnosis . | Source of Stem Cells . | Conditioning Regimen . | MNCs (108/kg) . | CD34+ (106/kg) . |
|---|---|---|---|---|---|---|
| 1 | 54/M | AA | Matched sibling BM | Cy/TBI | 2.1 | 2.9 |
| 2 | 44/M | CML | Matched sibling BM | Bu/Cy | 1.4 | 2.4 |
| 3 | 50/M | NHL | Matched sibling BM | CBV | 1.9 | 2.1 |
| 4 | 51/F | AML | Matched sibling BM | Bu/Cy | 1.9 | 2.3 |
| 5 | 30/M | AML | Matched sibling PB | Flud/Ara-C | 10.5 | 6 |
| 6 | 32/M | NHL | Autologous BM + PB | CBV | 8.7 | 1 |
| 7 | 53/M | HD | Autologous BM | CBV | 2.7 | 1 |
| 8 | 47/M | NHL | Autologous BM | CBV | 2.2 | ND |
| 9 | 36/F | Breast Ca | Autologous PB | Cy/Carbo/TT | 9.1 | 2.9 |
| 10 | 66/F | Breast Ca | Autologous PB | Cy/Carbo/TT | 18 | 1.3 |
| 11 | 51/F | MM | Autologous PB CD34+ | Bu/Cy | 0.3 | 3.4 |
Abbreviations: AA, aplastic anemia; CML, chronic myelogenous leukemia; NHL, non-Hodgkin’s lymphoma; AML, acute myelogenous leukemia; HD, Hodgkin’s disease; Ca, cancer; MM, multiple myeloma; BM, bone marrow; PB, peripheral blood; Cy/TBI, cyclophosphamide/total body irradiation; Bu/Cy, busulfan/cyclophosphamide; CBV, cyclophosphamide/BCNU/VP-16; Flud/Ara-c, fludarabine/Ara-c; Cy/Carbo/TT, cyclophosphamide/carboplatin/thiotepa; ND, not done.
Cell collection, separation, and RNA isolation.
Baseline peripheral blood (PB) samples were collected before transplant from patients and donors. Posttransplant samples (minimum of 6) were collected from patients at various times up to 9 months or until the patients returned to their referring institutions. PB mononuclear cells (PBMCs) were separated by Ficoll-Hypaque density gradient (Organon Teknika, Durham, NC) and washed twice with phosphate-buffered saline (PBS). RNA was isolated from approximately 1 × 107PBMCs using TRIzol total RNA isolation reagent (Life Technologies, Gaithersburg, MD) according to the protocol supplied with the reagent.
cDNA synthesis and polymerase chain reaction (PCR) amplification.
One microgram of RNA isolated from blood samples was used for first-strand cDNA synthesis using a combination of oligo(dT) and random hexamers and SuperScript II Reverse transcriptase in a final volume of 20 μL. The protocol supplied with the cDNA synthesis kit was used (Life Technologies).
The amount of Ig constant region transcripts was estimated for each cDNA sample in semiquantitative PCRs to control for variations in B-cell numbers and transcriptional activity. This method has been previously used in TCR repertoire studies.31-34 The Ig constant region transcripts were amplified in PCRs using 0.25, 0.5, 1, and 2 μL of the cDNA together with the Cμ15 and Cμ2RC primers (Table 2). PCR cycles consisted of 30 seconds of denaturation at 92°C, 35 seconds of annealing at 68°C, and 1 minute of extension at 72°C. PCR conditions were otherwise identical to that described below. Samples were obtained after 20, 23, 26, 29, and 32 PCR cycles to visualize the products. Based on the amount of the Ig constant region transcripts estimated for each sample, the amount of cDNA used in subsequent analyses was adjusted in a final volume of 5 μL per PCR. In this way, it was assured that samples with similar amount of constant region transcripts were being compared.
PCR Primers
| 5′ Primers | |
| Pan VH-consensus primer (VH-FR1) | 5′-CAGGTGCAGCTGGTGGAGTCTGGG-3′ |
| VH3 (leader sequence) | 5′-GGGTCGACTCCATGGAGTTTGGGCTGAGC-3′ |
| VH6 (leader sequence) | 5′-GGGTCGACTGTCTCCTTCCTCATCTTCCT-3′ |
| Pan VH-FR3 | 5′-GGGTCGACACGGCCGTGTATTACTGT-3′ |
| 3′ Primers | |
| Cμ 15 | 5′-GACGAAGACGCTCACTTTGGG-3′ |
| Cγ 15 | 5′-GTGATTCACGTTGCAGGTGTAGGT-3′ |
| Cμ 5 | 5′-GGGAATTCCTGCTGATGTCAGAGTTGTT-3′ |
| Cγ 5 | 5′-GGGAATTCGCTGGTCAGGGCGCCTGAGTT-3′ |
| Cμ 2 | 5′-GGAATTCTCACAGGAGACG-3′ |
| Cγ 2 | 5′-GGAATTCCTGGAGCAGGGCGCC-3′ |
| Cμ 2-RC* | 5′-CGTCTCCTGTGAGAATTCC-3′ |
| 5′ Primers | |
| Pan VH-consensus primer (VH-FR1) | 5′-CAGGTGCAGCTGGTGGAGTCTGGG-3′ |
| VH3 (leader sequence) | 5′-GGGTCGACTCCATGGAGTTTGGGCTGAGC-3′ |
| VH6 (leader sequence) | 5′-GGGTCGACTGTCTCCTTCCTCATCTTCCT-3′ |
| Pan VH-FR3 | 5′-GGGTCGACACGGCCGTGTATTACTGT-3′ |
| 3′ Primers | |
| Cμ 15 | 5′-GACGAAGACGCTCACTTTGGG-3′ |
| Cγ 15 | 5′-GTGATTCACGTTGCAGGTGTAGGT-3′ |
| Cμ 5 | 5′-GGGAATTCCTGCTGATGTCAGAGTTGTT-3′ |
| Cγ 5 | 5′-GGGAATTCGCTGGTCAGGGCGCCTGAGTT-3′ |
| Cμ 2 | 5′-GGAATTCTCACAGGAGACG-3′ |
| Cγ 2 | 5′-GGAATTCCTGGAGCAGGGCGCC-3′ |
| Cμ 2-RC* | 5′-CGTCTCCTGTGAGAATTCC-3′ |
Abbreviation: FR, framework.
Complementary to the Cμ 2 primer sequence. This primer is used in combination with Cμ 15 to amplify constant region transcripts.
Ig H CDR3s were amplified in nested PCRs to generate a sufficient amount of product for fingerprinting and SSCP. Primers for PCR are shown in Table 2. Ig H panVH (consensus) or family-specific VH3 or VH6 primers were used in combination with Cμ15 or Cγ15 in the primary and Cμ5 or Cγ5 in the first nested PCRs. Primary and first nested PCRs consisted of 20 cycles composed of 30 seconds of denaturation at 92°C, 35 seconds of annealing at 68°C, and 1 minute of extension at 72°C (final cycle, 10 minutes). The panVH-FR3 5′primer was used in combination with Cμ2 or Cγ2 in the second nested PCRs. This primer is designed to recognize framework 3 (FR3) regions irrespective of the VH family. The second nested PCRs consisted of 30 cycles of 30 seconds of denaturation at 92°C, 35 seconds of annealing at 58°C, and 1 minute of extension at 72°C (final cycle, 10 minutes). PCRs were performed in a final volume of 25 μL, including 20 pmol of both 5′ and 3′ primers, 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 9.0), 0.1% Triton X-100, 1.5 mmol/L MgCl2, 0.2 mmol/L of each dNTPs, and 2.5 U of TaqDNA Polymerase (Promega, Madison, WI).
H CDR3 fingerprinting and SSCP.
For fingerprinting, PCR products were purified by phenol extraction, separated on 5% denaturing polyacrylamide gels containing 7 mol/L urea, and visualized by silver staining as described previously.27,35 Briefly, samples were heated at 92°C for 5 minutes and chilled on ice before loading. The gels were run for approximately 2 hours at 65 W and fingerprinting profiles were visualized using Promega silver staining system according to the supplier’s instructions. For SSCP,36 37 7% polyacrylamide gels containing 5% ultrapure glycerol (J.T. Baker, Phillipsburg, NJ) were run for approximately 7 hours at 25 W while being cooled with an electric fan. SSCP images were obtained by silver staining (Promega).
The new technologies of H CDR3 fingerprinting and SSCP were used, because they have overcome several shortcomings of the other methods used in B-cell repertoire analysis. Hybridization with VHfamily-specific probes, a method used in the previous analysis of posttransplant repertoire,9 does not show the diversity of clones within VH families. Flow cytometry has similar limitations. Cloning and sequencing is technically laborious and, unless the sample size is very large, the less frequently used genes are overlooked. Fingerprinting of CDR3s, on the other hand, generates a snapshot of the total repertoire.27,33 35 It also provides information regarding the clonality within VH families. There are also limitations of the fingerprinting method. Although clones within a VH family that differ in CDR3 size are visualized as separate bands, fingerprinting does not show information regarding the number of clones within a particular band. SSCP analysis was incorporated in this study to overcome this limitation and to estimate the number of clones within a family.
Effect of B-cell numbers on H CDR3 fingerprinting.
To determine the influence of B-cell numbers on fingerprinting profiles, we analyzed normal donor samples using serial dilutions of PBMCs. B-cell numbers were determined by counting anti-CD19 antibody (Becton Dickinson, San Jose, CA) labeled cells using fluorescence-activated cell sorting (Becton Dickinson). In these experiments, as few as 2 × 104 normal donor CD19+ cells exhibited a diverse panVH, VH3 and in some samples VH6 fingerprinting pattern. In other samples, 3 to 7 × 104CD19+ cells were needed for a diverse VH6 fingerprinting pattern. A threshold of 1 × 104 PBMCs has been described for TCR repertoire analysis using fingerprinting.33
To determine the influence of B-cell numbers on the VH6 repertoire posttransplant, IgM VH6 H CDR3 fingerprinting profiles of 3 normal adults and 3 stem cell recipients were compared in a limiting dilution analysis using equal numbers of B cells. Samples from patients no. 1, 9, and 11 obtained at 36, 36, and 26 weeks, respectively, were studied. These samples were chosen for analysis based on the availability of the flow cytometric data. B-cell numbers were determined by counting anti-CD19 antibody (Becton Dickinson) labeled cells using fluorescence-activated cell sorting (Becton Dickinson). PBMCs containing 5 × 105CD19+ cells were used for each patient and normal adult. RNA isolation and cDNA synthesis were performed as above. Before the PCR amplification of the VH6 transcripts, the total amount of constant region transcripts was estimated for each sample using PCR amplification with Cμ15 and Cμ2RC primers as described above. Similar to that described in T-cell receptor repertoire studies,31-34 potential differences in the transcriptional activity between samples can be detected in this way. All 6 samples exhibited similar transcriptional activity in these studies; therefore, no further adjustment was necessary. PCRs were performed using 5 μL cDNA and 10-, 100-, and 1,000-fold dilutions in an attempt to simulate the low B-cell counts of the posttransplant period. PCRs and fingerprinting were performed as described above. In these experiments, dilutions of cDNA instead of PBMCs were used, because patient samples stored in TRIzol reagent were analyzed retrospectively.
Cloning, sequencing, and analysis of H CDR3s.
Nested PCR products were used for random cloning and sequencing as described elsewhere.27,35 The H CDR3 sequences were analyzed using DNASTAR software (DNASTAR Inc, Madison, WI). Kabat’s nomenclature38 was used for identification of H CDR3 regions. V BASE directory and the published germline DHgene sequences were used to search for homology with known human germline DH and JH sequences.39-46Straight and inverted orientation of DH sequences were analyzed using DNASTAR COMPARE and SEQCOMP programs. For H CDR3s >12 amino acids (aa), a minimum of 9 consecutive base pair (bp) identity and for H CDR3s ≤12 aa, a minimum of 6 consecutive bp identity was required with known germline DH genes for DHgene assignment. The sequences analyzed in this study can be found in the GeneBank Database under accession nos. AF028092-AF028121.
RESULTS
B-cell repertoire is oligoclonal early posttransplant.
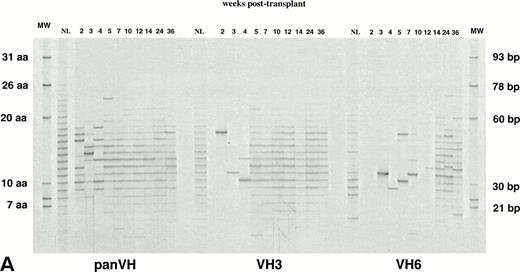
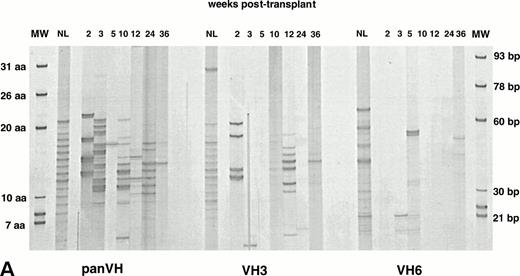
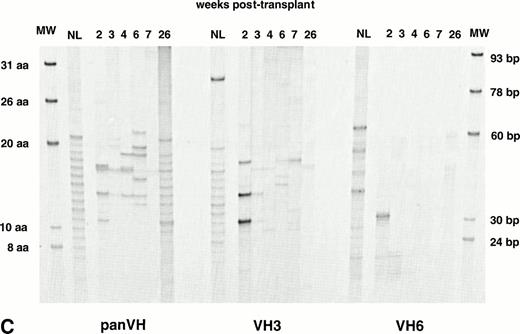
Figure 1A, B, and C show PB IgM H CDR3 fingerprinting profiles of a normal donor (NL) and recipients of allogeneic BMT (patient no. 1), autologous PBSCT (patient no. 9), and CD34 selected autologous PBSCT (patient no. 11), respectively. IgM panVH profiles were generated at various times posttransplant by PCR amplification of H CDR3s using the Ig H consensus primer together with the Cμ primers. As shown in lanes NL, a typically diverse PB repertoire of a normal donor is characterized by 16 to 20 bands separated from each other by 3 bp. Each band corresponds to a certain H CDR3 length. The bands show a Gaussian distribution in intensity, with central (average size) bands being darker than the larger and shorter bands at both ends. Intensity of a band correlates directly with the total amount of H CDR3s present within the band.33
IgM H CDR3 fingerprinting after (A) allogeneic BMT (patient no. 1), (B) autologous PBSCT (patient no. 9), and (C) CD34 selected autologous PBSCT (patient no. 11). IgM H CDR3s were amplified in PCRs using PBMC cDNA as described in Materials and Methods. The Ig H panVH (consensus) primer, VH3 primer, or VH6 primer was used in combination with the Cμ primers to obtain panVH, VH3, or VH6 fingerprinting profiles, respectively. PCR products were run on 5% polyacrylamide sequencing gels that result in resolution of CDR3s according to basepair size. Therefore, each band in fingerprinting corresponds to a certain CDR3 size. Molecular markers (MW) are provided of known basepair size and the corresponding number of encoded aa. Bands are separated from each other by 3 bp because of the abundance of in-frame Ig transcripts in total RNA. Nevertheless, violation of 3-bp spacing of bands can be seen occasionally. The numbers above the lanes indicate the number of weeks when the blood samples were collected posttransplant. Lanes marked NL show the H CDR3 profiles of a normal donor. The number of bands in each lane correlates directly with the repertoire diversity. The band intensity reflects the total amount of CDR3 transcripts that comprise the band. The band intensities are comparable to each other within a lane. Absolute lymphocyte counts per microliter at corresponding weeks were as follows: (A) 10 (2 weeks), 136 (3 weeks), 196 (4 weeks), 398 (5 weeks), 1,175 (7 weeks), 1,479 (10 weeks), 1,295 (12 weeks), 627 (14 weeks), 910 (24 weeks), and 1,240 (36 weeks); (B) 200 (2 weeks), 500 (4 weeks), 1,918 (6 weeks), 1,500 (19 weeks), and 1,231 (36 weeks); (C) 19 (2 weeks), 200 (3 weeks), 700 (4 weeks), 400 (6 weeks), 1,000 (7 weeks), and 1,100 (26 weeks). Shown are IgM fingerprinting profiles representative of 11 patients and normal donors of allogeneic BMT recipients.
IgM H CDR3 fingerprinting after (A) allogeneic BMT (patient no. 1), (B) autologous PBSCT (patient no. 9), and (C) CD34 selected autologous PBSCT (patient no. 11). IgM H CDR3s were amplified in PCRs using PBMC cDNA as described in Materials and Methods. The Ig H panVH (consensus) primer, VH3 primer, or VH6 primer was used in combination with the Cμ primers to obtain panVH, VH3, or VH6 fingerprinting profiles, respectively. PCR products were run on 5% polyacrylamide sequencing gels that result in resolution of CDR3s according to basepair size. Therefore, each band in fingerprinting corresponds to a certain CDR3 size. Molecular markers (MW) are provided of known basepair size and the corresponding number of encoded aa. Bands are separated from each other by 3 bp because of the abundance of in-frame Ig transcripts in total RNA. Nevertheless, violation of 3-bp spacing of bands can be seen occasionally. The numbers above the lanes indicate the number of weeks when the blood samples were collected posttransplant. Lanes marked NL show the H CDR3 profiles of a normal donor. The number of bands in each lane correlates directly with the repertoire diversity. The band intensity reflects the total amount of CDR3 transcripts that comprise the band. The band intensities are comparable to each other within a lane. Absolute lymphocyte counts per microliter at corresponding weeks were as follows: (A) 10 (2 weeks), 136 (3 weeks), 196 (4 weeks), 398 (5 weeks), 1,175 (7 weeks), 1,479 (10 weeks), 1,295 (12 weeks), 627 (14 weeks), 910 (24 weeks), and 1,240 (36 weeks); (B) 200 (2 weeks), 500 (4 weeks), 1,918 (6 weeks), 1,500 (19 weeks), and 1,231 (36 weeks); (C) 19 (2 weeks), 200 (3 weeks), 700 (4 weeks), 400 (6 weeks), 1,000 (7 weeks), and 1,100 (26 weeks). Shown are IgM fingerprinting profiles representative of 11 patients and normal donors of allogeneic BMT recipients.
When patients were analyzed at various times posttransplant, fewer bands were obtained for up to 6 to 10 weeks after transplantation compared with the normal donor sample. This suggests a more restricted B-cell repertoire during this time (panVHs in Fig 1A, B, and C). Primary and nested PCRs were performed in triplicates to ascertain that fewer bands observed early posttransplant were not simply due to preferential amplification of certain H CDR3s. The same results were obtained in these experiments, consistent with a more restricted repertoire early posttransplant. A few of the bands in some samples appeared darker in intensity than expected from the Gaussian distribution when compared with the other bands in the same sample. This observation suggests clonal expansions. At around 3 to 4 months posttransplant, the number of bands reached that seen in the normal adult repertoire.
Contribution of VH3 and VH6 families to posttransplant Ig H CDR3 repertoire.
To determine the contribution of individual families to posttransplant repertoire, fingerprinting analysis was performed at several times posttransplant using the Cμ primers together with either the VH3 or VH6 specific primers instead of the consensus Ig H primer (Table 2). VH3 is the largest VH family, with approximately 22 functional members that account for 50% to 60% of rearrangements in the normal adult repertoire.20-22 Limited number of bands were observed in IgM VH3 H CDR3 profiles within the first 6 to 10 weeks (Fig1A, B, and C). This would be predicted from the restricted panVH H CDR3 repertoire seen during the same time period. However, by 3 to 4 months, the IgM VH3 repertoire in all recipients was as diverse as in healthy adults.
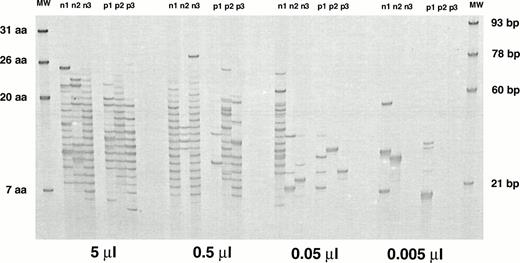
VH6 is a single-member family that has been reported to be overly expressed after BMT.9,11,30 The IgM VH6 repertoire showed a limited number of bands for as long as 6 to 9 months posttransplant (Fig 1A, B, and C). This suggests a relatively small contribution of this family and a limited diversity. In light of the fact that VH6 represents less that 5% of transcripts in normal individuals,19-22 it was important to determine the influence on these results of overall B-cell numbers, which are known to be low during this period. To this end, IgM VH6 H CDR3 repertoires from PB of 3 normal adults and 3 stem cell recipients were compared using 5 × 105 CD19+ cells. VH6 profiles showed missing bands in specimens from 2 of 3 normals and 2 of 3 patients (Fig 2). This would be supportive of the smaller contribution of VH6 to the repertoire compared with VH3 but fails to support a greater contribution of VH6 in reconstituted patients. In addition, fingerprinting profiles for VH6 were generated using 10-, 100-, and 1,000-fold dilutions of the cDNA from all 6 individuals in an attempt to simulate the low B-cell counts of the posttransplant period (Fig 2). Under conditions representing limiting B-cell numbers, the differences in profile complexity between normals and patients became more obvious, with patients, if anything, being less complex. Taken together, these results suggest a limited but variable contribution of VH6 in both normal and patient H CDR3 repertoires, with no apparent evidence of a greater contribution of VH6 in the patient population.
Comparison of the IgM VH6 H CDR3 fingerprinting profiles of 3 normal adults and 3 stem cell transplant recipients using equal numbers of B cells for cDNA synthesis. Fingerprinting profiles were obtained using 5, 0.5, 0.05, and 0.005 μL of the cDNA. p1, 36-week sample from patient no. 1; p2, 36-week sample from patient no. 9; p3, 26-week sample from patient no. 11 (patient numbers are according to Table 1). n, normal donor.
Comparison of the IgM VH6 H CDR3 fingerprinting profiles of 3 normal adults and 3 stem cell transplant recipients using equal numbers of B cells for cDNA synthesis. Fingerprinting profiles were obtained using 5, 0.5, 0.05, and 0.005 μL of the cDNA. p1, 36-week sample from patient no. 1; p2, 36-week sample from patient no. 9; p3, 26-week sample from patient no. 11 (patient numbers are according to Table 1). n, normal donor.
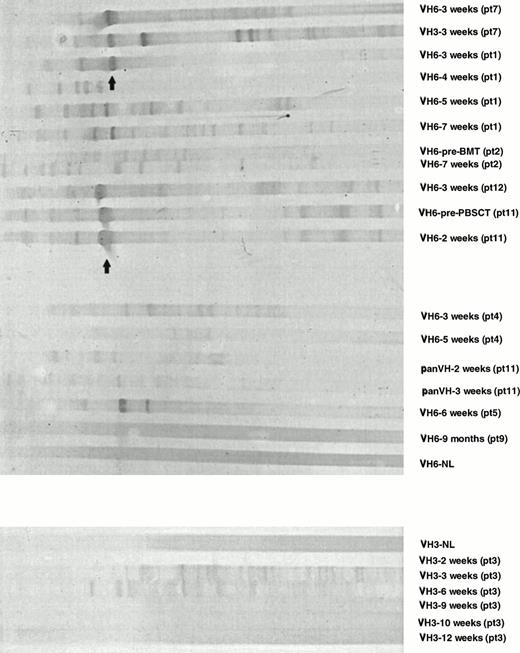
Analysis of H CDR3s by SSCP.
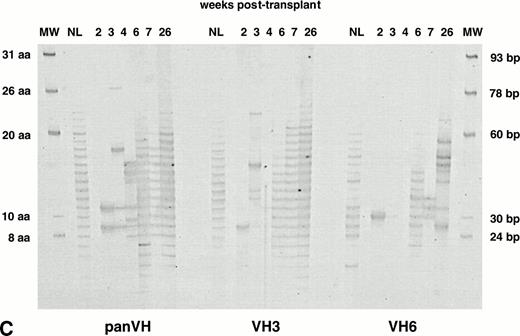
With the technique of fingerprinting, the excessively dark bands could be interpreted as a clonal population or many different clones with H CDR3 sequences of the same size.33 To distinguish between these possibilities, SSCP studies were performed. Using SSCP analysis, different clones sharing the same H CDR3 size can be separated as individual bands based on differences in tertiary structures.36,37 IgM H CDR3s from PB of normal adults showed a smear-like pattern in SSCP analysis, indicating the presence of polyclonal H CDR3s (lane NL in Fig 3). In contrast, samples obtained within the first 7 weeks posttransplant showed several bands with less intense background (Fig 3). SSCP studies were performed in duplicate to confirm the reproducibility of the observed patterns. These data indicate that the limited number of bands early posttransplant in fingerprinting profiles reflect relatively few clones. Expansion of the IgM H CDR3 repertoire to normal adult levels by 10 weeks posttransplant was shown by SSCP analyses that showed a smear-like pattern typical of a polyclonal repertoire (Fig 3).
Analysis of IgM H CDR3s posttransplant by SSCP. IgM panVH, VH3, and VH6 H CDR3s were amplified in nested PCRs as described in Materials and Methods. Two times the amount of PCR product used in fingerprinting experiments was loaded on to 5% polyacrylamide gels containing 5% glycerol. Gels were run at room temperature for 7 hours while being cooled by an electrical fan. Images were obtained by silver staining. In SSCP, CDR3s of the same basepair size can be separated due to differences in their mobility based on their tertiary structures. NL, normal donor. Pt, patient. Patient numbers are according to Table 1. Patient no. 12, who died of refractory NHL 2 months after an autologous PBSCT, was not included in other analyses. Bands indicated by the arrows represent renatured DNA determined by the simultaneously run control samples that were not denatured before loading (data not shown).
Analysis of IgM H CDR3s posttransplant by SSCP. IgM panVH, VH3, and VH6 H CDR3s were amplified in nested PCRs as described in Materials and Methods. Two times the amount of PCR product used in fingerprinting experiments was loaded on to 5% polyacrylamide gels containing 5% glycerol. Gels were run at room temperature for 7 hours while being cooled by an electrical fan. Images were obtained by silver staining. In SSCP, CDR3s of the same basepair size can be separated due to differences in their mobility based on their tertiary structures. NL, normal donor. Pt, patient. Patient numbers are according to Table 1. Patient no. 12, who died of refractory NHL 2 months after an autologous PBSCT, was not included in other analyses. Bands indicated by the arrows represent renatured DNA determined by the simultaneously run control samples that were not denatured before loading (data not shown).
H CDR3s posttransplant are adult size.
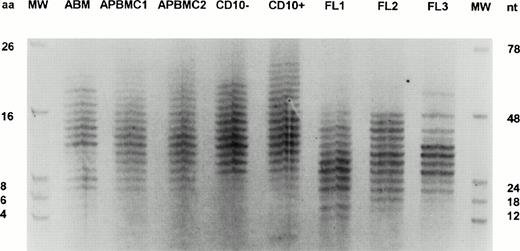
The size of a H CDR3 is determined by the size of DH and JH elements used in the rearrangement and the number of nucleotides inserted or deleted at VH-DH and DH-JH junctions. Adult human H CDR3s range between 4 and 30 aa in size, with an average of 15 aa (lanes NL in Figs1 and 4). Fetal H CDR3s, on the other hand, are significantly shorter in length. This is depicted in Fig 5, which compares the size distribution of fetal liver H CDR3s with those of adult PB and BM. In this study performed by F.M.R. using the same fingerprinting methodology, the 12-week-old fetal liver H CDR3s ranged in length from 4 to 16 aa in two samples and 6 to 18 aa in the third sample.27 Shorter sizes of fetal H CDR3s have also been shown by direct nucleotide sequencing.14,15,17,18,23 25 In all patients analyzed in this study, the size of H CDR3s posttransplant fell within the adult range. Most H CDR3s were longer than the average fetal H CDR3 size of 10 aa. This was observed even in the earliest stages posttransplant when the repertoire was limited in diversity (Figs 1 and 4). H CDR3s as long as 20 aa were not uncommon. It should be noted that adult size range includes those seen in the fetal repertoire; however, H CDR3s longer than 16 to 18 aa are absent in the fetal repertoire, although they are common in the adult and posttransplant repertoire. These results suggested extensive N-nucleotide additions to the recombination joints and/or absence of a bias towards utilization of shorter DH and JH elements posttransplant, unlike that seen in the fetal repertoire.
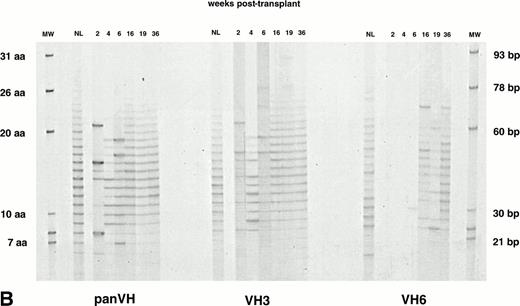
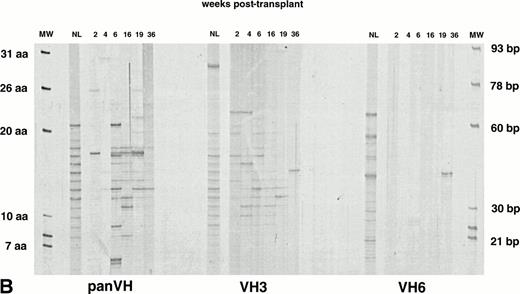
IgG H CDR3 fingerprinting after (A) allogeneic BMT (patient no. 1), (B) autologous PBSCT (patient no. 9), and (C) CD34 selected autologous PBSCT (patient no. 11). IgG panVH, VH3, and VH6 H CDR3 fingerprinting profiles were obtained as described in Fig 1 and Materials and Methods. Cγ primers were used instead of Cμ primers to amplify H CDR3s in PCRs. MWs are provided of known basepair size and the corresponding number of encoded aa. The numbers above the lanes indicate the number of weeks when the blood samples were collected posttransplant. Absolute lymphocyte counts at corresponding weeks are as shown in Fig 1. Lanes marked NL show the IgG H CDR3 profiles of a normal donor. IgG H CDR3 repertoire reconstitution was significantly delayed posttransplant. Shown are IgG fingerprinting profiles representative of 11 patients and normal donors of allogeneic BMT recipients.
IgG H CDR3 fingerprinting after (A) allogeneic BMT (patient no. 1), (B) autologous PBSCT (patient no. 9), and (C) CD34 selected autologous PBSCT (patient no. 11). IgG panVH, VH3, and VH6 H CDR3 fingerprinting profiles were obtained as described in Fig 1 and Materials and Methods. Cγ primers were used instead of Cμ primers to amplify H CDR3s in PCRs. MWs are provided of known basepair size and the corresponding number of encoded aa. The numbers above the lanes indicate the number of weeks when the blood samples were collected posttransplant. Absolute lymphocyte counts at corresponding weeks are as shown in Fig 1. Lanes marked NL show the IgG H CDR3 profiles of a normal donor. IgG H CDR3 repertoire reconstitution was significantly delayed posttransplant. Shown are IgG fingerprinting profiles representative of 11 patients and normal donors of allogeneic BMT recipients.
IgM panVH H CDR3 fingerprinting on human fetal liver and adult B cells. Fingerprinting methodology is identical to that described in the Materials and Methods. Abbreviations: ABM, adult BM; APBMC, adult peripheral blood mononuclear cells; CD10−, CD10−CD20+ mature B-cell population sorted from ABM by FACS; CD10+, CD10+ CD20+ pre-B cells sorted from ABM by FACS; FL, 12-week-old human fetal liver; MW, molecular markers; nt, nucleotides; aa, amino acids. Reprinted with permission from International Immunology, Vol 9, No 10, pp 1503-1515 by permission of Oxford University Press.
IgM panVH H CDR3 fingerprinting on human fetal liver and adult B cells. Fingerprinting methodology is identical to that described in the Materials and Methods. Abbreviations: ABM, adult BM; APBMC, adult peripheral blood mononuclear cells; CD10−, CD10−CD20+ mature B-cell population sorted from ABM by FACS; CD10+, CD10+ CD20+ pre-B cells sorted from ABM by FACS; FL, 12-week-old human fetal liver; MW, molecular markers; nt, nucleotides; aa, amino acids. Reprinted with permission from International Immunology, Vol 9, No 10, pp 1503-1515 by permission of Oxford University Press.
Direct nucleotide sequencing of H CDR3s confirms an adult repertoire pattern.
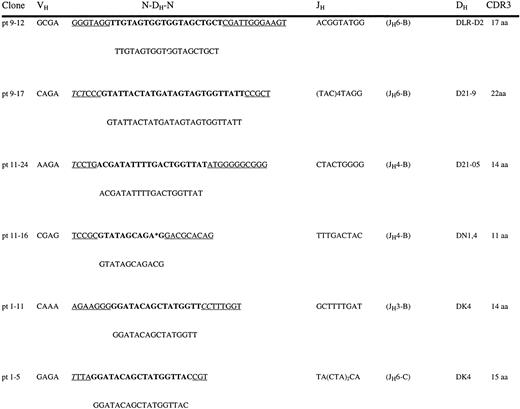
A nucleotide sequence analysis of 32 panVH H CDR3 clones obtained from patients no. 1, 9, and 11 at 7, 16, and 6 weeks posttransplant, respectively, was performed to further investigate whether adult size H CDR3s observed posttransplant contained adult-like N-nucleotide insertion and DH-JH gene utilization (Fig 6). These time points were chosen because they roughly corresponded to the expansion of H CDR3 repertoire in fingerprinting experiments. Two clones each from patients no. 1 and 11 had identical sequences. All rearrangements were in-frame. The average H CDR3 size was 13.8 aa (range, 8 to 22 aa). Only four clones were shorter than the average fetal H CDR3 size of 10 aa. All clones exhibited N-region additions. Two clones lacked N-nucleotides at either VH-DH or DH-JHjoint but not at both. The remaining 30 clones contained N-nucleotides at both recombination sites. The mean numbers of N-or P-nucleotides in VH-DH and DH-JH joints were 5.6 (range, 0 to 21) and 8.2 (range, 0 to 26). DH gene assignment was not possible in 7 clones according to the sequence homology required by established criteria. Members of the DXP family were used in 9 rearrangements, whereas the remaining clones used members of DLR (3 clones), DK (3 clones), DN (5 clones), and DM (1 clone). In 3 clones, there were convincing sequence homologies with DIR genes alone. One clone was best explained with a DN-DIR fusion. DHQ52 gene utilization, which is well documented in up to 40% of the fetal transcripts by several groups14,15,17,18,24,25 and by one of the authors of this study (F.M.R.)23 was not observed in any of the 32 clones. Eleven clones used JH3, 15 clones JH4, and 6 clones JH6. These characteristics of the posttransplant PB H CDR3s are adult-like and similar to that described for adult BM pre-B and mature B cells.27
Direct nucleotide sequencing of posttransplant H CDR3s. Shown are the 6 representative panVH CDR3 sequences from patients no. 1, 9, and 11. JH family assignments are shown in parentheses following the sequences. Also shown are the germline (GL) DH sequences that match CDR3s. Asterisks represent gaps introduced to optimize homology. N- and P-nucleotides are underlined. Italic letters represent P-nucleotides. CDR3 aa sizes are shown.
Direct nucleotide sequencing of posttransplant H CDR3s. Shown are the 6 representative panVH CDR3 sequences from patients no. 1, 9, and 11. JH family assignments are shown in parentheses following the sequences. Also shown are the germline (GL) DH sequences that match CDR3s. Asterisks represent gaps introduced to optimize homology. N- and P-nucleotides are underlined. Italic letters represent P-nucleotides. CDR3 aa sizes are shown.
IgG repertoire reconstitution significantly lags behind IgM repertoire reconstitution.
To gain insight into the memory compartment of B-cell immunity posttransplant, IgG H CDR3 fingerprinting profiles were obtained. Shown in Fig 4A, B, and C are IgG panVH, VH3, and VH6 H CDR3 fingerprinting profiles of an allogeneic BMT recipient (patient no. 1), an autologous PBSCT recipient (patient no. 9), and a CD34-selected autologous PBSCT recipient (patient no. 11), respectively.
The IgG panVH, VH3, and VH6 H CDR3 repertoires in patients were strikingly less complex when compared with normal donor counterparts (Fig 4A, B, and C). The number of bands in most patient samples did not reach adult levels as late as 9 months posttransplant. Nevertheless, an improvement in IgG panVHrepertoire was seen in most patients, as demonstrated by the increasing number of bands observed over time (panVHs in Fig 4B and C). The expansion of the IgG panVH repertoire in these patients was likely due to usage of many VH families, because the expansion observed could not be attributed solely to the IgG VH3 and/or VH6 H CDR3 repertoires (Fig 4B and C). The IgG panVH, VH3, and VH6 H CDR3 repertoires were significantly less complex also when compared with the IgM counterparts of the same patient (Figs 1 and4). The IgG panVH H CDR3 repertoire of a representative patient shown in Fig 4A showed 4 bands at 12 weeks, whereas the IgM counterpart of the same patient shown in Fig 1A contained 18 bands at 12 weeks.
Typical IgG panVH and VH3 H CDR3 repertoires of a normal donor (NL in Fig 4) are characterized by 16 to 20 bands, similar to that seen in the normal IgM counterparts. However, the distribution of band intensities in IgG repertoires did not show a clear pattern. In contrast to the average size bands (11 to 16 aa) being darker in IgM panVH and VH3 H CDR3 repertoires, the darker bands in the IgG panVH and VH3 H CDR3 repertoires of the normal donor in Fig 4 were 21 and 31 aa in size. IgG VH6 H CDR3 repertoire of the donor showed approximately 15 bands, 4 of which were significantly darker than others. The darker bands were not clustered according to size. These results suggest clonal expansions. Violation of the 3-bp spacing of the bands was not uncommon in the IgG VH6 H CDR3 repertoire. This may reflect PCR artifacts resulting from limiting amounts of template. These observations suggest that there is low abundance of functional IgG VH6 transcripts in the normal adult repertoire in general, but that expansion of a few clones is likely.
DISCUSSION
In this study, B-cell repertoire reconstitution after hematopoietic stem cell transplantation was analyzed by monitoring Ig H CDR3 diversification using fingerprinting and SSCP in combination with sequencing. In contrast to previous reports,6,7,9,11 30 it is shown that the emerging repertoire posttransplant is not fetal-like when assessed at the molecular level. This conclusion is based on the demonstration of adult-size H CDR3s and absence of a fetal-like Ig gene utilization posttransplant.
Several studies have examined B-cell reconstitution posttransplant in terms of quantitative recovery, morphology, phenotype, and function, whereby both similarities and differences with fetal B cells were observed (reviewed in Storek and Saxon7 and Storek et al8). In the few studies in which posttransplant B-cell repertoire was examined at the molecular level, 9,11,29,30the characteristics that typify fetal and adult repertoires were partially addressed. Despite only modest similarities between fetal and posttransplant repertoires, it has been concluded and generally accepted that a fetal-like pathway is followed in B-cell recovery posttransplant recapitulating ontogeny.
One of the most striking characteristics of the fetal repertoire is the relatively small size of H CDR3 regions.14,15,17,18,23,25This is in part due to N-nucleotide insertions either being absent or limited in the fetus. Sequencing the expressed H CDR3s was particularly informative relative to the question of an adult versus fetal-like N-region diversification. Consistent with the adult-like size of the H CDR3s detected by fingerprinting, all of the sequences analyzed (n = 32) exhibited N-insertions indicative of an adult repertoire. Interestingly, in a study in which VH6 sequences from 2 patients were examined posttransplant, all clones also showed N-region additions in either VH-DH or DH-JH joints or in both.30 This contrasts a fetal pattern of diversity in which a significant portion of VH-DH or DH-JHjoints lack N-regions.14,15,17,18,23,25 Because the VH6 H CDR3 sizes were found to be close to the length of the relatively small sample size of fetal H CDR3,47 it was concluded that the posttransplant sequences were fetal-like despite adult-like N-nucleotide insertions.30
We found no evidence posttransplant for a biased expression of Ig gene segments that typify the fetal repertoire. The DH genes expressed are characteristic of an adult repertoire. The DHQ52 gene, which is expressed in nearly half of fetal Ig H chain transcripts,14,15,17,18,23-25 was not found in any of the sequenced transcripts from patients. JH genes identified in sequenced clones in this study (JH3, JH4, and JH6) are common in both fetal and adult repertoires.14,15,17,18,23-25 Several studies have suggested preferred expression of VH6 in the fetal repertoire,13-19 although this is controversial.48 Our results suggest that the contribution of VH6 is not increased posttransplant relative to that seen in normal adults. This is in contrast to that shown by Storek et al,11 which indicated up to 10-fold higher utilization of VH6 at 6 to 12 months posttransplant. In another study by Fumoux et al,9 increased utilization of VH6 was shown at 60 days but not at 30 or 90 days after allogeneic BMT. In the same study, VH6 utilization was not found to be different after autologous BMT than in normal adults. Because we examined 26- and 36-week samples in the limiting dilution analysis, we cannot exclude an increased contribution of VH6 during earlier time points posttransplant, as previously suggested.9 However, it is important to note that in the previous studies the largest proportion of B cells expressing VH6 was 7% and was usually in the 0.1% to 2% range. VH6-containing rearrangements were shown previously to have a relatively high frequency of replacement mutations, suggesting positive selection of these clones posttransplant.30 Moreover, identical VH6-containing rearrangements were identified several times, suggesting clonal expansion. Therefore, it is possible that the marginal increases in VH6 usage reported in previous studies of posttransplant patients9,11 30 represents oligoclonal proliferations rather than preferred usage of VH6 in clonally diverse B cells.
Conclusions drawn in previous studies regarding a fetal-like emergence of the lymphocyte repertoire after transplantation of adult stem cells to patients seem counterintuitive based on the results of the previous preclinical studies addressing this issue. There is considerable evidence, particularly in mice, that fetal lymphoid precursors are distinct from adult precursors in terms of morphological characteristics, surface marker expression, TdT expression, and antigen-specific receptor repertoires.49-51 Studies have been performed to address the question of whether the source of stem cells (fetal v adult) or the host microenvironment (fetalv adult) is more important in shaping the characteristics of the emerging lymphocyte repertoires. In our own laboratory, it was found that VH utilization in developing fetal B cells remained fetal-like when propagated in the presence of an adult BM stromal layer, arguing against the fetal phenotype resulting from microenvironmental differences.52 Similarly, in mouse fetal thymic lobe repopulation studies using T-cell precursors from either fetal liver or adult BM, it was found that the extent of TCR N-nucleotide insertions was determined by the progenitor source rather than the microenvironment.53,54 However, in another study, reconstitution of adult mice with either fetal liver or adult BM precursors gave rise to lymphocytes with adult characteristics.55 In any event, adult- but not fetal-like lymphocyte characteristics at the molecular level were observed in these studies when either adult progenitor cells or adult hosts were used. Based on these studies, an adult-type repertoire would be predicted after transplantation of adult progenitors to adult hosts.
In the study presented here, an adult-like repertoire was evident in all patients analyzed, including the patient receiving CD34+ selected stem cells, whereby CD34−B-cell precursors are largely eliminated. This observation provides evidence against the possibility that adult-like repertoire posttransplant could be simply due to passive transfer of mature B cells with established adult characteristics. The patient population in this study was diverse in terms of stem cell source (allogeneic [n = 5], autologous [n = 6], BMT [n = 6], PBSCT [n = 4], and BM + PBSC [n = 1]), preparative regimen, and disease type. Adult-like B-cell repertoires observed in all patients suggest that results are not likely to be biased by any one of these variables.
Our results show a significant lag in diversification of IgG H CDR3s posttransplant, suggesting limitations in secondary B-cell responses and memory formation. Previous studies indicate that serum IgM levels return to normal within 3 to 6 months, whereas recovery of serum IgG levels can be delayed up to 1 year or longer (reviewed in Lum,2 Atkinson,5 and Storek and Saxon7). Other criteria for secondary B-cell responses and memory development include loss of membrane IgD on antigen-specific B cells after stimulation and increasing somatic mutation of the CDRs. In an analysis of 59 BMT recipients, Storek et al56 showed significantly fewer IgD− B cells at 1 year posttransplant than in normal controls. In addition, Suzuki et al29 showed that VH genes exhibited fewer somatic mutations posttransplant in 4 patients. This is consistent with the data shown here indicating restricted diversity in the IgG H CDR3s being expressed. Taken together, these studies show a significant impairment in the memory compartment posttransplant. Given that IgG and memory responses are dependent on germinal center formation, the kinetics and quality of restructuring the architecture of the secondary lymphoid organs after transplantation will have a significant impact on reconstitution of the primary B-cell repertoire. In this regard, histological analyses of spleen and lymph nodes from nonsurviving patients of allogeneic BMT showed reduced cellularity and an absence of lymphoid follicules and germinal centers.57-60 Another important factor in Ig class switch and memory response is the help provided by T cells. Failure of T cells has been shown posttransplant.61-66 Therefore, successful recovery of secondary B-cell responses will also be dependent on the recovery of the T-cell compartment.
The data presented here expand the knowledge of B-cell immune reconstitution posttransplant and demonstrate that it does not recapitulate fetal ontogeny at the molecular level, as previously suggested. Therefore, the delay in B-cell reconstitution is unlikely due to the need for fetal reprogramming. Posttransplant immune deficiency appears to be rather due to functional B-cell defects described in several studies (reviewed in Lum,2Atkinson,5 and Storek and Saxon7) and an impairment in the memory compartment that is dependent on germinal center formation and proper T-cell help. Recent studies have demonstrated improved numerical and functional recovery of B cells after allogeneic PBSCT.67 68 One technique to improve memory compartment reconstitution posttransplant may be adoptive transfer of donor memory cells. To that end, we are currently investigating the H CDR3 repertoire reconstitution in correlation with numerical and functional B-cell recovery after PBSCT and also exploring the expansion of the B-cell compartment in ex vivo expansion systems for potential posttransplant use.
ACKNOWLEDGMENT
The authors thank Dr Manuel Santiago for his help in preparation of the figures and for his support of fellow research training. We also thank Oxford University Press for the permission granted to reprint Fig 5.
Supported by Grants No. A I 19896, A I 33221, and N S 35974 from the National Institutes of Health. E.G. is supported by a fellowship grant by Amgen.
Address reprint requests to Judy M. Teale, PhD, Department of Microbiology, The University of Texas Health Science Center at San Antonio, 7703 Floyd Curl Dr, San Antonio, TX 78284; e-mail: Teale@uthscsa.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal