Abstract
The general utility of a novel, fluorescence-based procedure for assessing gene transfer and expression has been demonstrated using hematopoietic stem and progenitor cells. Lineage-depleted hematopoietic cells were isolated from the bone marrow or fetal livers of acid sphingomyelinase–deficient mice, and retrovirally transduced with amphotropic or ecotropic vectors encoding a normal acid sphingomyelinase (ASM) cDNA. Anti–c-Kit antibodies were then used to label stem- and progenitor-enriched cell populations, and the Bodipy fluorescence was analyzed in each group after incubation with a Bodipy-conjugated sphingomyelin. Only cells expressing the functional ASM (ie, transduced) could degrade the sphingomyelin, thereby reducing their Bodipy fluorescence as compared with nontransduced cells. The usefulness of this procedure for the in vitro assessment of gene transfer into hematopoietic stem cells was evaluated, as well as its ability to provide an enrichment of transduced stem cells in vivo. To show the value of this method for in vitro analysis, the effects of retroviral transduction using ecotropic versus amphotropic vectors, various growth factor combinations, and adult bone marrow versus fetal liver stem cells were assessed. The results of these studies confirmed the fact that ecotropic vectors were much more efficient at transducing murine stem cells than amphotropic vectors, and that among the three most commonly used growth factors (stem cell factor [SCF] and interleukins 3 and 6 [IL-3 and IL-6]), SCF had the most significant effect on the transduction of stem cells, whereas IL-6 had the most significant effect on progenitor cells. In addition, it was determined that fetal liver stem cells were only approximately twofold more “transducible” than stem cells from adult bone marrow. Transplantation of Bodipy-selected bone marrow cells into lethally irradiated mice showed that the number of spleen colony-forming units that were positive for the retroviral vector (as determined by polymerase chain reaction) was 76%, as compared with 32% in animals that were transplanted with cells that were nonselected. The methods described within this manuscript are particularly useful for evaluating hematopoietic stem cell gene transfer in vivo because the marker gene used in the procedure (ASM) encodes a naturally occurring mammalian enzyme that has no known adverse effects, and the fluorescent compound used for selection (Bodipy sphingomyelin) is removed from the cells before transplantation.
HEMATOPOIETIC STEM cells (HSCs) are an important target for gene therapy.1-4 However, until now they have remained refractable to most gene transfer techniques because of their low numbers and lack of proliferation. Thus, before HSCs can be widely used in a clinical setting, new vectors must be developed and improved methods of HSC enrichment and transformation must be obtained. As these new gene transfer techniques are developed, ways to assess their usefulness will be needed.
Towards this end, we have developed a new method to quantitatively determine gene transfer and expression in hematopoietic stem and progenitor cells. The technique takes advantage of the fact that cells expressing a functional acid sphingomyelinase (ASM; sphingomyelin phosphodiesterase) can metabolize fluorescent sphingomyelin derivatives, whereas those that lack ASM activity (eg, from acid sphingomyelinase–deficient [ASMKO] mice5 or human patients with the genetic disorder Niemann-Pick disease [NPD]6) cannot. Thus, after labeling with fluorescent sphingomyelin, normal cells (expressing ASM) and NPD cells (lacking ASM activity) can be readily discriminated by fluorescence microscopy or flow cytometry.7 8 The same is true for NPD cells and such cells that have been enzymatically corrected by gene transfer.
This manuscript shows the utility of this method using hematopoietic stem and progenitor cells obtained from the bone marrow or fetal livers of ASMKO mice. The advantages of this technique include the facts that (1) the target cells do not need to be proliferating (such as HSCs), and any gene transfer system can be analyzed after inserting ASM into the vector; (2) it is fluorescence based and highly sensitive, permiting analysis of rare cell populations; and (3) it can be easily used for long-term in vivo analysis because it requires only 2 days to complete, does not involve extensive cell manipulation, and the transplanted cells do not express a foreign protein with potentially adverse effects.
MATERIALS AND METHODS
Cell preparations.
To obtain adult nucleated bone marrow cells, the tibia and femurs of 12- to 16-week-old C57BL/SV129 normal and ASMKO mice were flushed with buffered Hanks solution (10 mmol/L Hepes, pH 7.5). Single-cell suspensions were obtained by passing the cells through a 0.4-micron mesh (Becton Dickinson Labware, Franklin Lakes, NJ). Low-density bone marrow cells (<1.085 g/cm3) were isolated by discontinuous density gradient centrifugation using Nycoprep (Nycomed Pharma AS, Oslo, Norway),9 and washed with buffered Hanks solution containing 5% heat-inactivated fetal calf serum (HSA). To obtain fetal liver cells, the livers of day 14.5 fetuses were isolated in buffered Hanks solution. Single-cell suspensions were prepared by gently pipetting the tissue up and down through the bore of a 5-mL pipette. The cells were then washed with HSA.
Lineage depletion.
To obtain Lin− cells, a modification of the method of Bertoncello et al10 was used. Isolated adult bone marrow and fetal liver cells were incubated for 30 minutes on ice with the biotinylated antibodies anti-TER-119, anti-CD45R/B220, and anti-Ly-6G (Pharmingen, San Diego, CA). The concentration of each antibody was 1 μg/106 cells. The cells were than washed with HSA, resuspended in buffered Hanks solution, and incubated with streptavidin-coated magnetic beads (Dynabeads M-280 Streptavidin; Dynal, Lake Success, NY) at 4°C for 30 minutes at a 10:1 bead:cell ratio. Magnetic force was then applied for 1 minute and the supernatant was collected. The cell pellet was washed with buffered Hanks solution three times using the same procedure, and the supernatants were combined.
Anti–c-Kit labeling.
Cultured or freshly collected cells were washed once with HSA and counted. The cells were than incubated with phycoerythrin (PE)-conjugated anti-CD117 (c-Kit) antibodies (Pharmingen) at a concentration of 1 μg/106 cells for 30 minutes on ice. After labeling, the cells were washed once with HSA and resuspended in buffered Hanks solution. Control labeling was performed with a rat IgG2b, kappa isotype (Pharmingen).
Synthesis of fluorescent sphingomyelin.
Sphingomyelin to which the fluorescent probe Bodipy was covalently linked via a 12-carbon spacer (Bodipy dodecanoyl sphingosyl phosphocholine; B12SPM) was synthesized as previously described for lissamine rhodamine sphingomyelin,7 except that Bodipy dodecanoic acid (Molecular Probes Inc, Eugene, OR) was condensed with sphingosyl phosphocholine. To incorporate B12SPM into liposomes, B12SPM was mixed with phosphatidyl choline (PC; Sigma, St Louis, MO) at a molar ratio of 1:4. The solvent was evaporated and the mixture was resuspended in buffered Hanks solution followed by a 1-minute sonication.
“Pulse-chase” labeling with B12SPM.
B12SPM/PC liposomes (final concentration 0.5 to 1 nmol/mL) were incubated at 37°C for 4 hours with Lin− cells that had been suspended in buffered Hanks solution. Labeling was terminated by centrifuging the cells (400g) and washing the pellets once with HSA. Fresh medium was then added and the cells were further incubated for 48 hours in standard culture media containing Iscove’s Modified Dulbecco’s Medium (IMDM; GIBCO-BRL, Gaithersburg, MD), 10% HSA (GIBCO-BRL), and antibiotics, but no B12SPM/PC liposomes.
Retroviral transduction.
To achieve retroviral transduction, adult bone marrow and fetal liver cells were cocultured for 48 hours with amphotropic or ecotropic retroviral-producing cells containing an ASM/MFG retroviral vector.8 Control untreated cells were cocultured with producer cells alone. Cocultures were performed in 0.4-mm Transwell dishes (Corning Costar, Cambridge, MA) containing the packaging cells in the upper compartment. Stem cell factor (SCF; 50 ng/mL), interleukin-3 (IL-3; 20 ng/mL), and IL-6 (10 ng/mL) (Genzyme, Cambridge, MA) were added to the media unless indicated otherwise.
Fluorescence-activated cell sorter (FACS) analysis and expansion of sorted cells.
Cells were analyzed using a FACScan instrument (Becton Dickinson Immunocytometry Systems, San Jose, CA) and the WinMDI program. Cells were sorted using a FACStar flow cytometer (Becton Dickinson Immunocytometry Systems). Sorted cells were resuspended in expansion media containing IMDM, 10% HSA, SCF (50 ng/mL), IL-6 (10 ng/mL), IL-3 (20 ng/mL), and antibiotics, and then grown at 37°C for 10 days.
Polymerase chain reaction (PCR) analysis.
A modification of our previously described procedure was used.8 Murine and human ASM sequences were amplified using one common sense primer, 5′-TGCTGAGGATCGAGGAGACAA-3′ (P1) constructed from human and murine ASM exon 3, and two species-specific antisense primers, 5′-GGGTAGAGTGACAGAAGATTGA-3′ (P2) and 5′-GGCACAAGAGTAGCCAGACG-3′ (P3), constructed from murine ASM intron 3 and human ASM exon 6, respectively. Primer pair P1 and P2 amplified a 211-bp genomic murine ASM product, whereas primer pair P1 and P3 amplified a 554-bp product from the human ASM/MFG sequence. Each amplification reaction (100 μL final volume) contained 200 pmol of primer P1, 100 pmol each of primers P2 and P3, 300 ng of genomic DNA, 1× PCR buffer (Promega, Madison, WI), 1.5 mmol/L MgCl2, 5 U of Taq polymerase (Promega) and 200 mmol/L each of dNTPs. A standard curve was generated using DNA mixtures as described in Yeyati et al.8 After amplification (30 cycles, each consisting of 1 minute at 93°C, 1 minute at 61°C, 1 minute at 72°C), PCR products were electrophoresed on 1.5% agarose gels and stained with ethidium bromide. The intensity of the bands was determined using the National Institutes of Health Imager software package (NIH, Bethesda, MD). By comparing the intensity of the two amplified bands, the number of transduced cells in the sorted populations could be estimated. This calculation was based on the assumption that each transduced cell contained one proviral genome. It should be noted that the murine-specific ASM band is present in all samples and serves as an internal control, and that while the ASMKO mice have no ASM activity, the murine ASM genomic sequences are still present.
Enzyme assays.
Fresh or cultured adult bone marrow cells were obtained, washed once with HSA, and incubated on ice for 15 minutes in 0.2% Triton X-100. Total protein was determined by the method of Stein et al.11 The standard 15-μL ASM assay mixture consisted of 10 μL of protein source and 2 nmol of B12SPM suspended in 0.1 mol/L sodium acetate buffer, pH 5.2, containing 0.6% Triton X-100 and 5 mmol/L EDTA. After incubating the assay mixture at 37°C (up to 3 hours), the samples were loaded onto thin layer chromatography plates (TLC LK6 D Silica gel; Whatman, Clifton, NJ) and resolved using chloroform/methanol (95:5 vol/vol). After resolution, the band containing the fluorescently labeled ceramide (the product of B12SPM hydrolysis) was scraped from the plates, extracted in chloroform/methanol/water (1:2:1 vol/vol) for 15 minutes at 55°C, and quantified in a spectrofluorometer (fluorescence spectrophotometer 204-A, Perkin-Elmer, Norwalk, CT). The instrument settings were excitation 505 nm and emission 530 nm.
Spleen colony-forming unit (CFU-S) assays.
Adult nucleated bone marrow cells were obtained from ASMKO mice that had been pretreated with 5-fluorouracil (5-FU) (150 mg/Kg) 2 days before harvesting, retrovirally transduced with the ecotropic vector, and “pulse-chase”-labeled with B12SPM as described above, and then sorted by FACS. Cells representing the least 25% fluorescent in FL-1 (B12low) were collected and 4 × 104 were injected into the tail veins of lethally irradiated (800 cGy) adult ASMKO mice. For comparison, the same number of nonselected, transduced cells were injected into another set of animals. After 14 days the mice were killed, and the spleens were removed and then fixed in a 70% solution of formalin:acetic acid:ethanol (1:1:20 vol/vol/vol) for 3 days.
To prepare DNA from the CFU-S colonies, a modification of the method of Frank et al12 was used. The CFU-S colonies were separated and washed individually three times overnight in 1× Tris-EDTA (TE) (pH 8.2) at 4°C, and then minced and incubated overnight again at 37°C in a solution containing 50 mmol/L Tris (pH 8.2) and 200 ng/μL Proteinase K (Boehringer Mannheim, Mannheim, Germany). The microcentrifuge tubes containing the digested materials were then immersed in boiling water for 8 minutes, and the extracted DNA was placed on ice. For PCR, the DNA solutions were diluted 1:100 and 40 μL was used. The PCR was performed as described above except that only primers P1 and P3 were used and the number of cycles was 40. A positive colony was defined as a colony in which the human ASM (hASM) transgene-specific PCR product was found in at least three independent amplification reactions.
RESULTS
Identification of stem- and progenitor-enriched cell populations.
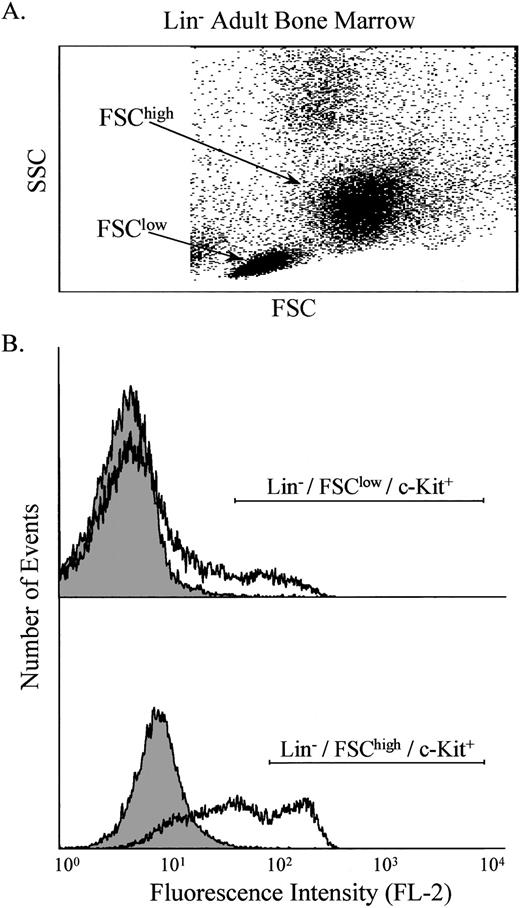
Bone marrow was obtained from normal and ASMKO adult mice and analyzed by FACS. After pre-enrichment by Nycodenz (Nycomed Pharma, Oslo, Norway) density gradient centrifugation and lineage depletion,9,10 two main populations of nucleated cells were identified, designated FSChigh and FSClow(Fig 1A). Labeling of these cells with antibodies against c-Kit showed that both groups contained c-Kit+ cells (Fig 1B), and that the number of lineage depleted (Lin−)/c-Kit+ cells was approximately 2% of the total nucleated cell population. c-Kit was used as a marker because previous work had shown that the Lin−/FSChigh/c-Kit+population is highly enriched for progenitor cells, whereas the Lin−/FSClow/c-Kit+ population is highly enriched for stem cells.13 14
Analysis of Lin− adult bone marrow cells. The light scatter properties of the cells were measured (A), and two populations were identified, designated FSClow and FSChigh. These populations were analyzed individually for the presence of c-Kit on the cell surface using PE-conjugated anti–c-Kit antibodies (B). The shaded area indicates control cells, whereas the open area indicates cells labeled with anti–c-Kit antibodies. Note that FL-2 measures PE.
Analysis of Lin− adult bone marrow cells. The light scatter properties of the cells were measured (A), and two populations were identified, designated FSClow and FSChigh. These populations were analyzed individually for the presence of c-Kit on the cell surface using PE-conjugated anti–c-Kit antibodies (B). The shaded area indicates control cells, whereas the open area indicates cells labeled with anti–c-Kit antibodies. Note that FL-2 measures PE.
“Pulse-chase” labeling of normal, ASMKO, and transduced cells.
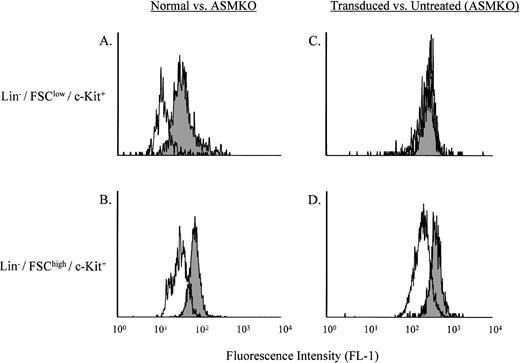
Lin− cells from normal or ASMKO mice were incubated for 4 hours with B12SPM, and then grown at 37°C for 48 hours in standard culture media without B12SPM. Analysis of the stem- and progenitor-enriched populations for B12 fluorescence is shown in Fig 2A and B. Comparison of the Bodipy fluorescence (FL-1) of normal and ASMKO adult bone marrow cells after labeling with B12SPM showed that in both the Lin−/FSClow/c-Kit+ (stem cell–enriched) and Lin−/FSChigh/c-Kit+(progenitor cell–enriched) populations, the normal cells were less fluorescent than those from ASMKO animals. These results showed that stem and progenitor cells could internalize B12SPM and that both types of normal cells expressed a functional ASM.
Analysis of B12SPM labeling of hematopoietic stem- and progenitor-enriched cell populations. (A) and (C) depict the Lin−/FSClow/c-Kit+ stem cell–enriched population, whereas (B) and (D) depict the Lin−/FSChigh/c-Kit+ progenitor cell–enriched population. (A) and (B) shaded area, ASMKO cells; open area, normal cells. (C) and (D) shaded area, nontransduced ASMKO cells; open area, transduced ASMKO cells. Note that FL-1 measures B12SPM. The experiment was repeated three times, and the representative data from one experiment are shown.
Analysis of B12SPM labeling of hematopoietic stem- and progenitor-enriched cell populations. (A) and (C) depict the Lin−/FSClow/c-Kit+ stem cell–enriched population, whereas (B) and (D) depict the Lin−/FSChigh/c-Kit+ progenitor cell–enriched population. (A) and (B) shaded area, ASMKO cells; open area, normal cells. (C) and (D) shaded area, nontransduced ASMKO cells; open area, transduced ASMKO cells. Note that FL-1 measures B12SPM. The experiment was repeated three times, and the representative data from one experiment are shown.
The same experiment was then repeated on ASMKO cells that had been retrovirally transduced with an amphotropic MFG retroviral vector expressing human ASM. The results of this experiment are shown in Fig2C and D. A significant shift to lower fluorescence was observed in the Lin−/FSChigh/c-Kit+ cells (progenitor cell–enriched), whereas only a very minor shift was observed in the Lin−/FSClow/c-Kit+ (stem cell–enriched) population. These results confirmed previous studies15 16 showing that murine progenitor cells (but not stem cells) could be efficiently transduced by amphotropic vectors.
PCR and enzyme analysis of sorted cells.
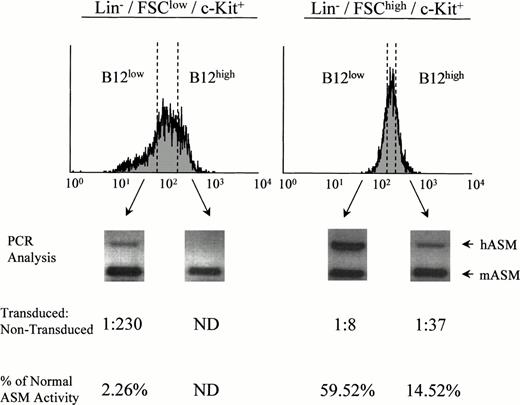
To further investigate this finding, retrovirally transduced Lin−/c-Kit+ ASMKO adult bone marrow cells were labeled with B12SPM and subjected to FACS sorting. Cells with the highest (B12high) and lowest (B12low) FL-1 were sorted from the stem- and progenitor-enriched groups (Lin−/FSClow/c-Kit+ and Lin−/FSChigh/c-Kit+, respectively), grown in expansion cultures, and subjected to semiquantitative PCR analysis and ASM activity assays (Fig 3). Note that within the sorted B12low group of Lin−/FSClow/c-Kit+ cells, a small number were transduced (1 in 230, based on the PCR assay), whereas in the B12low group of Lin−/FSChigh/c-Kit+ cells, significantly more were transduced (1 in 8). By comparison, without selection for FL-1, among the Lin−/FSClow/c-Kit+ cells no transduced cells could be identified, whereas among the Lin−/FSChigh/c-Kit+ cells the frequency of transduced cells was approximately 1 in 20 (not shown). These data confirmed the fact that the stem cell–enriched population from adult bone marrow was very difficult to transduce with amphotropic vectors compared with the progenitor-enriched population, but also showed that the Bodipy selection technique provided a significant enrichment of transduced cells. Indeed, by using this method, transduced cells from both groups could be identified and collected.
Sorting and analysis of retrovirally transduced ASMKO bone marrow cells. PCR analysis and ASM enzyme assays were conducted in triplicate on sorted B12low and B12high cells from the Lin−/FSClow/c-Kit+population (left; stem cell–enriched) and Lin−/FSChigh/c-Kit+ population (right; progenitor cell–enriched) as described in the text. ND, not detected. The experiment was repeated three times, and the representative data from one experiment are shown.
Sorting and analysis of retrovirally transduced ASMKO bone marrow cells. PCR analysis and ASM enzyme assays were conducted in triplicate on sorted B12low and B12high cells from the Lin−/FSClow/c-Kit+population (left; stem cell–enriched) and Lin−/FSChigh/c-Kit+ population (right; progenitor cell–enriched) as described in the text. ND, not detected. The experiment was repeated three times, and the representative data from one experiment are shown.
Analysis of fetal liver stem and progenitor cells.
We next used this system to assess the transduction of fetal liver c-Kit+ cells by the amphotropic vector (Fig 4). Lin− fetal liver cells were isolated, labeled with PE-conjugated anti–c-Kit antibodies, and analyzed on FACS. The light scatter properties of the cells were measured, and in contrast to adult bone marrow, only one population of Lin− cells was identified. This population was analyzed for the presence of the c-Kit molecule (FL-2) on the cell surface, and it was determined that the number of c-Kit+cells was approximately 5% of the total nucleated fetal liver cells. The Lin− ASMKO fetal liver cells were either retrovirally transduced with the ASM/MFG amphotropic vector or untreated, labeled with B12SPM, and the FL-1 of the two groups was compared. Only a small shift to lower fluorescence was observed in the retrovirally transduced population, corresponding to a low transduction efficiency of approximately 1 in 50 to 1 in 100 cells.
Analysis of retroviraly transduced Lin−fetal liver cells. The light scatter properties of the cells were measured (A), and the major population was identified and gated for further analysis (shown in box). This population was then analyzed for the presence of c-Kit on the cell surface ([B] shaded area, control cells; open area; cells labeled with anti–c-Kit antibodies). Note that FL-2 measures PE. The c-Kit+ cells were further analyzed for B12 fluorescence ([C] shaded area, control cells; open area, transduced cells). Note that FL-1 measures B12SPM. The experiment was repeated twice, and the representative data from one experiment are shown.
Analysis of retroviraly transduced Lin−fetal liver cells. The light scatter properties of the cells were measured (A), and the major population was identified and gated for further analysis (shown in box). This population was then analyzed for the presence of c-Kit on the cell surface ([B] shaded area, control cells; open area; cells labeled with anti–c-Kit antibodies). Note that FL-2 measures PE. The c-Kit+ cells were further analyzed for B12 fluorescence ([C] shaded area, control cells; open area, transduced cells). Note that FL-1 measures B12SPM. The experiment was repeated twice, and the representative data from one experiment are shown.
Comparison of the retroviral transduction efficiencies using amphotropic versus ecotropic viruses.
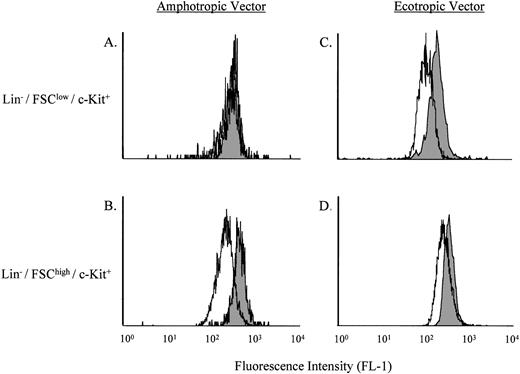
Because the transduction of stem cell–enriched bone marrow or fetal liver cells using the amphotropic vector was low, we next sought to determine if this could be improved using an ecotropic vector. Several recent reports have shown increased transduction of murine hematopoietic stem cells using ecotropic versus amphotrophic retroviral vectors.15 16 As shown in Fig5, by using the B12SPM selection technique the transduction efficiencies of Lin−/FSClow/c-Kit+ stem cells with the amphotropic versus ecotropic ASM/MFG vectors could be directly compared. Of note, the ecotropic vector led to a more than 50-fold increase in transduction over that found with the amphotropic vector.
Comparison of the transduction efficiencies using amphotropic versus ecotropic retroviral vectors. (A) and (C) depict the Lin−/FSClow/c-Kit+ stem cell–enriched population, whereas (B) and (D) depict the Lin−/FSChigh/c-Kit+ progenitor cell–enriched population. The shaded areas represent nontransduced ASMKO cells, whereas the open areas depict transduced cells. Note that FL-1 measures B12SPM. The experiment was repeated three times, and the representative data from one experiment are shown.
Comparison of the transduction efficiencies using amphotropic versus ecotropic retroviral vectors. (A) and (C) depict the Lin−/FSClow/c-Kit+ stem cell–enriched population, whereas (B) and (D) depict the Lin−/FSChigh/c-Kit+ progenitor cell–enriched population. The shaded areas represent nontransduced ASMKO cells, whereas the open areas depict transduced cells. Note that FL-1 measures B12SPM. The experiment was repeated three times, and the representative data from one experiment are shown.
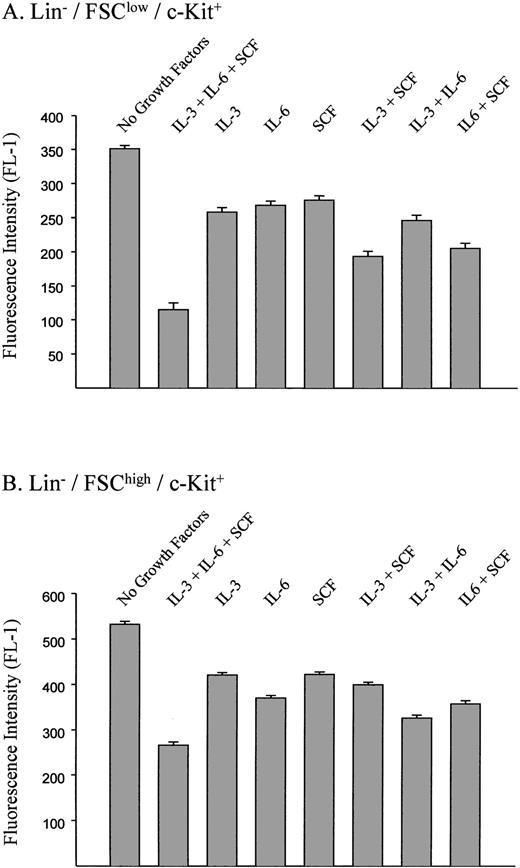
Effects of individual growth factors on the transduction of murine stem and progenitor cells.
To evaluate the specific contribution of growth factors to the transduction of hematopoietic cells, various combinations of SCF, IL-3, and IL-6 were used during the retroviral transduction of B12SPM-labeled cells with the ecotropic vector (Fig 6). This was followed by FACS analysis to determine the B12 fluorescence of the cells. In the Lin−/FSClow/c-Kit+ population (stem cell–enriched), SCF had the biggest effect on the transduction efficiency, whereas among the Lin−/FSChigh/c-Kit+population (progenitor-enriched), the largest contribution was provided by IL-6. In both groups of cells, the combination of all three growth factors led to the highest transduction efficiencies.
Effects of growth factors on transduction efficiences using ecotropic vectors. The standard retroviral transduction methods and concentrations of each growth factor are described in the Materials and Methods. FL-1 measures B12SPM fluorescence. The bars indicate ±1 standard deviation using data derived from three independent experiments.
Effects of growth factors on transduction efficiences using ecotropic vectors. The standard retroviral transduction methods and concentrations of each growth factor are described in the Materials and Methods. FL-1 measures B12SPM fluorescence. The bars indicate ±1 standard deviation using data derived from three independent experiments.
CFU-S analysis of sorted cells.
To ensure that cells selected with these techniques were enriched for hematopoietic stem cells, ASMKO bone marrow cells were transduced with the ecotropic ASM/MFG vector in the prescence of SCF, IL-3, and IL-6; B12low cells were collected by flow cytometry; and then transplanted into lethally irradiated adult ASMKO mice. Fourteen days later the spleens were obtained and the number of CFUs-S positive for the retroviral vector was determined by PCR. As shown in Table 1, in the absence of selection, 32% of the CFU-S colonies were vector positive, whereas among animals transplanted with B12SPM-selected cells, 76% of the colonies were vector positive. Thus, these results showed that a significant enrichment of transduced repopulating stem cells can be obtained using this procedure.
PCR Analysis of CFU-S Colonies
| Cell Type Injected . | Total Colonies Analyzed . | PCR-Positive Colonies . |
|---|---|---|
| Bodipy-selected | 37 | 28 (76%) |
| Nonselected | 28 | 9 (32%) |
| Cell Type Injected . | Total Colonies Analyzed . | PCR-Positive Colonies . |
|---|---|---|
| Bodipy-selected | 37 | 28 (76%) |
| Nonselected | 28 | 9 (32%) |
CFU-S assays were performed as described in Materials and Methods. The number of total and PCR-positive CFU-S colonies found after injection of either Bodipy-selected (25% least fluorescent) or nonselected transduced cells are shown. The cells selected for lower Bodipy fluorescence produced a significantly higher number of PCR-positive colonies than cells that were nonselected (χ2 = 10.607; P = .001).
DISCUSSION
A novel, fluorescence-based method is presented for the analysis of gene transfer into hematopoietic stem and progenitor cells. In our view there are two applications of this technology, one specific for the treatment of NPD, and the other more general. With regards to NPD, the data clearly show that by using these techniques, enriched populations of retrovirally transduced NPD hematopoietic stem and progenitor cells can be identified and collected for subsequent transplantation into patients. Such cells are currently being transplanted into ASMKO mice so that their engraftment potential and clinical usefulness can be compared with transduced hematopoietic cells that have not undergone the selection procedure. It is hypothesized that in a competitive repopulation setting, particularly in young animals that have not been lethally ablated, such selected cells may be clinically advantageous because the likelihood of engrafting a transduced, long-term repopulating stem cell will be greater due to the fluorescent enrichment procedure. Indeed, the results of the CFU-S experiment suggest that this may be the case. If successful, similar methods can be applied to CD34+ cells obtained from human NPD patients. Although obtaining sufficient numbers of such B12low/CD34+ cells for human transplantation may be problematic at the present time, the development of high-speed flow cytometers17-19 promises to overcome this limitation.
We also propose that this simple analytical system can be broadly used in a variety of gene therapy and basic research settings. The target cells do not need to be proliferating to take up B12SPM (S.E. and E.H.S., unpublished data), making these techniques amenable to HSCs and other quiescent cells. We have shown the usefulness of this method by evaluating transduction with retroviral vectors; however, other gene transfer systems can be easily studied by inserting the ASM cDNA as a marker. This would include viral- or nonviral-based systems. Importantly, the system relies on expression of the transferred gene as its final endpoint, and is fluorescence based, making it highly sensitive. Also of note, the corrected cells can be isolated from the noncorrected ones using a FACS, even when they represent 1% or less of the total cell population, and the properties of the two groups can be directly compared in vitro or in vivo, as long as ASMKO mice are used as a source of target cells.
Several findings were presented in this paper regarding the transduction of murine hematopoietic stem and progenitor cells with retroviral vectors to show the usefulness of these techniques. First, we confirmed the fact that amphotropic vectors could not transduce murine hematopoietic stem cells efficiently as compared with ecotropic vectors.15,16 Indeed, the transduction frequency of the stem cell–enriched population was increased more than 50-fold using ecotropic versus amphotropic vectors. Of note, we also found that among the three most commonly used growth factors, SCF contributed the most to stem cell transduction, whereas IL-6 contributed most to progenitor cell transduction. Again, these results confirmed previous reports20 and showed the usefulness of these techniques. Finally, we documented that c-Kit+ fetal cells, which are approximately twofold more prevelant than c-Kit+ bone marrow cells, were transduced with amphotropic vectors at an intermediate frequency (approximately 1 in 100 in the Bodipy-selected populations) when compared with bone marrow stem or progenitor cells.
Clearly, new vector systems, such as the Lentivirus-based retroviral vectors21,22 as well as new growth factor cocktails and packaging cell lines,23 24 can be readily analyzed by these methods. Before the development of these and similar fluorescence-based techniques (see below), data such as these could only be obtained by transduction with vectors containing marker genes such as those encoding neomycin resistance or β-galactosidase activity, followed by labor-intensive and indirect colony-forming assays. Direct isolation of the transduced cells would have been impossible.
Several alternative fluorescence-based selection systems have been developed in recent years to enrich for transduced hematopoietic stem cells.25-27 Among these, the use of the green fluorescence protein (GFP) has attracted considerable interest.28However, one important difference between our system and those which use GFP is that in our system the fluorescent marker (B12SPM) is not encoded by the gene transfer vector. Identification of the transduced cells relies on enzymatic expression of ASM, leading to degradation of the fluorescent molecule and a subsequent loss of fluorescence. Indeed, the target cells are only exposed to B12SPM for a short period of time, resulting in little or no toxicity. In the GFP system, the GFP gene is inserted into the vector of interest and the cells are selected for an increase in fluorescence. Stably transduced cells will continue to produce GFP and remain fluorescent, possibly leading to increased toxicity or abnormal metabolism if the transduced cells are followed for long periods of time in vivo.
Thus, we believe that the selection approach described in this manuscript will be of interest to a wide range of scientists, and is particularly advantageous for the development of hematopoietic stem cell gene therapy for NPD and the analysis of HSC gene transfer in general.
ACKNOWLEDGMENT
We thank Dr Kevin Kelly for his expert assistance in the preparation of the fetal liver cells.
Supported by research grants from the National Institutes of Health (HD 28607 and HD 32654), March of Dimes Birth Defects Foundation (1-2224), a grant (RR 0071) from the National Center for Research Resources for the Mount Sinai General Clinical Research Center, and a grant (93-00015) from the US-Israel Binational Science Foundation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Edward Schuchman, PhD, Department of Human Genetics, Box 1498, Mount Sinai School of Medicine, 1425 Madison Ave, Room 14-20A, New York, NY 10029; e-mail: Schuchman@msvax.mssm.edu.

![Fig. 4. Analysis of retroviraly transduced Lin−fetal liver cells. The light scatter properties of the cells were measured (A), and the major population was identified and gated for further analysis (shown in box). This population was then analyzed for the presence of c-Kit on the cell surface ([B] shaded area, control cells; open area; cells labeled with anti–c-Kit antibodies). Note that FL-2 measures PE. The c-Kit+ cells were further analyzed for B12 fluorescence ([C] shaded area, control cells; open area, transduced cells). Note that FL-1 measures B12SPM. The experiment was repeated twice, and the representative data from one experiment are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/1/10.1182_blood.v93.1.80/5/m_blod40128004x.jpeg?Expires=1769122166&Signature=L8nz4Emi7FHl3o0Z73HRdgJY70aqERcXO~9-evI1a5Cv4bLLVrCEA85Z~uIbUh7iOCO~VTsTysisdSXz9b5OwS8XGO5iQacYLflkbVqYodg8Y-A35kPoQon2wHW3L7VdXJ9vZnpQ9faQ2qlMPrhc3Mes5AFtjRmAC3y5RTuqiB0fC7kZGMLOA3BaXHEMXWeBjdD0KejHOOG5RgcTyXlPKzgMwrTIRR0ca-Hp2hjKaF-X0alMRcM4b~RS6CXUGFwtgpFOQZQxmZMArBO~T7qY7Dph6QfEwKA4PI-hI1BM3gdvn7rRKOoD3mgPrKKdB8e5XIxyMpi7XHYuo~aGX5Zxuw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal