Abstract
To determine the cellular basis for the excellent clinical outcome of hyperdiploid acute lymphoblastic leukemia (ALL), defined by a modal chromosome number of 51 to 65, we assessed the growth potential of leukemic cells from 129 children with newly diagnosed ALL. Flow cytometric analysis was used to compare leukemic cell recoveries at the beginning and at the end of 7-day cultures on allogeneic bone marrow–derived stromal layers. The median percentage of cell recovery after culture was 91% (range, <1% to 550%). Among the 25 hyperdiploid cases, only two had cell recoveries above the median value, compared with 63 of 104 cases with different ploidies (P< .001); 21 had recoveries within the first quartile, in contrast to only 12 of the 104 other cases. Cell recoveries in the 16 cases with duplications of chromosomes 4 and 10, a feature previously associated with a superior outcome, were all within the first quartile. Flow cytometric studies indicated that rapid induction of apoptosis was the underlying cause of low cell recoveries in cases with hyperdiploidy. The demise of hyperdiploid cells on stroma was not due to failure to adhere with stromal elements (as shown by electron microscopy) or to deficiencies of interleukin-1 (IL-1), IL-2, IL-3, IL-4, IL-6, IL-7, IL-11, stem-cell factor, interferon- (IFN-), tumor necrosis factor- (TNF-), or to combinations of these cytokines. Inactivation of IL-4, IFN- and TNF-, which if secreted by stromal layers could be toxic to ALL cells, failed to improve the survival of hyperdiploid blasts. We conclude that leukemic cells bearing 51 to 65 chromosomes have a marked propensity to undergo apoptosis. The stringent survival requirements of these cells, together with their potentially higher sensitivity to antileukemic drugs, may well account for the high cure rates achieved in patients with this form of ALL.
AMONG THE DISTINGUISHING cellular features of acute lymphoblastic leukemia (ALL), hyperdiploidy of 51 to 65 chromosomes is one of the most reliable predictors of a favorable clinical outcome.1-9 Approximately 85% of children with this form of ALL can be cured with contemporary treatments.10 Several hypotheses have emerged to account for this outstanding result. One suggestion is that hyperdiploid cases have relatively higher percentages of cells in S phase,11resulting in greater sensitivity to cell cycle–specific drugs. Another is that hyperdiploid blasts accumulate significantly higher levels of methotrexate polyglutamates than do cells of lower ploidy,12,13 rendering them more sensitive to antimetabolites.14 An intriguing idea is that such cells lack the ability to expand rapidly and cannot grow outside the bone marrow microenvironment. Support for this hypothesis comes from the lower leukocyte counts and the general lack of bulky extramedullary disease typically seen at diagnosis in hyperdiploid cases.1,2,8 15
Underlying our research is the hypothesis that leukemic cell growth requirements strongly influence treatment outcome. Thus, cells with less stringent requirements may have an enhanced ability to grow outside the bone marrow microenvironment, enabling them to infiltrate pharmacologic sanctuaries with consequent adverse effects on treatment response. Methods to culture leukemic lymphoblasts on bone marrow–derived stromal layers provide a unique opportunity to measure the growth potential of leukemic lymphoblasts. Stromal feeder layers secrete factors that prevent apoptosis in vitro in most cases of ALL.16,17 In a previous study of 70 cases of childhood ALL, we found a strong correlation between in vitro growth potential and treatment outcome, and observed that hyperdiploid cases were among those least able to grow in stromal cultures.18 In the present study, we investigated in greater detail the relation between ploidy classification and the ability of leukemic cells to survive in vitro, using bone marrow–derived stromal cells as a source of survival factors. The findings suggest that hyperdiploid 51-to-65 ALL is unique among commonly recognized immunophenotypic, karyotypic, and genotypic subtypes of this disease, by virtue of its exquisitely stringent survival requirements and its marked propensity to undergo apoptosis.
MATERIALS AND METHODS
Characterization of leukemic cells.
Bone marrow samples were obtained at diagnosis from 129 patients with ALL, aged 3 months to 18 years (median, 5 years). These studies were approved by the institutional review board, with informed consent obtained from patients, their parents, or their guardians. Immunophenotyping was performed by standard methods, as previously described.19 One hundred fourteen cases were classified as B-lineage ALL because greater than 80% of the blast cells were CD19+ and CD22+. In the remaining 15 cases, the blast cells expressed CD7 and cytoplasmic or surface CD3, diagnostic of T-ALL. Karyotyping was done by standard banding techniques. DNA content was measured by flow cytometry after propidium iodide staining.11 Mononuclear cells were collected after centrifugation on a Lymphoprep density gradient (Nycomed, Oslo, Norway) and washed three times in phosphate-buffered saline (PBS).
Preparation of stromal layers and cell culture experiments.
To obtain bone marrow stromal cells, we collected mononuclear cells from healthy bone marrow donors. In previous studies, the source of stromal layers was not a factor in the survival and proliferation of leukemic lymphoblasts in vitro.20 The cells were cultured in RPMI 1640 (Whittaker, Walkersville, MD) with 10% fetal calf serum (FCS; Whittaker), 10-6 mol/L hydrocortisone (Sigma, St Louis, MO), 2 mmol/L L-glutamine (Whittaker), and antibiotics in an incubator set at 33°C and 5% CO2 in 96-well flat-bottomed microtiter plates (Costar, Cambridge, MA), as described.18 21-26 Immediately before initiating the cell culture experiments, we removed the media of bone marrow stromal cultures and washed the adherent cells seven times with AIM-V medium (Gibco, Grand Island, NY). Leukemic cells were resuspended in AIM-V medium at a final concentration of 1.5 × 106/mL. Two hundred microliters of the suspension were then placed in a 96-well tissue culture plate or seeded onto bone marrow stromal cells. In 22 cases (including all T-ALL samples) fresh leukemic cells were placed in culture within 5 hours of collection; the remaining 107 samples were cryopreserved and used immediately after thawing. In all samples, cell viability exceeded 80% by Trypan blue dye exclusion. In preliminary experiments with nine B-lineage ALL cases, in which both fresh and cryopreserved samples were cultured on stromal layers, cryopreservation did not significantly affect the survival and growth of lymphoblasts in culture (not shown). Recombinant human interleukin (IL)-1α, IL-1β, IL-2, IL-3, IL-6, IL-7, IL-11, and stem cell factor (SCF) were purchased from R & D Systems (Minneapolis, MN); IL-4 and tumor necrosis factor-α (TNF-α) were from Genzyme (Cambridge, MA); and interferon-α (IFN-α) was from Schering (Kenilworth, NJ). Cytokines were used at the final concentrations indicated in the Results, in excess of the active concentrations measured in the manufacturer’s tests. Neutralizing antibodies to IL-4, TNF-α, and IFN-α (from R & D) were used at the concentrations recommended by the manufacturer. All cell cultures were performed at 37°C under 5% CO2.
At the termination of cultures, cells were obtained by vigorous pipetting. B-lineage ALL samples were incubated with CD19 monoclonal antibody conjugated to fluorescein isothiocyanate (FITC); T-ALL samples were incubated with FITC-conjugated CD7. All antibodies were from Becton Dickinson (San Jose, CA). Samples were analyzed with a FACScan flow cytometer with Lysis II or CellQuest softwares (Becton Dickinson), as previously described.18 21-26 After 7 days of culture the percentage of cell recovery was calculated as follows: (No. of CD19+ or CD7+ Lymphoblasts After 7 Days of Culture) × 100 / (No. of CD19+ or CD7+ Lymphoblasts After 1 Hour of Culture). All results are reported as the mean of at least duplicate experiments. Leukemic cells were counted without knowledge of the patient’s clinicobiologic features or treatment response.
Determination of apoptosis.
To detect phosphatidylserine residues exposed on the cell surface (a marker of apoptosis),27 we labeled cells with FITC-conjugated Annexin-V (Trevigen, Gaithersburg, MD), following the manufacturer’s instructions. In these experiments, cell membrane permeabilization was revealed by labeling cells with 5 μg/mL of propidium iodide (Trevigen) for 15 minutes at 20°C. To detect DNA fragmentation, cells were fixed in 1% wt/vol paraformaldehyde, permeabilized with 70% vol/vol ice-cold ethanol, and incubated with bromodeoxyuridin (BrdU)-labeled dUTP plus terminal deoxynucleotidyl transferase.28 BrdU-dUTP incorporated into fragmented DNA was visualized with a FITC-labeled anti-BrdU antibody. At the same time, DNA content was measured after treating the permeabilized cells with RNAse and propidium iodide (5 μg/mL). All reagents were purchased from Phoenix (San Diego, CA), and the staining procedures followed the manufacturer’s instructions.
Electron microscopy.
For scanning electron microscopy, stromal layers were prepared on membrane cell culture inserts (Falcon Cyclopore; Becton Dickinson) as previously described17 and seeded with leukemic lymphoblasts from four cases of ALL. After 1 to 3 days of culture, the inserts were washed with PBS, fixed in 2.5% glutaraldehyde in PBS, rinsed with PBS containing 7% sucrose, postfixed in 2% osmium tetroxide, rinsed in water, passed through ethanol gradients (30%, 70%, 85%, 95%, and 100%) for 5 minutes each, and placed in fresh 100% ethanol for an additional 5 minutes. The preparations were dried by the critical point method in an Autosamdri-840 (Tousimis Research, Rockville, MD), mounted on a specimen holder, and coated with gold in a sputter-coater (Denton Vacuum, Cherry Hill, NJ). Cells were examined in the scanning mode of a 1200 EXII TEMSCAN electron microscope (JEOL, Tokyo, Japan).
Statistical analysis.
Distributions of commonly measured presenting features according to the percentage of viable leukemic cells (above or below the median value) recovered after 7 days of culture were compared by Fisher’s exact test.
RESULTS
Relation of immunophenotypic and genetic features to cell survival on stromal feeder layers.
Among the 129 diagnostic ALL samples studied, the median percentage of cell recovery after 7 days of culture on bone marrow–derived stromal layers was 91% (range, <1% to 550%), compared with 1% (range, <1% to 151%) among 125 parallel cultures without stroma. Table 1 compares cell recovery on stroma according to the presenting immunophenotypic features of the cases. The median percentage of recovered cells did not differ significantly between B-lineage ALL (n = 114) and T-lineage ALL (n = 15), nor was there any significant difference in cell recovery between the groups with an early pre-B (n = 73) or pre-B (cytoplasmic μ+; n = 41) immunophenotype.
Relation of Presenting Cellular Features With Leukemic Cell Recovery After Culture on Stroma
| Feature . | Category . | Percent Cell Recovery* . | P Value† . | |
|---|---|---|---|---|
| ≤91% . | >91% . | |||
| Lineage | T | 7 | 8 | .791 |
| B | 57 | 57 | ||
| Differentiation | Early B | 41 | 32 | .175 |
| Pre B | 17 | 23 | ||
| Adverse genetic features‡ | Yes | 8 | 17 | .047 |
| No | 56 | 46 | ||
| TEL rearrangements | Yes | 12 | 19 | .206 |
| No | 42 | 37 | ||
| Hyperdiploidy 51-to-65 | Yes | 23 | 2 | <.001 |
| No | 41 | 63 | ||
| Feature . | Category . | Percent Cell Recovery* . | P Value† . | |
|---|---|---|---|---|
| ≤91% . | >91% . | |||
| Lineage | T | 7 | 8 | .791 |
| B | 57 | 57 | ||
| Differentiation | Early B | 41 | 32 | .175 |
| Pre B | 17 | 23 | ||
| Adverse genetic features‡ | Yes | 8 | 17 | .047 |
| No | 56 | 46 | ||
| TEL rearrangements | Yes | 12 | 19 | .206 |
| No | 42 | 37 | ||
| Hyperdiploidy 51-to-65 | Yes | 23 | 2 | <.001 |
| No | 41 | 63 | ||
The median percentage of leukemic cells recovered after 7 days of culture on stromal layers relative to the number of cells originally seeded was 91% (range, <1% to 550%).
By Fisher’s exact test.
Cases with t(9;22), t(1;19) or MLL gene rearrangements.
We also determined the chromosomal and molecular genetic changes in these cases and attempted to relate the findings to survival on stromal layers. Although not statistically significant, there was a clear trend toward higher cell recoveries among cases with unfavorable genetic features. Thus, of 25 patients with the Philadelphia (Ph) chromosome (n = 6), MLL gene rearrangements (n = 14), or the t(1;19) (n = 5), 17 had a cell recovery above median value (Table 1). Importantly, in the 31 cases with a TEL gene rearrangement, an abnormality generally associated with favorable responses to treatment,29 cell recovery values showed essentially the same distribution as those in the remaining cases studied (Table 1).
Relation of ploidy to cell survival on stromal layers.
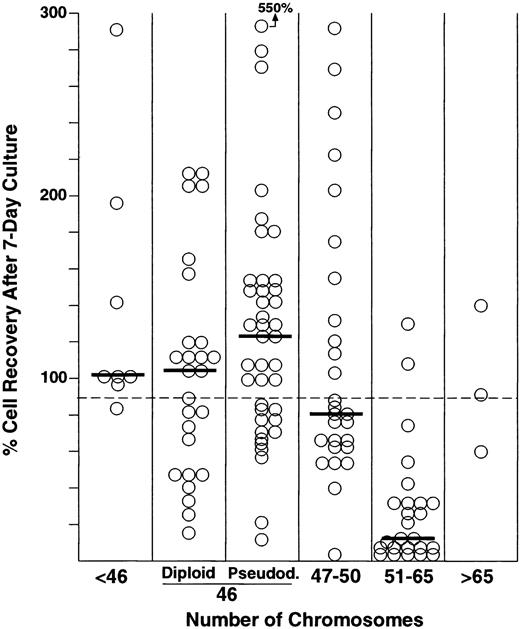
Complete information about ploidy was available for all the 129 cases studied. In addition to the 25 hyperdiploid cases with modal chromosome numbers of 51 to 65, 8 cases were classified as hypodiploid, 26 as having 46 chromosomes or constitutive trisomy 21 with no detectable abnormalities, and 40 as pseudodiploid. Among the 30 remaining cases, 27 were hyperdiploid with modal numbers of 47 to 50, and 3 were near-tetraploid. Figure 1 shows the percentage of cell recovery according to ploidy. The most striking finding was the significantly decreased survival of the hyperdiploid 51-to-65 cases compared with other ploidy groups. That is, cell recoveries above the median value of 91% were noted in only two of 25 cases, in contrast to 63 of the remaining 104 cases (P < .001; Table 1). Remarkably, all but four of the recovery values for the hyperdiploid 51-to-65 group were within the first quartile (<49%), compared with only 12 of the 104 cases with different ploidy classification.
Percentage of cell recovery after 7 days of culture on stroma according to ploidy. Numbers of leukemic lymphoblasts before and after culture were counted by flow cytometry as described in Materials and Methods. Horizontal bars indicate the median cell recovery in each ploidy group. The broken horizontal line indicates the overall median cell recovery.
Percentage of cell recovery after 7 days of culture on stroma according to ploidy. Numbers of leukemic lymphoblasts before and after culture were counted by flow cytometry as described in Materials and Methods. Horizontal bars indicate the median cell recovery in each ploidy group. The broken horizontal line indicates the overall median cell recovery.
The survival disadvantage of hyperdiploid cells was confirmed by flow cytometric analysis of cellular DNA content. Of the 128 cases in which this parameter was studied, 21 had a DNA index (DI) of 1.16 to 1.6, corresponding to a modal chromosome number of 51 to 65. Only 2 of these cases had cell recoveries above 91%, in contrast to 62 of the 107 cases with a DI less than 1.16 or greater than 1.6 (P < .001).
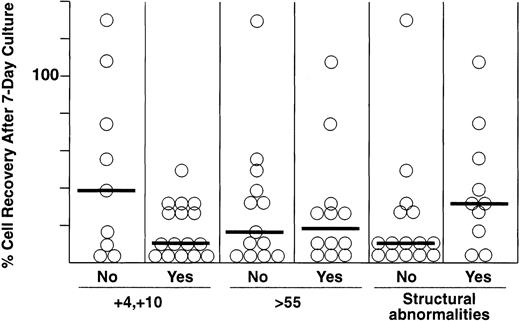
Within the hyperdiploid group of ALL cases, a duplication of chromosomes 4 and 10, a modal chromosome number greater than 55, and the absence of structural abnormalities have been associated with especially good treatment outcome.30-32 We therefore tested whether these features influenced recovery after culture on stromal feeder layers. As shown in Fig 2, cell recoveries did not differ significantly among these subsets of patients, although each of the 16 cases with a +4 and +10 karyotype had cell recoveries within the first quartile.
Percentage of cell recovery after 7 days of culture on stroma among different subgroups of hyperdiploid 51-to-65 ALL, including cases without and with duplications of chromosomes 4 and 10, ≤ or >55 chromosomes, and without and with structural chromosomal abnormalities. Numbers of leukemic lymphoblasts before and after culture were counted by flow cytometry as described in Materials and Methods. Horizontal bars indicate the median cell recovery in each group.
Percentage of cell recovery after 7 days of culture on stroma among different subgroups of hyperdiploid 51-to-65 ALL, including cases without and with duplications of chromosomes 4 and 10, ≤ or >55 chromosomes, and without and with structural chromosomal abnormalities. Numbers of leukemic lymphoblasts before and after culture were counted by flow cytometry as described in Materials and Methods. Horizontal bars indicate the median cell recovery in each group.
Mechanism of cell death of hyperdiploid blasts and effects of exogenous cytokines.
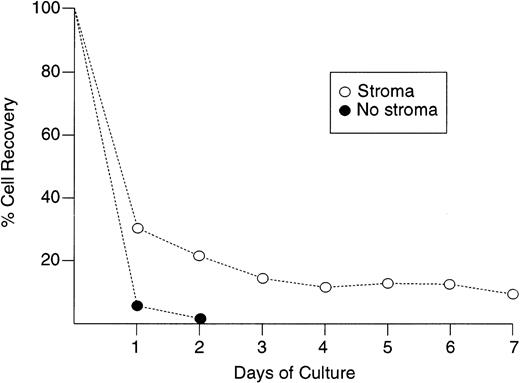
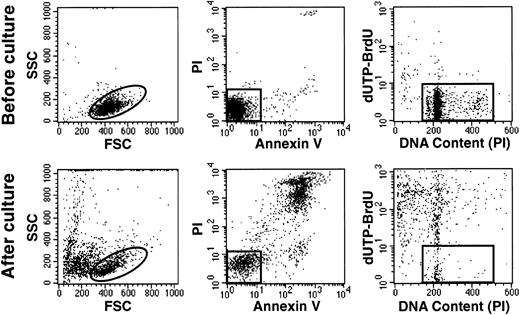
Hyperdiploid cells died rapidly in stromal cultures, as shown by the prominent terminal morphologic changes (eg, cell shrinkage, nuclear fragmentation) observed in situ with an inverted microscope within 48 to 72 hours of culture. In one experiment with a hyperdiploid sample (55 chromosomes), we counted viable cells after each day of culture for 7 days. After a single day of culture, only 27% of the originally seeded cells could be recovered. This value decreased progressively to 9% after 7 days of culture (Fig 3). A mechanism of hyperdiploid cell death was suggested by shifts in light-scattering properties similar to those of cells undergoing apoptosis after treatment with cytotoxic drugs.28 These included a reduction in forward scatter (FSC), indicating a reduction in cell size, and an increase in side scatter (SSC) indicating an increase in cell granularity (Fig 4). Occurrence of apoptosis was confirmed by binding of Annexin-V, loss of DNA, and prominent appearance of DNA fragmentation (Fig 4).
Kinetics of cell recovery during culture with and without stroma in one case of hyperdiploid ALL (55 chromosomes). Numbers of leukemic lymphoblasts before and after culture were counted by flow cytometry as described in Materials and Methods. Each point indicate the means of two measurements.
Kinetics of cell recovery during culture with and without stroma in one case of hyperdiploid ALL (55 chromosomes). Numbers of leukemic lymphoblasts before and after culture were counted by flow cytometry as described in Materials and Methods. Each point indicate the means of two measurements.
Hyperdiploid lymphoblasts undergo apoptosis despite the presence of stromal layers. Flow cytometric dot plots represent assessment of apoptosis before (top panels) and after 2 days of culture (bottom panels) in one case of hyperdiploid ALL (55 chromosomes). Left panels illustrate FSC (a measurement of cell size) versus SSC (a measurement of cell granularity). Center panels illustrate binding of Annexin V FITC (an indicator of apoptosis) and staining with propidium iodide (PI; a sign of membrane permeability). Right panels illustrate the cells’ DNA content, measured by staining with PI after cell membrane permeabilization, and labeling with dUTP-BrdU, which is incorporated into cells with fragmented DNA. In all panels, events outside the gates correspond to cells at various stages of apopotic death.
Hyperdiploid lymphoblasts undergo apoptosis despite the presence of stromal layers. Flow cytometric dot plots represent assessment of apoptosis before (top panels) and after 2 days of culture (bottom panels) in one case of hyperdiploid ALL (55 chromosomes). Left panels illustrate FSC (a measurement of cell size) versus SSC (a measurement of cell granularity). Center panels illustrate binding of Annexin V FITC (an indicator of apoptosis) and staining with propidium iodide (PI; a sign of membrane permeability). Right panels illustrate the cells’ DNA content, measured by staining with PI after cell membrane permeabilization, and labeling with dUTP-BrdU, which is incorporated into cells with fragmented DNA. In all panels, events outside the gates correspond to cells at various stages of apopotic death.
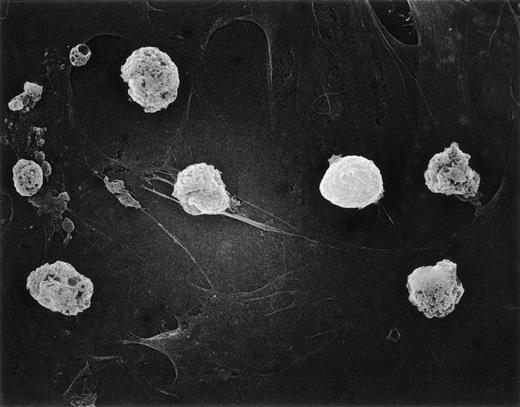
We previously showed an intimate relationship between immature B cells and stromal elements in which close contact of both normal and leukemic cells with stromal layers was a prerequisite for continued survival.17 20 Thus, it seemed reasonable to suspect that loss of adhesive interactions with stromal layers could account for the poor viability of hyperdiploid cells in such cultures. This prediction was tested by scanning electron microscopy studies of two hyperdiploid cases with low cell recoveries (<50%) after 7-day cultures. There was no indication that the hyperdiploid cells differed from nonhyperdiploid cells (from 2 cases studied in parallel) in their adherence to stromal layers (Fig 5). Thus, this factor does not appear to be related to the early demise of hyperdiploid lymphoblasts.
Hyperdiploid lymphoblasts adhere to stroma. Scanning electron microscopy of hyperdiploid lymphoblasts after 3 days of culture on bone marrow–derived stroma. Most cells are dying despite tight adhesion to stromal elements.
Hyperdiploid lymphoblasts adhere to stroma. Scanning electron microscopy of hyperdiploid lymphoblasts after 3 days of culture on bone marrow–derived stroma. Most cells are dying despite tight adhesion to stromal elements.
Alternatively, stromal cultures may lack some of the growth factors essential for the support of hyperdiploid leukemic cells. Therefore, in experiments representing eight cases of hyperdiploid ALL with 7-day cell recoveries of less than 1% to 46%, we added a variety of cytokines to the cultures: IL-1α at 50 to 200 pg/mL (n = 4), IL-1β at 50 to 400 pg/mL (n = 4), IL-2 at 200 ng/mL (n = 4), IL-3 at 2 to 10 ng/mL (n = 5), IL-4 at 100 U/mL (n = 5), IL-6 at 2 to 7.5 ng/mL (n = 6), IL-7 at 25 ng/mL (n = 8), IL-11 at 10 to 50 ng/mL (n = 1), SCF at 10 to 50 ng/mL (n = 6), TNF-α at 50 U/mL (n = 5), IFN-α at 2,000 U/mL (n = 3), or fetal calf serum 10% (n = 5). None of these exogenous factors suppressed apoptosis, whether added singly or in combination (data not shown). In fact, IL-4, IFN-α, and TNF-α proved toxic to the cells, as in previous studies.21,33 34
We examined the possibility that low concentrations of toxic cytokines may have been present in stromal cultures. Thus, neutralizing antibodies to IL-4, IFN-α, and TNF-α were added either alone or in combination to cultures supporting one case of hyperdiploid ALL (chromosome number, 57; cell recovery after stroma-supported culture, 7%). None of these additions significantly affected 7-day cell recoveries.
Finally, we considered whether allopurinol, which is often administered to leukemia patients before collection of diagnostic bone marrow, could have selectively impaired the survival of hyperdiploid cells in culture. Among the 25 hyperdiploid patients, however, 14 (11 of which with cell recoveries of <50%) either did not receive allopurinol or received it after the bone marrow sampling, thus excluding this possibility.
DISCUSSION
In this study, we show that hyperdiploid ALL with a modal chromosome number of 51 to 65 constitutes a biologically distinct subtype of ALL, characterized by a high rate of spontaneous cell death in vitro. Unlike results obtained with other ploidy groups, apoptosis in these cases was not suppressed by culture on bone marrow–derived stromal layers. Moreover, supplementation of the cultures with exogenous cytokines known to stimulate hematopoietic cells lacked any discernible effect on cell recovery, indicating that hyperdiploid lymphoblasts have exceedingly stringent survival requirements. Finally, it should be emphasized that extensive apoptosis in culture was not a feature of hyperdiploidy of less than 51 chromosomes or of other cases generally acknowledged to have a favorable prognosis, such as those with TEL gene rearrangement.
The molecular basis underlying the high propensity of hyperdiploid 51-to-65 to undergo apoptosis is still unknown. Tsuchiya et al postulated that the extra chromosomes of hyperdiploid ALL may control its tumorigenicity, possibly through increased gene dosage.35 Although plausible, this explanation does not account for the unfavorable prognostic features of near-triploid and near-tetraploid ALL, which may represent clinically distinct entities within the overall category of hyperdiploid ALL.36Alternatively, the pathogenesis of hyperdiploid ALL could involve molecular defects leading to both DNA content abnormalities and a propensity to undergo apoptosis. This idea is suggested by the observation that ectopic expression of the protein kinase PITSLREβ1 induces telophase delay, abnormal chromosome segregation, and an increase in the rate of apoptosis in mammalian cells.37Although preliminary studies have failed to detect PITSLREβ1 overexpression in hyperdiploid lymphoblasts (J. Lahti, V. Kidd, and D. Campana, unpublished results, March 1996), the possible involvement of similar mechanisms in the pathogenesis of this leukemia warrants further careful investigation.
The culture system used in this study simulates the in vivo growth conditions of leukemic lymphoblasts in that both survival and growth factors are derived from cells constituting the bone marrow microenvironment. However, failure of the system to support hyperdiploid cells (which thrive in vivo) clearly shows that the culture conditions are not optimal for all lymphoid progenitors. Nonetheless, our culture system appears to provide a reliable estimate of leukemic cell growth potential. Indeed, in a previous study with samples from children enrolled in a single program of chemotherapy, cell recovery after 7 days of culture on stromal layers emerged as an independent predictor of treatment outcome.18 Moreover, among the 25 patients with hyperdiploid ALL included in the present study, only two have relapsed. Cell recoveries in these cases were 133% and 74%, the first and the third highest values measured in this group.
In summary, our findings help explain the paradox of higher percentage of proliferating cells and lower white blood cell counts generally observed at diagnosis in ALL with 51 to 65 chromosomes.1,2,8,11,15 Hyperdiploid lymphoblasts appear to be ideal targets for innovative, potentially less toxic therapies designed to decrease microenvironmental support to leukemic cells.38 39
ACKNOWLEDGMENT
We thank Michael Hancock for statistical analysis and John Gilbert for editorial suggestions.
Supported by grants RO1-CA58297, P30-CA21765 (CORE), and CA-20180 from the National Cancer Institute; and by the American Lebanese Syrian Associated Charities (ALSAC).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Dario Campana, MD, PhD, Department of Hematology-Oncology, St Jude Children’s Research Hospital, 332 North Lauderdale, Memphis TN 38105; e-mail:dario.campana@stjude.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal