Abstract
Immunoglobulin class switching usually involves deletion of part of the immunoglobulin CH region. By DNA fiber fluorescence in situ hybridization (FISH) with a barcode of probes covering the DH, JH, and CH genes, the configuration of the entire CH region can be visualized on single DNA molecules. Using this technique, we have studied class switching in three types of B-cell neoplasia, mantle-cell lymphoma (MCL), follicular lymphoma (FL) and hairy cell leukemia (HCL), representing B cells in, respectively, pregerminal center, germinal center, and postgerminal center stages of development. In MCL and FL, simultaneous detection of the t(11;14) and t(14;18) breakpoint with probes for the BCL-1 and BCL-2 loci, respectively, allowed differentiation between productive and nonproductive alleles. In none of 10 MCL cases was class switching detected. In 21 HCL, all nonimmunoglobulin M (IgM) cases had class-switch deletion consistent with the expressed isotype on at least one allele. In FL, however, a peculiar pattern of CH rearrangement was observed. In IgM expressing FL, the translocated alleles had switched in 11 of 13 cases, and the nontranslocated allele showed complex rearrangements downstream from the Cμ-Cδ genes in 9 of 13 cases. These downstream rearrangements may reflect tumor-specific deregulation of the class-switch machinery. All seven immunoglobulin G (IgG) expressing FL showed class switching on both alleles. Fiber FISH analysis also showed several polymorphisms. The most frequent one, present on 38% of all analyzed alleles, consisted of an extra Cγ gene or pseudogene in the 3′ cluster.
© 1998 by The American Society of Hematology.
IMMUNOGLOBULIN HEAVY-CHAIN class switching in B cells is generally mediated by looping out of genomic DNA between the recombined variable-diversity joining (VDJ) genes and a target-constant region (CH) gene,1-3 as has been shown by the detection of circular, excised DNA fragments containing intervening CH sequences. In mouse B-cell lines, the frequent presence of identical switch deletions on productive and nonproductive alleles indicate a certain specificity in the class-switch process.4,5 Results obtained in human B-cell tumors and cell lines point to a less-specific mechanism,6-9 in which the nonproductive alleles frequently remain unswitched or have switched to one of the neighboring CH genes. However, the available information is limited, and no comparative studies on class switching in different types of B-cell neoplasia have been performed.
Methods for the detection of class-switch deletions include Southern blotting,10 polymerase chain reaction (PCR),11and inverse PCR.12 However, these techniques have a very narrow detection window, viewing only the recombination between two switch sites. If one would like to know the configuration of the entire immunoglobulin H (IgH) CH region on both alleles, fiber fluorescence in situ hybridization (FISH)13 14 is a more appropriate technique. We used fiber FISH with a barcode of probes labeled in two colors covering the CH, the JH, and part of the DH genes. By hybridization of this barcode, the entire CH region could be visualized and rearrangements could be analyzed on both alleles separately.
First a fiber FISH map of the germline CH region was constructed, using probes specific for Cμ, Cγ, and Cα genes. We then analyzed the CH region configuration in three types of human B-cell malignancies: mantle-cell lymphoma (MCL), follicular lymphoma (FL), and hairy cell leukemia (HCL). Each of these tumor-types is thought to be derived from B cells in a particular stage of development; MCL from immature follicle mantle cells, FL from follicle-center cells that have encountered antigen and are in the process of affinity maturation and immunoglobulin class switching, and HCL from mature B cells, probably preplasma cells. To be able to differentiate between functional and nonfunctional alleles, MCL and FL were selected for the presence of a BCL-1 or BCL-2 translocation breakpoint, respectively. These translocations could be detected by hybridizing IgH probes together with probes from the BCL-1 or BCL-2 region14 (Vaandrager et al, in preparation). Juxtaposition of BCL-1 or BCL-2 probe signals to IgH probe signals indicated that this IgH allele was translocated and thus could not be the functional allele.
MATERIALS AND METHODS
Tissue samples and cell line.
The foreskin fibroblast cell line VH25 was obtained from the Department of Radiation Genetics, Leiden University Medical Centre ([LUMC] Leiden, The Netherlands). Peripheral blood leukocytes (including granulocytes) of 10 normal donors were obtained from the Blood Bank, LUMC. Frozen tissue of 10 MCL and 23 FL, selected for the presence of a BCL-1 or BCL-2 translocation breakpoint, respectively, was obtained from the Department of Pathology, LUMC. The tumor involved a lymph node in all but two cases (thyroid, salivary gland). By estimation in hematoxylin and eosin-stained paraffin sections, the percentage of tumor cells varied between 10% and 50% for FLs and higher than 50% for MCLs. Mononuclear cell suspensions, isolated from spleen (n = 15) or blood (n = 6) of 21 HCL patients by Ficoll density-gradient centrifugation, were obtained from the Department of Hematology, LUMC. All samples containing greater than 80% hairy cells15 were directly used. Samples initially containing less hairy cells were T-cell depleted before analysis, using anti-CD3 monoclonal antibodies (MoAbs) (anti-leu4; Becton Dickinson, Mountain View, CA) and sheep-antimouse–coated magnetic beads (Dynabeads; Dynal, Oslo, Norway). All HCL patients except HCL20 have been karyotyped before, and no 14q32 abnormalities have been found.16 For all tumors Ig expression was assessed by two-color immunofluorescence on cell suspensions or frozen tissue sections, as described before.15 The IgH subclasses were detected with the following MoAbs: anti-IgG1 (MH 161-1-MO2; CLB, Amsterdam, The Netherlands), anti-IgG2 (MH 162-1-MO3; CLB), anti-IgG3 (MH 163-1-MO3; CLB), anti-IgG4 (MH 164-1-MO4; CLB), and anti-IgA1 (NI 69-11; Nordic, Tilburg, The Netherlands), followed by fluorescein isothiocyanate (FITC)-conjugated goat-antimouse immunoglobulins (GAM-FITC; CLB). To obtain T-cell–enriched fractions of HCL samples, samples were cultured for 72 hours in RPMI medium with 10% human serum, 180 μg/mL phytohemagglutinin (PHA) (PHA HA 15; Murex, Dartford, UK), and 120 U/mL IL-2. After 1 more week of culture with IL-2 but without PHA, cells were harvested. The percentage of T cells before and after PHA/IL-2 stimulation was determined by immunostaining with OKT3 mouse–anti-CD3 MoAb and goat-antimouse–FITC (DAKO, Glostrup, Denmark) and subsequent FACS analysis, using a FACSCAN (Becton Dickinson, San Jose, CA).
DNA fiber preparation.
Preparations of DNA fibers were made according to the halo technique13 14 with some modifications. Cells were suspended in phosphate-buffered saline (PBS) with 1% bovine serum albumin (BSA). Fifty μL of suspension was applied to 3-aminopropyltrietoxysilane–treated object slides. Nuclei were allowed to adhere for 2 minutes. Slides were dipped for 30 seconds in the following ice-cold solutions: solution 2 (25 mmol/L Tris.HCl pH 8.0, 0.2 mmol/L MgCl2, 2 mol/L NaCl) and solution 3 (solution 2 + 40 μg/mL propidium iodide). Slides were irradiated with ultraviolet light (UV) light (254 nm, 7000 μW/cm2) for 7 minutes, dipped in ice-cold solution 4 (25 mmol/L Tris.H Cl pH 8.0, 0.2 mmol/L MgCl2, 0.2 mol/L NaCl), solution 5 (25 mmol/L Tris.HCl pH 8.0, 0.2 mmol/L MgCl2), and twice in H2O, and air-dried. When cells were not sufficiently lysed using this protocol, the dip in solution 2 was preceded by a dip in 0.25% sodium dodecyl sulfate (SDS) and a dip in solution 1 (25 mmol/L Tris.HCl pH8.0, 10 mmol/L MgCl2, 0.5 mmol/L CaCl2, 0.5% Nonidet NP-40).
Probes and in situ hybridization.
IgH cosmids U2-2 and 3/64 have been described.17 Cosmid U2-2 contains the JH and part of the DH region, cosmid 3/64 contains the JH, Cμ, and Cδ genes. Cosmid cosIg6,18 containing the Cγ3 gene and surrounding sequences, was provided by T.H. Rabbitts (MRC Centre, Cambridge, UK). The Cα probe, a 16 kb BamHI-BamHI fragment19 containing switch α1 (Sα1) and Cα1, and the Cγ4 probe, a 6.2 kb HindIII-BamHI fragment20 containing the Cγ4 gene, were provided by P. Leder (NIH, Bethesda, MD). Probes for the detection of 11q13/BCL-1 translocation breakpoints have been described previously.14A contig of seven cosmid probes for detection of 18q21/BCL-2 breakpoints was made by subcloning YAC YA153A6 (provided by G.A. Silverman, Washington University School of Medicine, St Louis, MO). This contig is described elsewhere in detail (Vaandrager et al, in preparation). Probes were labeled by standard nick-translation with biotin-16-deoxyuridintriphosphate or digoxygenin-11-deoxyuridintriphosphate (Boehringer Mannheim, Mannheim, Germany). The hybridization solution consisted of 30% formamide, 10% dextransulphate, 50 mmol/L sodiumphosphate pH 7.0, 2 × SCC, 3 ng/μL of each probe, and a 50-fold excess of human Cot-1 DNA. Hybridization and immunofluorescence detection were performed as described previously.13 14
Fluorescence microscopy.
Images were captured using a COHU 4910 series monochrome CCD camera (COHU, San Diego, CA) attached to a DM fluorescence microscope (Leica, Wetzlar, Germany) equipped with a PL Fluotar 100×, NA 1.30 to 0.60 objective and I3 and N2.1 filters (Leica), and Leica QFISH software (Leica Imaging Systems, Cambridge, UK). Images were processed with Paintshop Pro (JASC Inc, Eden Prairy, CA) and Harvard Graphics (Software Publishing Corp, Santa Clara, CA).
RESULTS
Mapping of the germline immunoglobulin CH region.
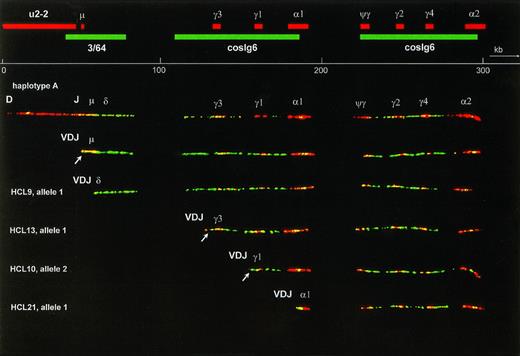
Several combinations of biotin- or digoxigenin-labeled probes were hybridized on DNA fibers from the fibroblast cell line VH25. From these experiments, the relative positions of the probes were derived. Cosmid cosIg6 gave two long hybridization signals, each almost twice as long as the insert size. Although the cosmid was cloned from the Cγ3 region, it hybridized with all CH genes and surrounding sequences. This cross-hybridization reflects the evolutionary duplications that have led to the current CH complex containing 4 highly homologous Cγ genes and 1 Cγ pseudogene. The Cα probe hybridized to both Cα genes and Sα sequences, and also to switch μ (Sμ), which is homologous to switch Sα19. The Cγ4 probe hybridized to all Cγ genes and Cψγ. One of the alleles of VH25 contained the expected number of 5 Cγ probe signals, grouped in two clusters that colocalized with the two cosIg6 signals (haplotype A, Fig 1). Because we could not identify the different Cγ and Cα genes on basis of the hybridization, the order of genes was inferred from a previously published map of the CH region.21 The other allele, which is shown in Fig 2, contained an extra Cγ signal in the 3′ CH cluster (haplotype B). Haplotype A was used to make a physical map (Fig 1). On five straight and optimally stretched signals, relative distances were measured. These measurements were averaged, and the relative distances were converted to kilobasepairs, using the distance between Cγ3 and Cγ1 as an internal standard of 26 kb.18
The upper part is a fiber FISH map of the immunoglobulin CH region, based on the hybridization pattern of the probes indicated as red (biotin-labeled) and green (digoxigenin-labeled) bars. Although cosmid probes U2-2 and 3/64 each showed a single, continuous signal, cosmid cosIg6 hybidized to two stretches of genomic DNA, each twice as long as a normal cosmid and separated by a gap. These two stretches represent the two evolutionary duplication units of the human CH region, illustrating the high degree of homology between and within these units. The 16 kb C probe hybridized to both C genes and S regions and also gave a small signal at the site of Sμ, because Sμ is highly homologous to S. The Cγ4 probe hybridized to all Cγ genes as well as to the Cψγ gene. Below the scale bar a fiber representing the germline configuration of the most common haplotype (A) is shown. The second fiber is an allele that has undergone VDJ recombination, resulting in loss of almost the whole cosmid U2-2 probe signal. The four fibers below are alleles, derived from HCL cases that have also recombined their V, D, and J genes, but in addition have undergone class-switch deletion to, from top to bottom, the Cδ, Cγ3, Cγ1, and C1 genes. In each case this deletion has led to loss of all probe signals between VDJ and the target CH gene. In the case with Cδ switching this deletion is recognized by shortening of cosmid 3/64 signal combined with absence of the Sμ signal. The probe signals representing the remains of cosmids U2-2 and 3/64 containing the JH-5′ Sμ region are often too small to be observed. In the Cγ3-and Cγ1-switched fibers they are visible, however, as a red and a green dot at the left end of the fiber (arrow). To facilitate interpretation, only fibers of haplotype A were selected for this figure, and the lengths of the class-switched fibers were normalized to the germline fiber. This was only done for Fig 1, allowing the inclusion of a scale bar. In the other figures no scale bars were given because the fibers have different degrees of magnification and stretching.
The upper part is a fiber FISH map of the immunoglobulin CH region, based on the hybridization pattern of the probes indicated as red (biotin-labeled) and green (digoxigenin-labeled) bars. Although cosmid probes U2-2 and 3/64 each showed a single, continuous signal, cosmid cosIg6 hybidized to two stretches of genomic DNA, each twice as long as a normal cosmid and separated by a gap. These two stretches represent the two evolutionary duplication units of the human CH region, illustrating the high degree of homology between and within these units. The 16 kb C probe hybridized to both C genes and S regions and also gave a small signal at the site of Sμ, because Sμ is highly homologous to S. The Cγ4 probe hybridized to all Cγ genes as well as to the Cψγ gene. Below the scale bar a fiber representing the germline configuration of the most common haplotype (A) is shown. The second fiber is an allele that has undergone VDJ recombination, resulting in loss of almost the whole cosmid U2-2 probe signal. The four fibers below are alleles, derived from HCL cases that have also recombined their V, D, and J genes, but in addition have undergone class-switch deletion to, from top to bottom, the Cδ, Cγ3, Cγ1, and C1 genes. In each case this deletion has led to loss of all probe signals between VDJ and the target CH gene. In the case with Cδ switching this deletion is recognized by shortening of cosmid 3/64 signal combined with absence of the Sμ signal. The probe signals representing the remains of cosmids U2-2 and 3/64 containing the JH-5′ Sμ region are often too small to be observed. In the Cγ3-and Cγ1-switched fibers they are visible, however, as a red and a green dot at the left end of the fiber (arrow). To facilitate interpretation, only fibers of haplotype A were selected for this figure, and the lengths of the class-switched fibers were normalized to the germline fiber. This was only done for Fig 1, allowing the inclusion of a scale bar. In the other figures no scale bars were given because the fibers have different degrees of magnification and stretching.
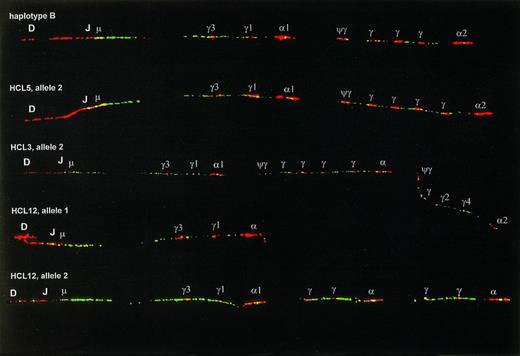
Examples of CH duplication and deletion polymorphisms. The upper fiber shows haplotype B, which has an extra Cγ signal in the 3′ cluster as compared with haplotype A (Fig1), and was present at a frequency of 37.5% in our series. The second fiber has two extra Cγ signals in the 3′ cluster. The third fiber shows an example of the second most frequent type of polymorphism, appearing as a multigene duplication. The lower two fibers are the two germline alleles of one HCL patient, allele 1 having a multigene deletion, allele 2 having a more complex pattern, probably the result of multiple deletion/duplication events. On the alleles in this figure most Cγ and C genes could not be identified and are indicated only with γ and .
Examples of CH duplication and deletion polymorphisms. The upper fiber shows haplotype B, which has an extra Cγ signal in the 3′ cluster as compared with haplotype A (Fig1), and was present at a frequency of 37.5% in our series. The second fiber has two extra Cγ signals in the 3′ cluster. The third fiber shows an example of the second most frequent type of polymorphism, appearing as a multigene duplication. The lower two fibers are the two germline alleles of one HCL patient, allele 1 having a multigene deletion, allele 2 having a more complex pattern, probably the result of multiple deletion/duplication events. On the alleles in this figure most Cγ and C genes could not be identified and are indicated only with γ and .
The probe combination described above was hybridized on DNA fibers prepared from peripheral blood leukocytes of 10 normal donors, spleen or blood samples of 21 HCL patients, and frozen tumor samples of 10 MCL and 23 FL. For the MCL the IgH probe set was supplemented with probes for the BCL-1 locus, for FL with probes for the BCL-2 locus. Using these probe sets we identified three different groups of IgH rearrangements: complete or almost complete deletion of the U2-2 cosmid signal, deletion of downstream probe signals consistent with class-switch deletion, and juxtaposition of IgH to BCL-1 or BCL-2 signals. Deletion of the cosmid U2-2 signal that covers the DJ region was exclusively observed in samples containing either polyclonal or monoclonal B cells, and hence was ascribed to VDJ or nonfunctional DJ rearrangement. Fibers without cosmid U2-2 deletion should be derived from non-B cells or occasional B cells with monoallelic rearrangement (with a possible exception for nonfunctional DJ rearrangement involving DQ52, which might be missed by this analysis). For the analysis of polymorphisms we exclusively tested DNA fibers without deletion of the U2-2 signal (Fig 2). Among 128 alleles analyzed, 69 (54%) had haplotype A, 48 (38%) had haplotype B, and 11 alleles (9%) had other haplotypes. Most of the latter could be matched with previously described duplications within the 3′ part of the CHregion.22 23 In cases HCL3, HCL5, and HCL12, a T-cell–enriched fraction was prepared by PHA-stimulation. This fraction showed the same polymorphism as found in crude spleen samples, further proving that these patterns represented real polymorphisms, and not B-cell–related rearrangements.
Absence of class-switch deletions in MCL.
In agreement with literature data,24 all MCL expressed IgM(D). In each MCL sample at least 10 fibers with juxtaposition of 11q13/BCL-1 probes were analyzed, and no deletions or other rearrangements in the CH region were found. Similarly, 10 to 20 fibers with deletion of the U2-2 signal but without juxtaposition of 11q13/BCL-1 probes were analyzed, and these fibers also lacked CH rearrangements. Because these fibers were found at approximately the same frequency as fibers with BCL-1 translocation, the majority should represent the functional IgH allele of the tumor cells. From these data we conclude that MCL lack class-switch recombination on both alleles.
Class-switch deletions in HCL.
Fiber FISH analysis of 21 cases of HCL showed several types of class-switch deletion (Fig 1). On basis of (V)DJ recombination, class-switch deletion, and polymorphisms, the two HCL-derived IgH alleles could be distinguished in all 21 cases. The fiber FISH results are shown in Table 1, and are aligned with IgH isotype expression as determined by immunofluorescence and the presence of class-switch deletions as previously determined by Southern blotting with JH and Cμ probes.25 In contrast to previous immunophenotyping15, we found in none of these 21 HCL cases expression of multiple isotypes. Among the IgM cases, only one of eight (HCL5) showed monoallelic class-switch deletion. In all non-IgM cases, at least one allele had class-switch deletion concordant with the expressed IgH isotype. These included two cases with expression of IgD only (HCL9 and HCL10).25
Class Switching in Hairy Cell Leukemia
| Case . | Ig Expression . | Fiber FISH . | Southern Blot . | |||||
|---|---|---|---|---|---|---|---|---|
| Allele 1 . | Allele 2 . | |||||||
| IgL . | IgH . | VDJ . | CS . | VDJ . | CS . | JH . | Cμ . | |
| HCL1 | λ | M | + | − | − | − | 1/2R | NE |
| HCL2 | κ | M | + | − | + | − | 1/2R | − |
| HCL3 | λ | MD | + | − | + | − | 2R | − |
| HCL4 | κ | MD | + | − | + | − | 3R | − |
| HCL5 | λ | MD | + | − | + | γ1 | 2R | −-150 |
| HCL6 | λ | MD | + | − | + | − | 2R | − |
| HCL7 | λ | MD | + | − | + | − | 2R | − |
| HCL8 | κ | MD | + | − | + | − | 2R | NE |
| HCL9 | λ | D | + | δ | + | − | 3R | −-150 |
| HCL10 | λ | D | + | δ | + | γ1 | 2R | 2D |
| HCL11 | λ | G3 | + | γ3 | + | − | 2R | 1D |
| HCL12 | λ | G3 | + | γ3 | − | γ1 | 2R | 2D |
| HCL13 | λ | G3 | + | γ3 | − | γ1 | 2R | 2D |
| HCL14 | λ | G3 | + | γ3 | + | γ1 | 3R | 2D |
| HCL15 | λ | G3 | + | γ3 | − | − | NT | NT |
| HCL16 | λ | G3 | + | γ3 | + | − | 2R | 1D |
| HCL17 | κ | G3 | + | γ3 | + | γ1 | 2R | 2D |
| HCL18 | λ | G1 | + | γ1 | + | γ1 | 2R | 2D |
| HCL19 | κ | G1 | + | γ1 | + | γ1 | NT | NT |
| HCL20 | λ | G1 | + | γ1 | + | − | NT | NT |
| HCL21 | λ | A | + | α1 | − | γ3 | 2R | 2D |
| Case . | Ig Expression . | Fiber FISH . | Southern Blot . | |||||
|---|---|---|---|---|---|---|---|---|
| Allele 1 . | Allele 2 . | |||||||
| IgL . | IgH . | VDJ . | CS . | VDJ . | CS . | JH . | Cμ . | |
| HCL1 | λ | M | + | − | − | − | 1/2R | NE |
| HCL2 | κ | M | + | − | + | − | 1/2R | − |
| HCL3 | λ | MD | + | − | + | − | 2R | − |
| HCL4 | κ | MD | + | − | + | − | 3R | − |
| HCL5 | λ | MD | + | − | + | γ1 | 2R | −-150 |
| HCL6 | λ | MD | + | − | + | − | 2R | − |
| HCL7 | λ | MD | + | − | + | − | 2R | − |
| HCL8 | κ | MD | + | − | + | − | 2R | NE |
| HCL9 | λ | D | + | δ | + | − | 3R | −-150 |
| HCL10 | λ | D | + | δ | + | γ1 | 2R | 2D |
| HCL11 | λ | G3 | + | γ3 | + | − | 2R | 1D |
| HCL12 | λ | G3 | + | γ3 | − | γ1 | 2R | 2D |
| HCL13 | λ | G3 | + | γ3 | − | γ1 | 2R | 2D |
| HCL14 | λ | G3 | + | γ3 | + | γ1 | 3R | 2D |
| HCL15 | λ | G3 | + | γ3 | − | − | NT | NT |
| HCL16 | λ | G3 | + | γ3 | + | − | 2R | 1D |
| HCL17 | κ | G3 | + | γ3 | + | γ1 | 2R | 2D |
| HCL18 | λ | G1 | + | γ1 | + | γ1 | 2R | 2D |
| HCL19 | κ | G1 | + | γ1 | + | γ1 | NT | NT |
| HCL20 | λ | G1 | + | γ1 | + | − | NT | NT |
| HCL21 | λ | A | + | α1 | − | γ3 | 2R | 2D |
The column “Ig expression” shows immunoglobulin class and subclass expression of the light and heavy chain, as determined by immunofluorescence. In column “Fiber FISH,” (V)DJ and class switch deletions are indicated for both alleles separately, indicated with VDJ and CS, respectively. In all cases an allele matching the IgH isotype expression is put in the column “Allele 1,” although in several cases both alleles are matching and the functional allele could not be identified. Presence of (V)DJ recombination, defined as deletion of cosmid U2-2 signal, is indicated with +. In cases HCL12, HCL13, and HCL21 in which the cosmid U2-2 signal is preserved, a deletion between JH and the proximal DQ52 gene may have occurred, which would be too small to be detected. This would be compatible with the presence of two rearranged JH bands on Southern blot. In the last column the interpretation of previously described Southern blot analysis25 is shown. Genomic DNA was digested withBamHI, EcoRI, HindIII, BglII, and Sac I and hybridized with probes for JHand Cμ. Column “JH” shows the number of rearranged JH alleles (R) and column “Cμ” the number of alleles with Cμ-deletions (D). In HCL1 and HCL2, a second rearranged JH band was found with only one restriction enzyme, which is indicated as 1/2R.
Abbreviations: NE, not evaluable; NT, not tested.
Discordance with fiber FISH results.
Class-switch deletions and other CH gene rearrangements in follicular lymphoma.
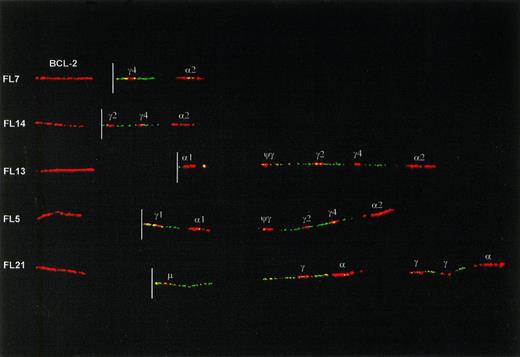
Fiber preparations of 23 cases of FL were hybridized with a combination of IgH probes and BCL-2 probes. The translocated, nonproductive allele could be identified in all cases on basis of juxtaposition of BCL-2 to IgH probe signals (Fig 3). In 3 of 23 FL (FL2, FL8, and FL21) the translocated IgH allele had retained the Cμ-Cδ region. In 2 of these cases the entire CH region was in the germline configuration, whereas in the third case (FL21) rearrangements downstream from Cδ were present. In the remaining 20 cases the translocated IgH allele had undergone class switching to one of the Cγ or Cα genes (Table2 and Fig 3). In one of these cases (FL16), in addition to a switch deletion between Sμ and Sγ1, a downstream deletion in the 3′ CH cluster had occurred.
Examples of class-switch rearrangements on the translocated allele of FLs, detected with a combination of IgH and BCL-2 probes. The red signal on the left side of each fiber is a BCL-2 cosmid probe located about 20 kb 5′ from the major breakpoint cluster (MBR) and 50 kb 5′ from the minor cluster region (MCR) of BCL-2. The location of the translocation breakpoint (derived from hybridizations with multiple BCL-2 probes, data not shown) is indicated with a white line. Originally, a more proximal (3′) BCL-2 cosmid was used that covers the breakpoint in case of a major breakpoint translocation. The more 5′ cosmid in this figure was used to create a gap between BCL-2 and IgH signals and hence facilitate interpretation of the IgH configuration. The upper 2 fibers show translocations with an MBR-breakpoint and with class switching to Cγ4 and Cγ2. The lower 3 fibers have translocation breakpoints in the MCR. Two of these have class switching to C1 and Cγ1, respectively. The bottom one has retained the Cμ-Cδ region but has downstream rearrangements, apparently consisting of deletion of a single Cγ gene from both the 5′ and the 3′ CHcluster.
Examples of class-switch rearrangements on the translocated allele of FLs, detected with a combination of IgH and BCL-2 probes. The red signal on the left side of each fiber is a BCL-2 cosmid probe located about 20 kb 5′ from the major breakpoint cluster (MBR) and 50 kb 5′ from the minor cluster region (MCR) of BCL-2. The location of the translocation breakpoint (derived from hybridizations with multiple BCL-2 probes, data not shown) is indicated with a white line. Originally, a more proximal (3′) BCL-2 cosmid was used that covers the breakpoint in case of a major breakpoint translocation. The more 5′ cosmid in this figure was used to create a gap between BCL-2 and IgH signals and hence facilitate interpretation of the IgH configuration. The upper 2 fibers show translocations with an MBR-breakpoint and with class switching to Cγ4 and Cγ2. The lower 3 fibers have translocation breakpoints in the MCR. Two of these have class switching to C1 and Cγ1, respectively. The bottom one has retained the Cμ-Cδ region but has downstream rearrangements, apparently consisting of deletion of a single Cγ gene from both the 5′ and the 3′ CHcluster.
Class Switching in Follicular Lymphoma
| Case . | Ig Expression . | Allele 1 . | Allele 2 t(14;18) . | |
|---|---|---|---|---|
| IgL . | IgH . | |||
| FL1 | κ | M | — | γ1 |
| FL2 | λ | M | — | — |
| FL3 | κ | MD | — | γ1 |
| FL4 | κ | M(D) | — | α1 |
| FL5 | κ | M | ds rear | γ1 |
| FL6 | κ | MD | ds rear | γ1 |
| FL7 | κ | M(D) | ds rear | γ4 |
| FL8 | κ | M | ds rear | — |
| FL9 | κ | M(D) | ds rear | γ1 |
| FL10 | λ | M(D) | ds rear\γ4? | γ4 |
| FL11 | κ | M | ds rear | γ1 |
| FL12 | λ | MD | ds rear | γ1 |
| FL13 | (κ) | M | ds rear | α1 |
| FL14 | λ | — | ds rear | γ2 |
| FL15 | κ | G2 | γ* | γ1 |
| FL16 | λ | G1 | γ1 | γ1 + ds rear |
| FL17 | κ | G1 | γ1 | γ2 |
| FL18 | λ | G2 | γ* | γ1 |
| FL19 | κ | G | γ2 | γ1 |
| FL20 | κ | G1 | γ1 + ds rear | α1 |
| FL21 | λ | G4 | γ* | ds rear |
| FL22 | — | — | γ1 | γ1 |
| FL23 | (κ) | — | γ* | γ1 |
| Case . | Ig Expression . | Allele 1 . | Allele 2 t(14;18) . | |
|---|---|---|---|---|
| IgL . | IgH . | |||
| FL1 | κ | M | — | γ1 |
| FL2 | λ | M | — | — |
| FL3 | κ | MD | — | γ1 |
| FL4 | κ | M(D) | — | α1 |
| FL5 | κ | M | ds rear | γ1 |
| FL6 | κ | MD | ds rear | γ1 |
| FL7 | κ | M(D) | ds rear | γ4 |
| FL8 | κ | M | ds rear | — |
| FL9 | κ | M(D) | ds rear | γ1 |
| FL10 | λ | M(D) | ds rear\γ4? | γ4 |
| FL11 | κ | M | ds rear | γ1 |
| FL12 | λ | MD | ds rear | γ1 |
| FL13 | (κ) | M | ds rear | α1 |
| FL14 | λ | — | ds rear | γ2 |
| FL15 | κ | G2 | γ* | γ1 |
| FL16 | λ | G1 | γ1 | γ1 + ds rear |
| FL17 | κ | G1 | γ1 | γ2 |
| FL18 | λ | G2 | γ* | γ1 |
| FL19 | κ | G | γ2 | γ1 |
| FL20 | κ | G1 | γ1 + ds rear | α1 |
| FL21 | λ | G4 | γ* | ds rear |
| FL22 | — | — | γ1 | γ1 |
| FL23 | (κ) | — | γ* | γ1 |
The second column shows immunoglobulin class and subclass expression as determined by immunofluorescence. Brackets were used to indicate weak expression. In case FL19, IgG subclass expression could not be determined. In the right two columns class-switch deletions on nontranslocated (allele 1) and translocated (allele 2) tumor alleles are indicated. In FL10 two nontranslocated alleles were identified, as shown. The question mark is explained in Results. Downstream rearrangements are indicated by ds rear. In cases FL5-14 these alleles had retained the Cμ-Cδ region, in FL16 and FL20 the downstream rearrangement was present together with an apparently normal γ1 switch.
The switch target gene could not be identified due to polymorphisms.
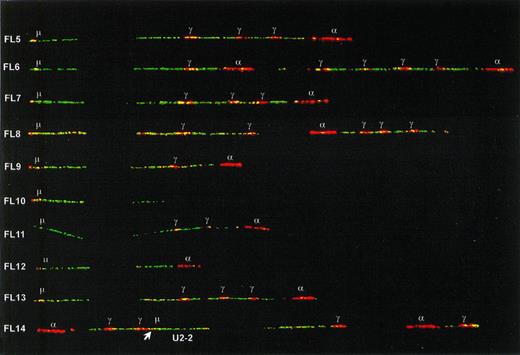
In all seven IgG expressing FL as well as in two nonexpressing cases, one type of fiber with both (V)DJ recombination and class-switch deletion was observed at a frequency comparable to the translocation (5% to 30%). We concluded that these fibers represented the functional tumor alleles. Switching had taken place to Cγ1 in four FL and to Cγ2 or Cγ4 in five FL (in some cases differentiation between Cγ2 and Cγ4 was not possible due to polymorphisms). One Cγ1-switched allele (in FL20) had only two Cγ genes in the 3′ cluster. Because the germline alleles had in their 3′ duplication units three and four Cγ genes, respectively, a downstream deletion must have occurred on this functional tumor allele. In 4 of the 13 IgM(D) expressing FL (FL1-4), no class-switch rearrangements were detected among the nontranslocated IgH fibers with deletion of cosmid U2-2. In contrast, in each of the nine other IgM(D) FL (FL5-13) and in one Ig-negative FL (FL14), an IgH allele was identified retaining its Cμ-Cδ region but with a rearrangement downstream of Cδ (Fig4). Because these downstream rearrangements were present exclusively in combination with (V)DJ recombination, we concluded that these IgH alleles represented somatic rearrangements in tumor cells. In most cases, the rearrangement consisted of a deletion of part of the CH locus. In cases FL5, FL8, and FL14, the configuration was more complex and was probably the result of multiple events including deletions and inversions. In some cases, for example cases FL6, FL10, FL12, and FL13, the CH configuration could not be reconstructed if the assumption was held that breakpoints occur solely in switch regions. For example, in FL10 the entire Cα probe signal was absent, though there is no known switch region located 3′ from Cα2. In the Ig-negative case FL14 the rearrangements also involved the JH-Cμ region, thereby probably disrupting the VDJ-Cμ transcription unit. In one case of FL (FL10), two different recurrent nontranslocated alleles were observed: one allele with retention of Cμ-Cδ and a downstream deletion (see Fig4), and one allele containing one Cγ and one Cα gene (not shown) suggesting a Cγ4-switched allele. Because the tumor exclusively expressed IgM, the latter allele may represent a remnant of a switch-circle that has been reintegrated somewhere in the genome,26 or the reciprocal product of a translocation in a Sγ site. Alternatively, it may represent a second tumor subclone that has lost IgH expression.
Class-switch rearrangements with retention of the Cμ-Cδ region on the nontranslocated, functional alleles of nine IgM expressing FL and one IgH-negative FL (FL14). The positions of Sμ and the Cγ and C plasmid probe signals are indicated. The differentiation between Cγ and C probe signals has been made by sequentially hybridizing probe mixes with and without the C probe. The position of the U2-2 cosmid in FL14 was determined by sequentially hybridizing the IgH probe mix with and without U2-2. Its orientation could be determined by the presence of a Sμ signal derived from the C probe (indicated with an arrow).
Class-switch rearrangements with retention of the Cμ-Cδ region on the nontranslocated, functional alleles of nine IgM expressing FL and one IgH-negative FL (FL14). The positions of Sμ and the Cγ and C plasmid probe signals are indicated. The differentiation between Cγ and C probe signals has been made by sequentially hybridizing probe mixes with and without the C probe. The position of the U2-2 cosmid in FL14 was determined by sequentially hybridizing the IgH probe mix with and without U2-2. Its orientation could be determined by the presence of a Sμ signal derived from the C probe (indicated with an arrow).
DISCUSSION
We have analyzed somatic rearrangements in B-cell malignancies using a technique that allows structural characterization of a large genomic region on single DNA molecules. We studied three types of B-cell malignancies that are supposed to represent three different stages in B-cell differentiation: MCL (pregerminal center B cells), FL (germinal center B cells), and HCL (postgerminal center B cells). Because germinal centers are major sites of class switching, switched IgH genes are expected to be present in FL and HCL but not in MCL. MCL is thought to be derived from follicle mantle B cells, always expresses IgM(D), and does not show hypermutation of the Ig genes.27 Indeed, in none of 10 MCL cases class-switch rearrangements were detected by fiber FISH on functional or nonfunctional alleles.
The immunophenotype of HCL, with relatively frequent expression of IgG or IgA, indicates an origin from postgerminal center B cells.15,28,29 Most (8 of 13) non-IgM expressing HCL had biallelic switch deletions. Although class switching to Cγ1 dominated when both alleles were taken into account, the expression pattern of HCL showed a strong preference for IgG3.15 Assuming that both alleles are subject to the same switch-regulating mechanism, the large proportion of IgG3 expressing HCL may well be the result of a selection process rather than induction of specific Cγ3 switching. In all HCL cases switching only involved CH genes in the 5′ cluster (Cγ3, Cγ1, Cα1) or Cδ. Within this region, distribution of switching on both alleles seems to be a stochastic process, with a high probability of switching to genes located in the middle of this region (Cγ1 and Cγ3) and a lower probability of switching to genes at the border (Cδ and Cα1). These results support a model in which probability of switching is determined by long-range activation of part of the CHregion.30 The distribution of switch target genes in HCL may be due to the influence of switch-inducing cytokines. For example IL-10, which has been shown to induce preferential switching to Cγ1 and Cγ3,31 is highly expressed in HCL cells.32
Follicular lymphoma is derived from follicle center cells and is characterized by ongoing hypermutation.33-35 Most cases express IgM and only a minority expresses IgG, IgA, or no IgH. However, at the ±2 kb resolution of fiber FISH, the translocated allele had switched normally in almost all IgG and IgM expressing cases, as has been observed before with Southern blotting, pulsed-field gel electrophoresis and reverse transcription-PCR techniques.36 37 On the functional alleles we found switch deletions correlating with IgH protein expression in the IgG expressing cases and retention of the Cμ-Cδ gene region in the IgM cases. However, a considerable proportion of the IgM(D) FL as well as one Ig-negative case had undergone CH rearrangement downstream from this Cμ-Cδ gene region on their functional allele.
Deletion of multiple CH genes with retention of the Cμ gene has been reported only once before,8 also in a case of follicular lymphoma. An obvious hypothesis to explain rearrangement of CH genes without involvement of Sμ is that Sμ is inactivated on this particular allele. This inactivation may be due to deletion of Sμ itself or to inactivation of parts of the Eμ-enhancer that are required for expression of Sμ germline transcripts. In mice with heterozygous deletion of JH-Eμ sequences, stimulation with LPS and IL-4 can induce rearrangement of Sγ1 without involvement of Sμ.38 To investigate in our FL with downstream rearrangements the presence of small genomic abnormalities that might be undetectable by fiber FISH, we recently performed Southern blotting on eight cases. On hybridization with a probe for Cμ, all cases showed a rearranged band in Bgl II,HindIII, and Xba I digests. In all cases but one (FL14), the rearranged fragment was 1 to 2kb smaller than the germline fragment in all 3 digests, strongly suggesting deletion of part of the Sμ sequences (data not shown).
A remaining question is why this Sμ inactivation occurs so frequently on the functional alleles of FL, whereas the translocated allele mostly shows normal class switching. This discrepancy between the two alleles would be compatible with selective pressure in favor of IgM or against IgG expression on a B-cell population that is at the same time permanently driven to switch. Such a process would lead to a high chance of developing a clone with either a functional IgM allele with downstream rearrangements, or a nonproductive allele like in FL14.
Using the fiber FISH technique we have obtained an unprecedentedly complete view of class switching in B-cell malignancies. We have found no evidence for class-switch mechanisms other than deletion of intervening sequences between Cμ and a downstream CHgene. In IgM expressing FL, we have found frequent CHrearrangements with retention of Cμ and Cδ. The lack of Sμ involvement in these cases is probably caused by deletion of part of the Sμ sequence. Fiber FISH analysis of the CH region in other types of B-cell malignancies and in subsets of normal B cells may tell whether this phenomenon is unique to follicular lymphoma.
Supported by grant NKB95-1047 from the Dutch Cancer Society.
Address correspondence to Jan-Willem Vaandrager, Department of Pathology, Leiden University Medical Centre, Bld 1, L1-Q, PO Box 9600, 2300 RC Leiden, The Netherlands; e-mail:Jvaandrage@Path_1.MedFac.LeidenUniv.NL.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal