Abstract
Splenic marginal-zone B cells, marginal-zone B cells of Peyer’s patches in the gut, and nodal marginal-zone B cells (also identified as monocytoid B cells) share a similar morphology and immunophenotype. These cells likely represent a distinct subset of B cells in humans and rodents, but their precise ontogenetic relationship as well as their origin from B cells of the germinal center is still debated. To study this, we performed a mutation analysis of the rearranged immunoglobulin variable genes (VH) of microdissected single nodal and splenic marginal-zone cells. In addition, we investigated the presence of proliferating cells and B-cell clones in the human splenic and nodal marginal zone as well as adjacent germinal centers. This was performed by immunohistochemical staining for the Ki-67 antigen and denaturing gradient gel analysis of amplified immunoglobulin heavy chain genes’ complementarity determining region 3 of microdissected cell clusters. A variable subset of nodal and splenic marginal-zone B cells showed somatic mutations in their rearranged VH genes, indicating that both virgin and memory B cells are present in the nodal and splenic marginal zone. Nodal and splenic marginal-zone B cells preferentially rearranged VH3 family genes such as DP47, DP49, DP54, and DP58. A preferential rearrangement of the same VH genes has been shown by others in the peripheral CD5− IgM+ B cells. These data suggest that the splenic and nodal marginal-zone B cells are closely related B-cell subsets. We also showed that marginal-zone B cells may cycle and that clones of B cells are frequently detected in the nodal as well as the splenic marginal zone. These clones are not related to those present in adjacent germinal centers. These data favor the hypothesis that clonal expansion occurs in the marginal zone. Whether the somatic hypermutation mechanism is activated during the clonal expansion in the marginal zone and which type of immune response triggers the clonal expansion need to be elucidated.
MARGINAL-ZONE B cells represent a distinct subset of B cells present in the spleen and ileal Peyer’s patches where an abundant influx of antigens occurs.1 The cells are medium sized with irregular nuclei and abundant pale cytoplasm, and typically express IgM and CD25 but not CD5, CD10, CD23, and usually not IgD.1,2 B lymphocytes with a similar morphology and phenotype are observed in the lymph node and are called monocytoid B cells or nodal marginal-zone B cells. These IgM+, IgD− B cells are inconspicuously intermingled with the small lymphocytes of the lymphocytic corona.2 However, in some reactive conditions such as toxoplasmic lymphadenitis or human immunodeficiency virus infections, a more prominent monocytoid B-cell proliferation previously known as “immature sinus histiocytosis” may be present.2
Studies in rodents on the migration of hapten-specific B cells provided evidence that memory B cells, generated in the immune response both to T-cell–dependent (TD) and T-cell–independent (TI) type 1 antigens, colonize the marginal zone of the spleen.3-6 Another subpopulation of marginal-zone B cells does not represent memory B cells and is involved in the immune response to TI-2 antigens. In addition, rodent marginal-zone B cells are noncirculating and do not cycle, indicating that clonal expansion does not occur in the rodent marginal zone.7,8 In humans, mutation analysis of rearranged immunoglobulin variable (VH) genes of microdissected marginal-zone B cells in the spleen and Peyer’s patches has shown mutated VH genes in the majority of the cells.9,10 A large population of marginal-zone B cells therefore likely represent memory B cells. Marginal-zone memory B cells may be related to the IgM-expressing B cells in the peripheral blood, in which somatic hypermutation of the rearranged Ig genes has been documented as well.11 The presence of somatic hypermutations, which until now have only been documented to occur in the germinal center in the course of TD immune reactions, may indicate that human marginal-zone memory B cells are generated in such an immune response. Indeed, there is as yet no evidence that the somatic hypermutation mechanism is activated in the TI immune reaction in humans. In addition, a minor population of marginal-zone B cells display rearranged VH genes without somatic mutations and therefore may represent the memory cells of a TI immune response or naive B cells.
The morphology and the immunophenotype of nodal marginal-zone B cells suggest that these cells are similar to those in the splenic marginal zone. To test this hypothesis, we performed a mutation analysis of rearranged VH genes of microdissected single monocytoid B cells. For comparison, a similar analysis was performed on splenic marginal-zone B cells.
In the human, it is not known whether nodal marginal-zone B cells may proliferate and clonally expand similar to the germinal-center B cells. Therefore, we investigated the presence of B-cell clones in the nodal marginal zone using denaturing gradient gel analysis of amplified immunoglobulin heavy chain genes’ complementarity determining region (IgH CDR3) products of microdissected nodal marginal-zones B-cell areas and compared these results with those obtained from adjacent germinal centers. A similar analysis was performed on splenic marginal-zone B cells for comparison. Cell proliferation was analyzed by the expression of the proliferation-associated nuclear antigen Ki67.
MATERIALS AND METHODS
Case selection.
Four lymph nodes with the morphology characteristic of lymphadenitis were selected for the analysis of monocytoid B cells.12 13Cases of reactive lymphadenitis with a prominent proliferation of monocytoid B cells have been selected. The cause of the lymphadenitis was not identified in three of the patients, whereas one showed serological evidence of toxoplasma infection. We studied spleen tissue obtained from two patients diagnosed with idiopathic thrombocytopenia and one patient in which the spleen was removed in the course of surgery for severe diverticulitis. Histological examination of these spleens showed a normal morphology including a prominent marginal zone. The patient characteristics are summarized in Table1.
Patient Characteristics
| Case . | Organ . | Morphological Findings . | Clinical Data . | Age (y)/Sex . |
|---|---|---|---|---|
| 1 | Lymph node | Reactive lymphadenitis | Single lymphadenopathy e causa ignota | 28/m |
| 2 | Lymph node | Reactive lymphadenitis | Toxoplasmosis (IgM+) | 41/f |
| 3 | Lymph node | Reactive lymphadenitis | Polyclonal gammopathy, acute angio-edema | 73/f |
| 4 | Lymph node | Reactive lymphadenitis | Chronic inflammatory disease, polyneuropathy, Borrelia+ | 44/m |
| 5 | Spleen | Normal | ITP | 66/m |
| 6 | Spleen | Normal | ITP | 32/f |
| 7 | Spleen | Normal | Diverticulitis | 83/f |
| Case . | Organ . | Morphological Findings . | Clinical Data . | Age (y)/Sex . |
|---|---|---|---|---|
| 1 | Lymph node | Reactive lymphadenitis | Single lymphadenopathy e causa ignota | 28/m |
| 2 | Lymph node | Reactive lymphadenitis | Toxoplasmosis (IgM+) | 41/f |
| 3 | Lymph node | Reactive lymphadenitis | Polyclonal gammopathy, acute angio-edema | 73/f |
| 4 | Lymph node | Reactive lymphadenitis | Chronic inflammatory disease, polyneuropathy, Borrelia+ | 44/m |
| 5 | Spleen | Normal | ITP | 66/m |
| 6 | Spleen | Normal | ITP | 32/f |
| 7 | Spleen | Normal | Diverticulitis | 83/f |
Abbreviations: ITP, idiopathic thrombocytopenic purpura; m, male; f, female.
Immunostaining of frozen sections.
Five-micrometer frozen tissue sections were stained with a mixture of anti-IgD (Dakopatts, Glostrup, Denmark) and anti-CD23 (Dakopatts) antibodies using the Avidin-Biotin-peroxidase complex (ABC) method as previously described.12 The slides were counterstained with hematoxylin. Splenic and nodal marginal-zone B cells were easily recognized in these tissue sections. Indeed, the marginal zone in the spleen is a distinct zone surrounding the white pulp. In the lymph nodes, a prominent marginal zone surrounded the lymphocytic corona or mantle zone and may locate along the marginal sinus. Immunostaining for IgD and CD23 highlighted the lymphocytic corona and the dendritic cells of follicle centers, respectively, and therefore unequivocally allowed the identification of the surrounding marginal zones.
To highlight the proliferating cells in the lymph node and spleen, 5-μm frozen tissue sections were stained with an antibody against the proliferation-associated nuclear antigen Ki67 (mib1; Biogenex, San Ramon, CA) using the ABC method.
Microdissection of cells.
The immunostained sections were digested with 10 mg/mL collagenase H (Boehringer Mannheim, Mannheim, Germany) in phosphate-buffered saline (PBS) for 1 hour at 37°C. Subsequently, nodal and splenic marginal-zone B cells were microdissected and aspirated using a hydraulic micromanipulator (Narishige MMW-202D, Tokyo, Japan) as previously described.13 Either single cells or cell clusters containing a similar number of cells (±500) were isolated. For the single-cell analysis and for the analysis of entire nodal and splenic marginal zones and germinal centers, cells or tissue were dropped in 10 μL of polymerase chain reaction (PCR) buffer (50 mmol/L KCL; 10 mmol/L Tris-HCL, pH 8.4; 0.01% gelatin) containing 200 μg/mL of proteinase K (Qiagen, Hilden, Germany). The isolated cells were covered with 50 μL of mineral oil, and digested at 37°C for 16 hours.
PCR amplification of the rearranged IgH genes.
The rearranged IgH genes from single cells were amplified using a seminested PCR method as previously described.13 14 In the first round of the PCR a mixture of six framework I (FR I) VH family-specific primers and three primers complementary to all JH genes were used. The second round of the PCR was performed in six separate reactions with one of the six VHFR I primers and internal JH primers.
After digestion of the cells, 30 μL of a mastermix (200 μmol/L dNTPs, 2.5 mmol/L MgCL2, 10 nmol/L of each primer in PCR buffer) was added and heated at 94°C for 10 minutes. While at 94°C, 1.5 U of Taq polymerase diluted in 10 μL of dH2O was added to the reaction mixtures. The PCR conditions of the first round consisted of 1 cycle at 95°C for 2 minutes, 59°C for 4 minutes, and 72°C for 80 seconds followed by 34 cycles at 95°C for 90 seconds, 59°C for 30 seconds, and 72°C for 80 seconds and 1 final cycle of 72°C for 5 minutes. In the seminested PCR reaction, which was performed in a 50-μL volume, 2 μL of the first round product were used as the template. In the second round of amplification the same reagent concentrations were used except for MgCL2, which was at 1.5 mmol/L, and for the primers, which were at 100 nmol/L. The second PCR consisted of a total of 35 cycles using annealing temperatures of 65°C and 61°C for VH3, VH4 and VH1, VH2, VH5, VH6 primers, respectively. The denaturing and extension temperatures as well as the cycling times were identical to those of the first round of PCR. All PCR reactions were performed in a Perkin Elmer 480 thermocycler (Perkin Elmer, Norwalk, CT). Positive controls consisted of the Namalwa cell DNA, whereas negative controls consisted of single T cells from CD3-stained serial sections of our cases, as well as DNA obtained from peripheral blood cells of healthy donors. An aliquot of 10 μL of the PCR products was size fractionated through a 1.5% agarose gel in 1 × TBE buffer.
For the analysis of dissected, entire nodal, and splenic marginal zones, and adjacent germinal centers, a seminested PCR method was used to amplify the IgH CDR3 of B cells, as previously described.15 16 In the first round of the PCR consensus primers to the 3′ end of the third framework region of VHgenes and to the 3′ end of the antisense JH segment were used. The second round of PCR was performed with the same VH primer and an internal JH primer with a 40-bp GC clamp attached to the 5′ end of the primer. After digestion of the cells, 40 μL of a mastermix (100 μmol/L dNTPs, 0.2 μmol/L of each primer, 2 mmol/L MgCL2, and 1 U Taq polymerase in PCR buffer) was added. The PCR conditions consisted of denaturing for 40 seconds at 94°C, annealing at 55°C for 40 seconds, and extension at 72°C for 40 seconds. Thirty cycles of PCR were performed. Two microliters of the PCR product of the first round was used as the template for the second round in a total volume of 100 μL. With the exception of the internal JH primer, the same reagents were used for the second round of PCR. The PCR conditions of the second round were identical to those of the first round. Positive controls were from the B-cell line Namalwa, whereas negative controls consisted of peripheral blood cell DNA.
Denaturing gradient gel electrophoresis (DGGE).
To sensitively detect small clones of B cells, IgH CDR3 PCR products were analyzed by DGGE.15
Forty microliters of the PCR GC-clamped products were concentrated by vacuum drying (DNA concentrator, Model 100; Savant Instruments, Holbrook, NY). The dried products were resuspended in 5 μL of loading buffer. These PCR products were then electrophoresed through a 40% to 70% denaturing gradient at 60°C and 160 V for 4.5 hours in TAE buffer (40 mmol/L Tris; 40 mmol/L sodium acetate; 1 mmol/L EDTA, pH 7.4) using the Biorad DGGE system (Biorad, Hercules, CA) as previously described.15 The products were visualized with ultraviolet light after staining with ethidium bromide.
Sequencing of the PCR products.
The PCR products obtained from the single-cell analysis were gel purified before sequencing. The PCR products were concentrated by vacuum drying. The dried products were resuspended in 10 μL of loading buffer and size fractionated through a 1.5% agarose gel. The appropriate bands were excised and the PCR products were extracted from the gel with Qiaquick spin-columns (Qiagen) following the manufacturer’s recommendations. The comigrating bands by DGGE analysis of IgH CDR3 PCR products in case 4 (see Fig 4) were eluted from the polyacrylamide gel and reamplified using the seminested PCR method previously described, except for the use of a nested consensus JH primer without GC-clamp. These PCR products were gel purified as outlined above, before sequencing.
An aliquot of the purified DNA was directly sequenced on both strands using Sanger’s chain-termination method and fluorescent dideoxynucleotide chain terminators.17 The sequencing primers were identical to the primers used for the second round of the seminested PCR of the rearranged VH genes. The JH primer used for the reverse sequencing reaction was determined based on the sequence obtained in the forward sequencing reaction. The products of the sequencing reaction were analyzed using the Applied Biosystems 373A sequenator (Applied Biosystems, Foster City, CA).
Sequence analysis.
The identification of the VH, IgH diversity genes (DH), and IgH junction genes (JH) germline sequences was performed by sequence comparison with the June 1997 update of V Base, which is a comprehensive database of human immunoglobulin germline gene sequences compiled from published sequences (V BASE sequence directory, Tomlinson I.M., MRC Centre for Protein Engineering, Cambridge, UK). Mac Vector 5.0 sequence analysis software (Oxford Molecular Group Inc, Campbell, CA) was used for the sequence analysis. For the germline DH gene attribution, the longest stretches with the highest homology were retained with a minimum of six successive matches or seven successive matches interrupted by one mismatch.
Somatic mutation analysis.
The probability that somatic mutations in the rearranged VH genes resulted from antigen selection was analyzed according to Chang and Casali.18 The probability that an excess of replacement (R) mutations in VH complementarity determining regions (CDRs) or FRs occurred by chance only was calculated using the binomial distribution model: P = (n!/(k!(n − k)!) × qk× (1 − q)n − k, where n is the total number of observed mutations, k is the number of observed R mutations in the CDRs or the FRs, and q is the probability that an R mutation will localize to CDRs or FRs (q = CDR rel (or FR rel) × CDR Rf [or FR Rf]). The inherent susceptibility of R mutations of the CDRs and FRs (CDR Rf and FR Rf, respectively) has been calculated for each of the identified germline genes and is based on the chances of the occurrence in each codon of an amino acid replacement given any single nucleotide change not resulting in a termination codon.
RESULTS
Sequence and mutation analysis of rearranged IgH genes of single marginal-zone cells.
A total of 76 nodal and splenic marginal-zone B cells were analyzed by PCR amplification, and 31 PCR products were obtained originating from 30 cells (Tables 2 and 3). Eight of the 31 rearranged genes (26%) likely represent unexpressed alleles because they are out of frame or contain a termination codon. Both nodal and splenic marginal-zone B cells preferentially rearranged genes of the VH3 family, and to a lesser extent, of the VH4 family. A marked overrepresentation of some VH genes such as DP 47 (5/31), DP 49 (5/31), and DP 54 (4/31) was observed. DP 58 was used in 2 cells. No preferential rearrangement involving any particular DH gene was observed except for the D21-9, a member of the DXP family, which was represented in 5 of the 31 sequences. In 16% (5/31) of the rearranged genes a recombination of 2 DH genes occurred. In addition, a marked predominance of JH4 and JH6 gene rearrangements (27/31) was observed.
Mutation Analysis of the Lymph Node Monocytoid B Cells
| Case/ Cells . | Germline VH (reference) . | N . | % Identity . | R/S . | Exp R CDR . | Exp R FR . | PCDR . | PFR . | Germline DH . | Germline JH . | Frame . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDR . | FR . | |||||||||||
| Case 1 | ||||||||||||
| 6-VH3 | DP77 (33) | 21 | 90.54 | 12/2 | 4/3 | — | — | — | — | D21-9 | JH6 | Out |
| 8-VH3 | DP54 (31) | 0 | 100 | 0/0 | 0/0 | — | — | — | — | DHQ52/DA1 | JH4 | In |
| 10-VH3 | DP47 (31) | 2 | 99.1 | 1/0 | 1/0 | — | — | — | — | DXP1/D21 | JH4 | In |
| 11-VH3 | COS 15 (33) | 1 | 99.5 | 0/0 | 0/1 | — | — | — | — | DM1 | JH6 | In |
| 21-VH3 | DP49 (31) | 1 | 99.5 | 0/0 | 0/1 | — | — | — | — | D2 | JH4 | Out |
| Case 2 | ||||||||||||
| 2-VH3 | DP54 (31) | 4 | 98.2 | 2/1 | 0/1 | — | — | — | — | DXP1 | JH4 | Out |
| 5-VH4 | 3d279 (32) | 10 | 95.5 | 6/1 | 0/3 | 2.5 | 5.1 | .017 | .0007 | D2 | JH4 | In |
| 11-VH4 | DP71 (31) | 1 | 99.5 | 0/0 | 0/1 | — | — | — | — | D4 | JH4 | In |
| 12-VH3 | DP47 (31) | 1 | 99.5 | 0/1 | 0/0 | — | — | — | — | DK4 | JH5 | In |
| 13-VH3 | DP54 (31) | 13 | 94.2 | 4/0 | 4/5 | 3.3 | 7 | .215 | .056 | DN4 + DXP′ 1 | JH6 | In |
| Case 3 | ||||||||||||
| 10-VH4 | DP70 (31) | 2 | 99.1 | 0/1 | 1/0 | — | — | — | — | DN1/D23-7 | JH4 | In |
| 14-VH4 | DP63/V4-34 (31) | 8 | 96.3 | 4/0 | 3/1 | — | — | — | — | DXP1 | JH4 | In (SC) |
| Case 4 | ||||||||||||
| 1-VH3 | DP35 (31) | 6 | 97.3 | 2/0 | 4/0 | 1.4 | 3.2 | .287 | .267 | NI | JH6 | In |
| 7-VH3 | DP49 (31) | 10 | 95.5 | 4/1 | 3/2 | — | — | — | — | D3 | JH4 | Out |
| 17-VH3 | DP58 (31) | 4 | 98.2 | 2/0 | 1/1 | — | — | — | — | DM1 | JH5 | Out |
| 20-VH4 | DP65/3d2773 (31) | 10 | 95.5 | 2/1 | 4/3 | 2.5 | 5.2 | .28 | .187 | DK4 | JH4 | In |
| Case/ Cells . | Germline VH (reference) . | N . | % Identity . | R/S . | Exp R CDR . | Exp R FR . | PCDR . | PFR . | Germline DH . | Germline JH . | Frame . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDR . | FR . | |||||||||||
| Case 1 | ||||||||||||
| 6-VH3 | DP77 (33) | 21 | 90.54 | 12/2 | 4/3 | — | — | — | — | D21-9 | JH6 | Out |
| 8-VH3 | DP54 (31) | 0 | 100 | 0/0 | 0/0 | — | — | — | — | DHQ52/DA1 | JH4 | In |
| 10-VH3 | DP47 (31) | 2 | 99.1 | 1/0 | 1/0 | — | — | — | — | DXP1/D21 | JH4 | In |
| 11-VH3 | COS 15 (33) | 1 | 99.5 | 0/0 | 0/1 | — | — | — | — | DM1 | JH6 | In |
| 21-VH3 | DP49 (31) | 1 | 99.5 | 0/0 | 0/1 | — | — | — | — | D2 | JH4 | Out |
| Case 2 | ||||||||||||
| 2-VH3 | DP54 (31) | 4 | 98.2 | 2/1 | 0/1 | — | — | — | — | DXP1 | JH4 | Out |
| 5-VH4 | 3d279 (32) | 10 | 95.5 | 6/1 | 0/3 | 2.5 | 5.1 | .017 | .0007 | D2 | JH4 | In |
| 11-VH4 | DP71 (31) | 1 | 99.5 | 0/0 | 0/1 | — | — | — | — | D4 | JH4 | In |
| 12-VH3 | DP47 (31) | 1 | 99.5 | 0/1 | 0/0 | — | — | — | — | DK4 | JH5 | In |
| 13-VH3 | DP54 (31) | 13 | 94.2 | 4/0 | 4/5 | 3.3 | 7 | .215 | .056 | DN4 + DXP′ 1 | JH6 | In |
| Case 3 | ||||||||||||
| 10-VH4 | DP70 (31) | 2 | 99.1 | 0/1 | 1/0 | — | — | — | — | DN1/D23-7 | JH4 | In |
| 14-VH4 | DP63/V4-34 (31) | 8 | 96.3 | 4/0 | 3/1 | — | — | — | — | DXP1 | JH4 | In (SC) |
| Case 4 | ||||||||||||
| 1-VH3 | DP35 (31) | 6 | 97.3 | 2/0 | 4/0 | 1.4 | 3.2 | .287 | .267 | NI | JH6 | In |
| 7-VH3 | DP49 (31) | 10 | 95.5 | 4/1 | 3/2 | — | — | — | — | D3 | JH4 | Out |
| 17-VH3 | DP58 (31) | 4 | 98.2 | 2/0 | 1/1 | — | — | — | — | DM1 | JH5 | Out |
| 20-VH4 | DP65/3d2773 (31) | 10 | 95.5 | 2/1 | 4/3 | 2.5 | 5.2 | .28 | .187 | DK4 | JH4 | In |
Abbreviations: VH gene, IgH variable gene; DH gene, IgH diversity gene; JH gene, IgH junction gene; N, total number of mutations observed; R, replacement mutations; S, silent mutations; CDR, complementarity determining region; FR, framework region; P CDR, probability that excess or scarcity of the R mutations in the VH gene CDRs resulted from chance only; P FR, probability that excess or scarcity of the R mutations in the VH gene FRs resulted from chance only; NI, no gene identified with certainty; SC, stop codon.
Mutation Analysis of the Splenic Marginal-Zone B Cells
| Case/ Cells . | VH (reference) . | N . | % Identity . | R/S . | Exp R CDR . | Exp R FR . | P CDR . | P FR . | DH . | JH . | Frame . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDR . | FR . | |||||||||||
| Case 5 | ||||||||||||
| 2-VH4 | V71-4 (32) | 7 | 96.8 | 1/2 | 4/0 | — | — | — | — | Dk5a | JH6 | Out |
| 2-VH3 | DP50 (31) | 13 | 94.1 | 5/0 | 6/2 | 3.2 | 7 | .13 | .18 | Da5 | JH4 | In |
| 4-VH3 | DP58 (31) | 9 | 95.9 | 1/1 | 2/5 | 2.2 | 4.9 | .24 | .04 | D21-9 | JH4 | In |
| 8-VH3 | DP51 (31) | 17 | 92.3 | 7/3 | 2/5 | 3.6 | 7.6 | .085 | .0003 | D23-7/Dxp4 | JH2 | In |
| 9-VH3 | DP47 (31) | 8 | 96.4 | 4/1 | 1/2 | 1.9 | 4.3 | .08 | .02 | DN4 + D23-7/Dxp4 | JH6 | In |
| 11-VH3 | DP47 (31) | 10 | 95.5 | 4/1 | 2/3 | 2.4 | 5.4 | .13 | .03 | DLR1 + D4/D4b | JH6 | In |
| 12-VH3 | DP49 (31) | 18 | 91.8 | 7/1 | 6/4 | 4.4 | 9.7 | .14 | .04 | Dxp1 | JH4 | In |
| Case 6 | ||||||||||||
| 6-VH3 | DP49 (31) | 4 | 98.2 | 2/1 | 1/0 | 0.9 | 2.1 | .03 | .21 | D21-9 | JH4 | In |
| 10-VH3 | DP54 (31) | 17 | 92.3 | 6/0 | 5/6 | — | — | — | — | DN4 | JH6 | In (SC) |
| Case 7 | ||||||||||||
| 6-VH3 | DP38 (31) | 5 | 97.8 | 3/0 | 1/1 | 1.3 | 2.6 | .09 | .02 | D21-9 | JH4 | In |
| 7-VH3 | DP38 (31) | 2 | 99.1 | 0/0 | 1/1 | — | — | — | — | DA1/DA4 | JH6 | In |
| 9-VH3 | VH3-8 (34) | 0 | 100 | 0/0 | 0/0 | — | — | — | — | DK4 | JH6 | In |
| 22-VH3 | DP49 (31) | 9 | 95.9 | 3/1 | 4/1 | 2.1 | 4.9 | .34 | .22 | Dxp4 | JH6 | In |
| 24-VH3 | DP47 (31) | 17 | 92.4 | 4/3 | 8/2 | 4.1 | 9.2 | .24 | .16 | D21-10 | JH4 | In |
| 29-VH3 | DP31 (31) | 3 | 98.6 | 0/0 | 1/2 | 1 | 1.6 | .43 | .11 | D21-10 + D4 | JH6 | In |
| Case/ Cells . | VH (reference) . | N . | % Identity . | R/S . | Exp R CDR . | Exp R FR . | P CDR . | P FR . | DH . | JH . | Frame . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDR . | FR . | |||||||||||
| Case 5 | ||||||||||||
| 2-VH4 | V71-4 (32) | 7 | 96.8 | 1/2 | 4/0 | — | — | — | — | Dk5a | JH6 | Out |
| 2-VH3 | DP50 (31) | 13 | 94.1 | 5/0 | 6/2 | 3.2 | 7 | .13 | .18 | Da5 | JH4 | In |
| 4-VH3 | DP58 (31) | 9 | 95.9 | 1/1 | 2/5 | 2.2 | 4.9 | .24 | .04 | D21-9 | JH4 | In |
| 8-VH3 | DP51 (31) | 17 | 92.3 | 7/3 | 2/5 | 3.6 | 7.6 | .085 | .0003 | D23-7/Dxp4 | JH2 | In |
| 9-VH3 | DP47 (31) | 8 | 96.4 | 4/1 | 1/2 | 1.9 | 4.3 | .08 | .02 | DN4 + D23-7/Dxp4 | JH6 | In |
| 11-VH3 | DP47 (31) | 10 | 95.5 | 4/1 | 2/3 | 2.4 | 5.4 | .13 | .03 | DLR1 + D4/D4b | JH6 | In |
| 12-VH3 | DP49 (31) | 18 | 91.8 | 7/1 | 6/4 | 4.4 | 9.7 | .14 | .04 | Dxp1 | JH4 | In |
| Case 6 | ||||||||||||
| 6-VH3 | DP49 (31) | 4 | 98.2 | 2/1 | 1/0 | 0.9 | 2.1 | .03 | .21 | D21-9 | JH4 | In |
| 10-VH3 | DP54 (31) | 17 | 92.3 | 6/0 | 5/6 | — | — | — | — | DN4 | JH6 | In (SC) |
| Case 7 | ||||||||||||
| 6-VH3 | DP38 (31) | 5 | 97.8 | 3/0 | 1/1 | 1.3 | 2.6 | .09 | .02 | D21-9 | JH4 | In |
| 7-VH3 | DP38 (31) | 2 | 99.1 | 0/0 | 1/1 | — | — | — | — | DA1/DA4 | JH6 | In |
| 9-VH3 | VH3-8 (34) | 0 | 100 | 0/0 | 0/0 | — | — | — | — | DK4 | JH6 | In |
| 22-VH3 | DP49 (31) | 9 | 95.9 | 3/1 | 4/1 | 2.1 | 4.9 | .34 | .22 | Dxp4 | JH6 | In |
| 24-VH3 | DP47 (31) | 17 | 92.4 | 4/3 | 8/2 | 4.1 | 9.2 | .24 | .16 | D21-10 | JH4 | In |
| 29-VH3 | DP31 (31) | 3 | 98.6 | 0/0 | 1/2 | 1 | 1.6 | .43 | .11 | D21-10 + D4 | JH6 | In |
Abbreviations: VH gene, IgH variable gene; DH gene, IgH diversity gene; JH gene, IgH junction gene; N, total number of mutations observed; R, replacement mutations; S, silent mutations; CDR, complementarity determining region; FR, framework region; P CDR, probability that excess or scarcity of the R mutations in the VH gene CDRs resulted from chance only; P FR, probability that excess or scarcity of the R mutations in the VH gene FRs resulted from chance only; SC, stop codon.
The mutation analysis data are summarized in Tables 2 and 3. The out-of-frame rearranged genes as well as those containing stop codons have not been analyzed for evidence of antigen selection. The deduced amino acid sequences of the rearranged VH genes are given in Figs 1 and 2.
Deduced VH protein sequences of monocytoid B cells. Lowercase letters indicate replacement mutations with regard to the germline sequences. These sequence data are available from the European Molecular Biology Laboratory (EMBL) under accession numbers AJ 227721 to 227725, AJ 227727, AJ 227728, AJ 227729, AJ 227730, AJ227745, AJ 227726, AJ 227750, AJ 227738, AJ 227741, AJ 227751, AJ 227749, respectively. FR, framework; CDR, complementarity determining region; *, stop codon.
Deduced VH protein sequences of monocytoid B cells. Lowercase letters indicate replacement mutations with regard to the germline sequences. These sequence data are available from the European Molecular Biology Laboratory (EMBL) under accession numbers AJ 227721 to 227725, AJ 227727, AJ 227728, AJ 227729, AJ 227730, AJ227745, AJ 227726, AJ 227750, AJ 227738, AJ 227741, AJ 227751, AJ 227749, respectively. FR, framework; CDR, complementarity determining region; *, stop codon.
Deduced VH protein sequences of splenic marginal-zone B cells. Lowercase letters indicate replacement mutations with regard to the germline sequences. These sequence data are available from EMBL under accession numbers AJ 227736, AJ 227742, AJ 227740, AJ 227739, AJ 227731, AJ 227746, AJ 227743, AJ 227735, AJ 227732, AJ 227733, AJ 227737, AJ 227734, AJ 227744, AJ 227747, AJ 227748, respectively. FR, framework; CDR, complementarity determining region; *, stop codon.
Deduced VH protein sequences of splenic marginal-zone B cells. Lowercase letters indicate replacement mutations with regard to the germline sequences. These sequence data are available from EMBL under accession numbers AJ 227736, AJ 227742, AJ 227740, AJ 227739, AJ 227731, AJ 227746, AJ 227743, AJ 227735, AJ 227732, AJ 227733, AJ 227737, AJ 227734, AJ 227744, AJ 227747, AJ 227748, respectively. FR, framework; CDR, complementarity determining region; *, stop codon.
In all but two cases, the sequences of the rearranged Ig VHgenes contained point mutations with respect to the closest germline gene sequences. It cannot be excluded that some of the so-called mutations may actually represent germline polymorphisms. However, these are likely few because most of the human VH germline genes have been identified.19 In addition, some of the so-called mutations could be attributed to Taq polymerase–induced errors. However, these errors are infrequent in our experience; in our lab, Taq polymerase–induced errors occur at a frequency of 0.095/1,000 bp.
Mutation analysis of the rearranged VH genes showed a mutation pattern that differed between the splenic marginal-zone B cells and the nodal marginal-zone B cells with regard to the mutation frequency and the pattern of somatic mutation. In nodal marginal-zone B cells, the mutation frequency of the rearranged VH genes ranged from 0% to 9.5% with a mean of 2.6%. One cell showed a germline configuration of its rearranged VH gene whereas six cells showed one or two point mutations in their VHgenes. All but one nodal marginal-zone B cell showed a random distribution pattern of mutations in their rearranged VHgenes.
In contrast, the marginal-zone B cells of the spleen displayed a mutation frequency of 4.2% in their rearranged VH genes, ranging from 0% to 8.2%. The rearranged Ig VH genes of eight splenic marginal-zone B cells showed a mutation pattern characteristic of antigen selection. Of these, seven rearranged VH genes showed fewer R mutations in the FRs than expected by chance only. This is suggestive of negative antigen selection. One rearranged VH gene displayed R mutations in the CDRs suggestive of positive antigen selective pressure, as reflected by the obtained P value (.03). A random pattern of somatic mutations was observed in the rearranged VH genes of four cells. Two cells (7-VH3 and 9-VH3 of case 7) showed zero or two point mutations in their VH genes, respectively. Codon 52b was missing in one sequence when compared with the closest germline gene (4-VH3 of case 4). This sequence may likely represent a germline polymorphism with two instead of three three ATG repeats at codons 52 to 52b with respect to the published closest germline sequence.
Immunohistochemistry for Ki67.
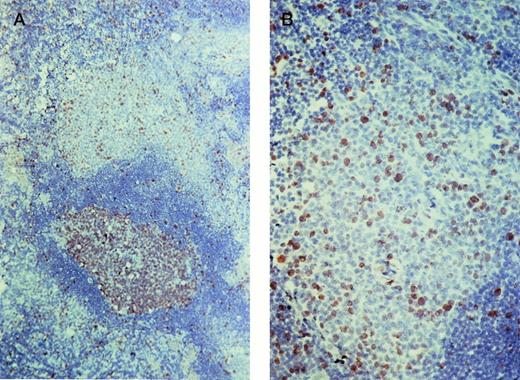
As expected, germinal-center B cells showed a prominent expression of the proliferation-associated nuclear antigen Ki67. In addition, clusters of B cells within the marginal zone of the lymph node also showed a distinct nuclear staining for Ki67, indicating that a large number of these cells are cycling (Fig3). In contrast, the splenic marginal zone contained only a few Ki67-expressing B cells. The absence of T cells or histiocytes as stained on serial sections showed that the Ki67-expressing cells were B cells.
Ki67 staining of a lymph node. (A) The germinal center of the secondary follicle (bottom) contains numerous Ki67-expressing cells. A smaller but impressive number of Ki67-expressing cells are also observed in the marginal zone (top). (B) Detail of the marginal zone.
Ki67 staining of a lymph node. (A) The germinal center of the secondary follicle (bottom) contains numerous Ki67-expressing cells. A smaller but impressive number of Ki67-expressing cells are also observed in the marginal zone (top). (B) Detail of the marginal zone.
DGGE analysis of IgH CDR3 products from microdissected nodal and splenic marginal-zone B-cell clusters.
We performed a DGGE analysis of the amplified IgH CDR3 products from microdissected clusters of splenic and Ki67-positive and -negative nodal marginal-zone B cells, and from adjacent germinal-center B cells. DGGE analysis showed the following two patterns according to the zone analyzed: a smear, indicating polyclonal IgH gene rearrangements, and distinct bands, indicating IgH gene rearrangements originating from clones of B cells in the marginal zone. The majority of the splenic and nodal marginal zones analyzed harbored small clones of B cells, whereas the minority of the marginal zones consisted purely of polyclonal B cells (Figs 4 and 5). Interestingly, when analyzing marginal zones containing numerous proliferating B lymphocytes, our results invariably showed the presence of clonal IgH gene rearrangements. However, clonal IgH gene rearrangements could also be observed when analyzing marginal zones containing only few or no cycling cells.
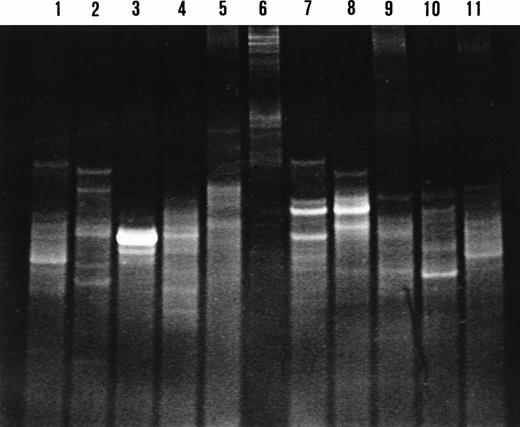
DGGE analysis of IgH CDR3 PCR products from microdissected nodal and splenic marginal zones. Lane 1, lanes 2 and 3, lanes 4 to 6, lanes 7 to 8, lanes 9 to 10, and lane 11 represent cases 5, 6, 2, 4, 7, and 1, respectively. Two patterns are observed on the denaturing gradient gel: a smear indicating polyclonal IgH gene rearrangements in lanes 1, 5, 6, and 9; and one or multiple bands indicating clonal IgH gene rearrangements in lanes 2 to 4, 7, 8, 10, and 11. Lanes 7 and 8 display comigrating bands indicating the presence of the same B-cell clone in the respective marginal zones. Sequence analysis of these IgH CDR3 products confirmed the presence of the same clone.
DGGE analysis of IgH CDR3 PCR products from microdissected nodal and splenic marginal zones. Lane 1, lanes 2 and 3, lanes 4 to 6, lanes 7 to 8, lanes 9 to 10, and lane 11 represent cases 5, 6, 2, 4, 7, and 1, respectively. Two patterns are observed on the denaturing gradient gel: a smear indicating polyclonal IgH gene rearrangements in lanes 1, 5, 6, and 9; and one or multiple bands indicating clonal IgH gene rearrangements in lanes 2 to 4, 7, 8, 10, and 11. Lanes 7 and 8 display comigrating bands indicating the presence of the same B-cell clone in the respective marginal zones. Sequence analysis of these IgH CDR3 products confirmed the presence of the same clone.
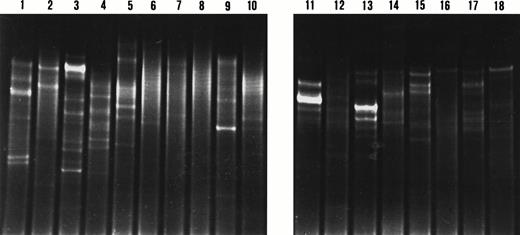
DGGE analysis of IgH CDR3 PCR products from microdissected marginal zones and adjacent germinal centers. Lanes 1 to 4, lanes 5 to 10, lanes 11 to 14, and lanes 15 to 18 represent cases 4, 1, 5, and 7, respectively. The even and odd numbers represent the analysis of the marginal zones and adjacent germinal centers, respectively. Two patterns are observed: a smear indicating polyclonal IgH rearrangements in lanes 6, 7, 8, 10, and 12; and a pattern with one or multiple bands indicating clonal IgH rearrangements in lanes 1 to 5, 9, 11, and 13 to 18. The analysis of the IgH CDR3 PCR products from Ki67-expressing marginal zones are observed in lanes 2, 4, 6, and 10.
DGGE analysis of IgH CDR3 PCR products from microdissected marginal zones and adjacent germinal centers. Lanes 1 to 4, lanes 5 to 10, lanes 11 to 14, and lanes 15 to 18 represent cases 4, 1, 5, and 7, respectively. The even and odd numbers represent the analysis of the marginal zones and adjacent germinal centers, respectively. Two patterns are observed: a smear indicating polyclonal IgH rearrangements in lanes 6, 7, 8, 10, and 12; and a pattern with one or multiple bands indicating clonal IgH rearrangements in lanes 1 to 5, 9, 11, and 13 to 18. The analysis of the IgH CDR3 PCR products from Ki67-expressing marginal zones are observed in lanes 2, 4, 6, and 10.
All but one of the microdissected germinal centers harbored clonal B-cell proliferations (Fig 5). Interestingly, DGGE analysis of the amplified IgH CDR3 products from microdissected germinal centers and adjacent splenic or nodal marginal zones never showed comigrating bands. In contrast, the IgH CDR3 products of two nodal marginal zones obtained from the same lymph node (case 4) displayed a comigrating band (Fig 4). The identical migration pattern of these bands on DGGE suggested the presence of the same B-cell clone in these monocytoid B-cell areas. Sequence analysis of these IgH CDR3 products confirmed the presence of the same clone (not shown).
DISCUSSION
Our study shows that the marginal-zone B cells of the lymph node and the marginal-zone B cells of the spleen predominantly rearrange VH3 family genes, and to a lesser extent, VH4 family genes. The preferential usage of VH3 family genes is accounted for by the frequent rearrangement of some VH3 family genes such as DP47, DP49, DP54, and DP58. These genes represented 39% (12/31) of the productively rearranged VHgenes. Similarly, there was a slight preferential rearrangement of the D21-9 gene, and a striking preferential rearrangement of the JH4 and JH6 genes. No difference in VH gene usage was noted between the splenic and nodal marginal-zone B cells. Why these particular genes are preferentially rearranged in nodal and splenic marginal-zone B cells is not clear. The restricted use of a small number of VH genes by the nodal and splenic marginal-zone B cells might reflect a case bias. However, this does not seem likely for nodal marginal-zone B cells because the lymph nodes selected for this study were obtained from patients presenting with different conditions. Although two of the three spleens studied were obtained from patients with idiopathic thrombocytopenic purpura (ITP), it is also unlikely that case bias may have accounted for the limited VH gene repertoire of splenic marginal-zone B cells. Indeed, in 90% of the patients diagnosed with ITP, antibodies against various platelet membrane glycoproteins have been shown.20 Because these antibodies are almost exclusively of the IgG isotype, the marginal-zone B cells are not likely the effector cells in this condition accounting for the VH gene bias.
Interestingly, a preferential usage of the same VH3 germline genes as observed in the nodal and splenic marginal-zone B cells studied here has been shown in circulating CD5−IgM+B cells.21 DP 47 is the most frequently rearranged VH gene segment, observed in 12.1% of the CD5− IgM-expressing B cells, whereas other genes of the VH3 family such as DP49, DP54, and DP58 are observed in 5.8%, 5.8%, and 4.4% of the productive rearrangements, respectively. In addition, a preferential usage of the D21-9 gene and the JH4 and JH6 genes is also a striking characteristic of the CD5−IgM+ B cells in peripheral blood. Taken together, these findings indicate that the splenic and nodal marginal-zone B cells as well as the circulating CD5−IgM+ B cells likely belong to the same subset of B cells.
The majority of the splenic and nodal marginal-zone B cells in our study showed somatic mutations in their rearranged VHgenes. This indicates that memory B cells constitute the majority of B cells in the splenic and nodal marginal zone. In addition, marginal zones also harbor naive B cells. These findings are in agreement with those previously published for the human marginal-zone B cells of the spleen and the Peyer’s patch.9,10 Interestingly, the lymph node marginal-zone B cells showed a lower mutation frequency of their rearranged VH genes than the splenic marginal-zone B cells. In contrast to only 2 out of 14 marginal-zone B cells in the spleen, 7 of the 16 nodal marginal-zone B cells showed a germline or near-germline configuration of their rearranged VH genes. In addition, all but 1 of the rearranged VH genes of the marginal-zone B cells in the lymph node showed a random distribution of R mutations in the CDRs and FRs. In contrast, 8 marginal-zone B cells of the spleen showed a mutation distribution pattern in their rearranged VH genes indicative of an antigen selective pressure. Thus, at least in our study, the majority of the nodal marginal-zone B cells seem to be naive or early memory B cells, whereas the majority of splenic marginal-zone B cells represent late memory B cells. Interestingly, the number of mutations in the rearranged VH genes of the marginal-zone B cells of Peyer’s patch is even higher than that of the marginal-zone B cells of the spleen.10 These differences in the number of mutations observed in nodal marginal-zone B cells and marginal-zone B cells of the spleen and Peyer’s patch are likely caused by differences in antigen exposure. Alternatively, these findings may indicate that the nodal and splenic B cells located in the marginal zone represent different subsets of B cells. However, the latter seems less likely in view of the similarities in morphology, immunophenotype, and VH gene usage of these B cells in the lymph node and the spleen. Interestingly, the majority of IgM-expressing peripheral blood B cells, representing 10% of the circulating B cells, also carry mutated VH genes.11,22,23 They do not express CD10, CD38, CD77, or activation markers such as interleukin-2 receptor (CD25) or transferrin receptor (CD71), and are usually negative for CD23, which is predominantly expressed by naive B cells.11 These data indicate that IgM+peripheral blood B cells also represent memory B cells. In addition to the pattern of VH gene usage, these data are also in agreement with the hypothesis that nodal and splenic marginal-zone B cells and circulating IgM+ B cells belong to the same B-cell compartment.11 Whether the memory B cells of this compartment result from a TD or TI immune reaction or both, or whether this may vary according to anatomic location is not known. It is clear that mutations arise in the rearranged VH genes of B cells generated through a TD immune reaction, but as yet there is no evidence that the hypermutation mechanism is activated in the TI immune reaction. However, somatic mutations may occur independent of a TD antigen stimulation in Xenopus and sheep,24 25 although in these species somatic mutations serve to diversify the primary antibody repertoire.
Interestingly, it is documented here for the first time that both the nodal marginal zone as well as the splenic marginal zone may harbor B cell clones. This is in agreement with the previous finding of the presence of B-cell clones in the marginal zone of Peyer’s patch.10 The presence of B-cell clones in the marginal zone may indicate either local clonal expansion or the migration of clonally related memory B cells, possibly from nearby germinal centers. The latter is less likely because no identical clones could be identified in the marginal zones or adjacent germinal centers of the lymph node and the spleen. In addition, our data show that an important fraction of the nodal but not the splenic marginal-zone B cells are cycling. Taken together, these findings favor the hypothesis that clonal expansion occurs in the marginal zone and that clones may persist thereafter. From our findings it seems likely that some of these cells may then enter the circulation as CD5−IgM+circulating B cells. The fact that cycling marginal-zone B cells were frequently observed in the lymph node but only rarely in the spleen may be explained by the different phases of the immune response in which the cells were analyzed. Indeed, marginal-zone B cells in the spleen are resident cells, whereas those in the lymph node are only conspicuous when the lymph node is acutely inflamed. Whether the somatic hypermutation mechanism is activated during clonal expansion in the marginal zone and whether clonal expansion follows triggering through TI antigenic stimulation is speculative and needs to be further elucidated.
In the mouse as well as in the human B-cell repertoire, three B-cell subsets, B-1a, B-1b, and B-2, have been recognized by their surface markers and the location, timing, and pathway of development.26-29 Conventional B-2 cells constitute the major population of the adult B-cell repertoire whereas B-1 cells represent 15% to 30% of the circulating, splenic, and tonsillar B lymphocytes. B-1 cells mainly produce naturally occurring polyreactive IgM antibodies whereas B-2 cells produce antigen-induced monoreactive IgG. Polyreactive B cells akin to B-1 cells may localize in the splenic marginal zone in rodents.30 In addition, B-1 cells display a phenotype similar to marginal-zone B cells as well as a similar range of somatic mutations in their rearranged VHgenes. However, marginal-zone B cells do not express surface CD5 as B-1a cells. Whether marginal-zone B cells belong to the B-1b cell subset, which is CD5− but CD5 mRNA+, seems possible but still needs to be shown by the expression of CD5 mRNA.
In conclusion, we have provided evidence in this study that marginal-zone B cells of the lymph node comprise virgin as well as memory B cells, similar to marginal-zone B cells of the spleen. In addition to similar morphological and immunophenotypical characteristics, these data indicate that nodal and splenic marginal-zone B cells belong to the same B-cell compartment.
Our study also shows that B-cell clones are present both in the nodal and splenic marginal zone that are unrelated to those present in adjacent germinal centers and that B cells may cycle in the marginal zone. These data indicate that clonal expansion takes place in the human marginal zone. Our combined data suggest that somatic hypermutation occurs during clonal expansion in the marginal zone. Alternatively, the somatic hypermutations may have been acquired in germinal centers at distant sites before migration of the cells to the marginal zone and the occurrence of clonal expansion.
ACKNOWLEDGMENT
We thank Monique Pattou, Suzanne Taelemans, and Erna Van Dessel for excellent technical assistance, and Michel Pipeleers for photography.
Supported by grant G.0311.97 of the Fonds voor Wetenschappelijk Onderzoek, Vlaanderen. J.D. and P.V. are postdoctoral research fellows of the Fonds voor Wetenschappelijk Onderzoek, Vlaanderen.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Jan Delabie, MD, PhD, Department of Pathology, University Hospitals of Leuven, Minderbroedersstraat 12, B-3000 Leuven, Belgium.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal