Abstract
Induction of urokinase plasminogen activator (uPA) by retinoic acid (RA) is the initial event preceding certain subsequent biological changes in vascular endothelial cells. We investigated the molecular mechanism by which RA stimulates the expression of uPA, which lacks a canonical RA receptor (RAR)-responsive element, in bovine and human aortic endothelial cells. Upon stimulation with RA, mRNA levels of RAR and β transiently increased in parallel with the induction of uPA, and this increase was inhibited by cycloheximide. Results of transient transfection of RAR/RXR cDNAs and experiments using specific agonists and antagonists suggested that uPA induction is dependent upon RAR (initially, RAR) with the help of RXR. Deletion analysis of the uPA promoter suggested that RAR/RXR acts on GC box region within the uPA promoter. This was further supported by inhibition of Sp1 binding to this region. Coimmunoprecipitation studies, glutathioneS-transferase pull-down experiment, and mammalian two-hybrid assays suggested a physical interaction between RAR/RXR and Sp1. Furthermore, gel shift studies showed that the binding of Sp1 to the uPA GC box is significantly potentiated in the presence of RARs/RXRs. Finally, Sp1 and RAR/RXR synergistically enhanced the transactivation activity of the uPA promoter. These results suggest that (1) RA induces RARs mainly via RAR and that (2) RAR/RXR physically and functionally interact with Sp1, resulting in a potentiation of uPA transcription.
RETINOL (VITAMIN A) and its derivatives (retinoids) exert profound effects on the regulation of cell growth and differentiation, mainly through two families of nuclear receptors, the retinoic acid (RA) receptors (RARs) and the retinoid X receptors (RXRs).1,2 These receptors are ligand-dependent transcription factors that bind to cis-acting DNA sequences, called RAR-responsive elements (RAREs) and RXR-responsive elements (RXREs), located in the promoter region of their target genes.1-3 The RAR and RXR gene families are each composed of three subtypes, named α, β, and γ. Expression of RARs increases in an autocrine manner immediately after stimulation with RA, because these receptors contain typical RARE sequences.1-3RARs bind to the RARE in response to both all-trans-RA (atRA) and 9-cis-RA (9cRA), whereas RXRs bind and activate transcription in response to only 9cRA. To recognize an RARE, RARs usually must form a heterodimer with RXRs under physiological condition.1,2 Recent studies have indicated that RARs/RXRs, upon binding to ligands, promote transcription through interaction with coactivators such as SRC-1, GRIP1, p/CIP, and p300/CBP after dissociation from corepressors such as NCoR and SMRT.4-7However, not all RA-inducible genes contain RARE sequence(s) within their promoter.
In many cell types, RA enhances the production of plasminogen activator (PA), an enzyme responsible for the conversion of plasminogen to plasmin.8,9 There are two immunologically distinct PAs, urokinase PA (uPA) and tissue PA (tPA). uPA is thought to be related to tissue remodeling and metastasis, whereas tPA has high affinity for fibrin and mainly participates in vascular thrombolysis.8,9In bovine aortic endothelial cells (BAECs) and rat liver stellate cells, RA enhances cellular fibrinolytic levels through rapid stimulation of the expression of uPA and related genes, resulting in the induction of active transforming growth factor-β (TGF-β), that subsequently mediates some of the actions of RA in these cells.10-15
These sequential changes, induced in BAECs by RA, are initiated by upregulation of the transcription of the uPA gene, which does not contain a canonical RARE sequence. In the present study, we investigated the molecular mechanism of this initial event and found that (1) RA induces RARs expression initially via RARα and that (2) RARs/RXRs directly interact with Sp1, culminating in the augmented binding of Sp1 to the uPA GC box and in the enhanced transcription of the uPA gene in endothelial cells.
MATERIALS AND METHODS
Materials.
Actinomycin D, cycloheximide, atRA, and 9cRA were purchased from Sigma Chemical Co (St Louis, MO). Mithramycin was obtained from Calbiochem (La Jolla, CA). The RARα-selective retinoid (Am580), pan-RAR-selective retinoid (Ch55), and RARβ-selective antagonist (LE135) were synthesized and characterized as described previously.16-18 The RARβ-selective retinoid (CD2019) and RARγ-selective retinoid (CD666) were kindly provided by Dr S. Michel (CIRD/GALDERMA, Sophia Antipolis, France).19,20 The RXRα-selective retinoid (Ro25-7386), pan-RXR-selective retinoid (Ro47-5944), and RARα-selective antagonist (Ro41-5253) were generous gifts from Dr M. Klaus (F. Hoffmann-La Roche, Basel, Switzerland).21-23 Retinoids were diluted in ethanol and serially diluted into culture medium to yield a final ethanol concentration of 0.5%. Human Sp1 was obtained from Promega (Madison, WI). RARs-glutathione S-transferase (GST) and RXRs-GST fusion proteins were purified from Escherichia coliBL21 transformed with the pGEX-6P (Pharmacia Biotech, Uppsala, Sweden) vector containing inserted human RAR and murine RXR cDNAs, respectively. Briefly, after induction by 2 mmol/L isopropyl β-D-thiogalactopyranoside (IPTG) for 4 hours, the cells were disrupted by sonication and the fusion proteins were purified using glutathione-Sepharose (Pharmacia) from cell homogenates. The glutathione used to elute the proteins was removed from protein fractions by dialysis before using them for the gel shift assays or immunoprecipitation studies. cDNAs for human RARs and murine RXRs were generous gifts from Dr P. Chambon (INSERM, University Louis Pasteur, Strasbourg, France).24-27
Treatment of cells with retinoids.
BAECs were isolated and grown in α minimal essential medium (αMEM) containing 10% newborn calf serum (Hyclone Laboratories, Logan, UT). Human aortic endothelial cells were purchased from Kurabo Biomedical Business (Osaka, Japan) and maintained in attached Humedia-EG2 medium. After the cells were grown to confluence, the cultures were rinsed with phosphate-buffered saline (PBS), pH 7.4, and incubated in either serum-free αMEM containing 0.1% bovine serum albumin (αMEM-BSA) or Humedia-EG2 medium containing 50 μmol/L L-ascorbic acid (Sigma), respectively, plus either 0.5% ethanol or various concentrations of retinoids. At the indicated time, the medium was aspirated, the cultures were washed with PBS, and (1) lysed with guanidium isothiocyanate solution and stored at −80°C until isolation of RNA, (2) lysed with 0.5% Triton X-100 in 0.1 mol/L Tris-HCl, pH 8.1, and stored at −20°C until assay of cellular PA levels, or (3) lysed with 0.6% NP-40 solution and stored at −80°C until the preparation of nuclear extracts.
Isolation of RNA and Northern blot analysis.
Total RNA was extracted from the cells using the acid guanidinium isothiocyanate-phenol-chloroform extraction method.28 Each RNA (20 μg) was separated through 1% agarose-formaldehyde gel electrophoresis and transferred to the Biodyne A nylon membranes (Pall Biosupport, Port Washington, NY) according to the published protocols.29 Membranes were hybridized with cDNA probes for either human RARα, RARβ, or RARγ; mouse RXRα, RXRβ, or RXRγ; or bovine uPA (a gift from Dr Wolf-Dieter Schleuning, Research Laboratories of Schering AG, Berlin, Germany)14 and were rehybridized with a probe for chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The cDNA probes were labeled with [α-32P]dCTP (Du Pont, Wilmington, DE) via random priming using the kit (Boehringer Mannheim, Mannheim, Germany). After hybridization for 1 hour at 68°C using QuickHyb reagent (Stratagene, La Jolla, CA), blots were washed twice with 2× SSC, 0.1% sodium dodecyl sulfate (SDS) at room temperature for 15 minutes. The final wash was performed with 0.1× SSC, 0.1% SDS at 65°C for 30 minutes. For rehybridizations, blots were washed twice in 0.1× SSC, 0.1% SDS at 97°C for 15 minutes and stripped. Autoradiography was performed using a Fujix BAS 2,000 Bio-imaging analyzer (Fuji Photo-Film, Tokyo, Japan). Each band was scanned, and the signal intensity was normalized to that obtained with GAPDH.
Assay of cellular PA activity.
The levels of cellular PA activity were measured using the chromogenic substrate S-2403, as described previously,11 and expressed as urokinase units per milligram of protein in the sample. Protein concentration was measured by BCA (Pierce, Rockford, IL) assay using BSA as the standard.
Transient transfection and luciferase assay.
BAECs were seeded and grown in 35-mm dishes using phenol-red free αMEM supplemented with 10% newborn calf serum until approximately 70% confluence on the day of the transfection. The cultures were rinsed with PBS and the cells were cotransfected using Lipofectamine Plus reagent (GIBCO BRL Life Technologies, Inc, Rockville, MD) in 1 mL of serum-free medium containing a reporter plasmid plus RAR and/or RXR expressing vectors (250 ng each/dish), along with pRL-CMV (Renilla luciferase, 100 ng/dish; Promega) as an internal standard to normalize transfection efficiency. Total DNA transfected was always adjusted to 1.225 μg/dish with empty pSG5 vector. After 4 hours of incubation, the cultures were overlaid with an additional 1 mL of medium containing 20% newborn calf serum and further incubated for 24 hours. The cells were then washed with PBS and treated or untreated with 1 μmol/L atRA or 9cRA for an additional 12 hours in serum-free medium. Thereafter, cell lysates were prepared by scraping cells into 120 μL of lysis buffer, and the luciferase activity of each cell lysate was measured using the Dual-Luciferase Reporter Assay System (Promega). Changes in firefly luciferase activity were calculated and plotted after normalization with changes in Renilla luciferase activity in the same sample. The uPA promoter-luciferase expressing vector, pGL2-2350, contains the human uPA promoter (−2345 to +32)30 cloned into the Kpn I/Bgl II site of pGL2-Basic (Promega). Its deletion mutant, pGL2-GC, containing the first 65 bp and GC box and TATA box region (−2345 to −2280 and −91 to +32, respectively) was constructed by digestion of pGL2-2350 with Tth111I and Blp I, followed by blunting and religation. Another deletion mutant, pGL2-ΔGC, which contains uPA promoter devoid of the GC box region, was constructed by ligation of the pGL2 vector containing the TATA box (−28 to +32) with the uPA promoter fragment deficient in the GC and TATA boxes (−2345 to −71). The TATA box-containing vector was prepared by digestion of pGL2-GC with Apa I, blunt-ending of the DNA, and successive digestion with Kpn I. The uPA promoter fragment deficient in the GC and TATA boxes was the Kpn I-fragment of a polymerase chain reaction (PCR) product that used pGL2-2350 as template. The following primers were used to generate this product: sense primer (based on the vector sequence located upstream of the cloning site in pGL2 vector), CTTCCCTTCCTTTCTCG CCAC; and antisense primer (based on a gene-specific sequence corresponding to −71 to −91 of the uPA promoter), TCCCCTGTCTTGCAGCGCTCA. The DR5-luciferase reporter plasmid, pGL3-DR5, was constructed by insertion of DR5 globin promoter fragment (a gift from Dr S. Kato, Tokyo University, Tokyo, Japan)31 into the Kpn I/Bgl II site of pGL3-Basic (Promega). Sp1-Gal4 expressing vector, which contains Sp1 fused with DNA binding domain of yeast Gal4 protein, and its reporter plasmid Gal4-UAS fused to luciferase, were kindly supplied from Dr S.L. Friedman (Mount Sinai Medical Center, New York, NY).32,33 The Sp1 expression vector, pCIneo-Sp1, was constructed as follows. Human Sp1 cDNA, obtained from pSp1-778C (a gift from Dr J.T. Kadonaga, UCSD, San Diego, CA),34 was subcloned first into NotI/HindIII site of pPROEX-HTb vector (GIBCO BRL), then intoSpe I/HindII site of pBluescript SK II (+) (Stratagene), and finally into the Xba I/Acc I site of pCIneo mammalian expression vector (Promega).
Preparation of nuclear extracts and gel shift assay.
Nuclear extracts were prepared in the following manner. Cells were detached from dishes, washed with Tris-buffered saline, and resuspended in ice-cold 10 mmol/L HEPES, pH 7.9, containing 10 mmol/L KCl, 0.1 mmol/L EDTA, 0.1 mmol/L EGTA, 1 mmol/L dithiothreitol, and 0.5 mmol/L phenylmethylsulfonyl fluoride. After being incubated on ice for 15 minutes, cells were ruptured in the presence of 0.6% Nonidet NP-40, and the nuclear fraction was collected by centrifugation at 15,000g for 30 seconds at 4°C. Nuclei were lysed by shearing through a 27-gauge needle in ice-cold 50 mmol/L Tris-HCl, pH 8, containing 150 mmol/L NaCl, 5 mmol/L 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS; Sigma), and the Complete protease inhibitor cocktail (Boehringer Mannheim), and nuclear extracts were obtained as supernatants after centrifugation at 105,000g for 1 hour at 4°C. The protein concentrations were determined by BCA assays. Oligonucleotides corresponding to uPA GC box (−63 to −32), its mutant (sequences presented in Fig 4B), and consensus GC box (ATTCGATCGGGGCGGGGCGAGC; Santa Cruz Biotechnology, Santa Cruz, CA) were synthesized, double-stranded, and end-labeled with [γ-32P]ATP by T4 polynucleotide kinase using the kit from Boehringer Mannheim. For binding reactions, either 1 μg of nuclear proteins or 10 to 20 ng of purified Sp1 (Promega) were preincubated with or without 200 ng of purified RAR-GST and/or RXR-GST for 15 minutes on ice in the absence and presence of unlabeled oligonucleotides, and then 40 fmol of labeled oligonucleotide (10,000 μCi/mol) was added in the presence of 1 μg of dI-dC (Pharmacia) in 20 to 40 μL of binding buffer (20 mmol/L HEPES, pH 7.4, containing 1 mmol/L MgCl2, 10 μmol/L ZnSO4, 20 mmol/L KCl, and 8% glycerol). The reaction mixture was incubated for 15 minutes on ice and separated on a 4% polyacrylamide gel at 4°C. The gel was dried and exposed on films for a Fujix BAS 2,000 Bio-imaging analyzer (Fuji Photo-Film).
Immunoprecipitation and Western blotting.
After a preincubation of 100 ng of Sp1 with 200 ng of each of RARs-and/or RXRs-GST proteins in PBS or in nuclear extract for 2 hours on ice, samples were combined with either anti-RARγ antibody, M-454 (final concentration, 10 μg/mL; Santa Cruz Biotechnology), which uniformly recognizes three subtypes of RARs, or anti-RXRα antibody, ΔN 197 (final concentration, 10 μg/mL; Santa Cruz Biotechnology), which uniformly recognizes three subtypes of RXRs, and were incubated for 2 hours at 4°C or overnight on a rotating mixer. Protein A-Sepharose beads (Pharmacia) were then added and the samples were incubated for another 2 hours at the same temperature. The beads were then washed three times with PBS containing 0.05% Tween 20, and immunoprecipitated proteins were eluted into 4× reducing sample buffer for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to electrophoresis. Western blotting was performed using rabbit polyclonal anti-Sp1 antibody, PEP2 (final concentration, 1 μg/mL; Santa Cruz Biotechnology) as described previously,11 except that peroxidase-conjugated protein A (1/2,000 dilution; Sigma) was used instead of peroxidase-conjugated second antibody. Protein bands were visualized using the Amersham ECL system (Amersham, Buckinghamshire, UK).
GST fusion protein interaction assay.
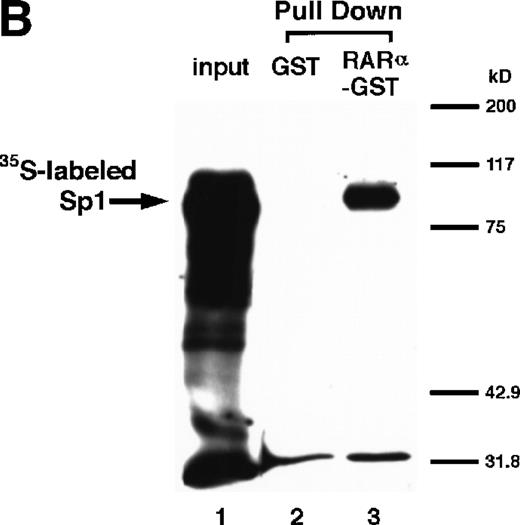
The [35S]methionine-radiolabeled Sp1 protein was prepared by in vitro translation reaction using the TNT T7 system (Promega) according to the manufacturer’s instructions. GST fusion proteins were mixed with glutathione-Sepharose 4B beads (Pharmacia) for 2 hours at 4°C in binding buffer (50 mmol/L Tris-HCl, pH 7.6, containing 50 mmol/L NaCl, 0.02% Tween 20, 0.02% BSA, and the Complete protease inhibitor cocktail). The glutathione beads, preincubated with RARα-GST, were washed three times with binding buffer and further incubated for 30 minutes at 4°C with [35S]methionine-radiolabeled Sp1 present in 15 μL of the reaction mixture after in vitro translation reaction, in a final volume of 300 μL with the binding buffer. After washing five times with the washing buffer (50 mmol/L Tris-HCl, pH 7.6, containing 150 mmol/L NaCl, 0.02% Tween 20, and the Complete protease inhibitor cocktail), the bound proteins were eluted into 4× reducing sample buffer for SDS-PAGE and subjected to electrophoresis followed by fluorography.
Statistics.
Each number in the figures represents the average ± SD (n = 3).
RESULTS
Induction of uPA through RAR.
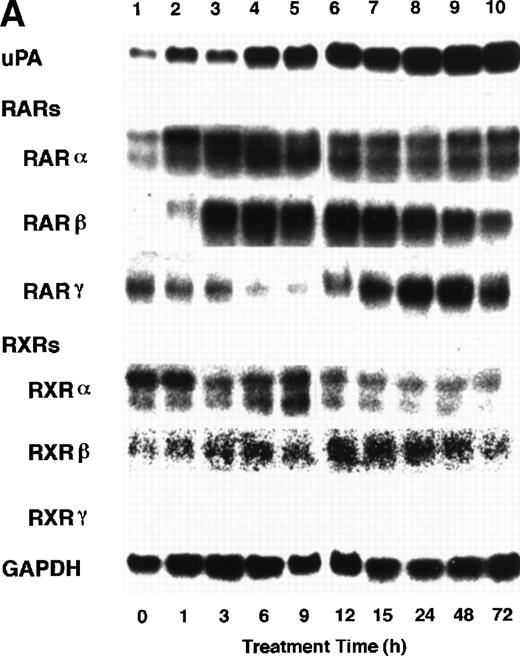
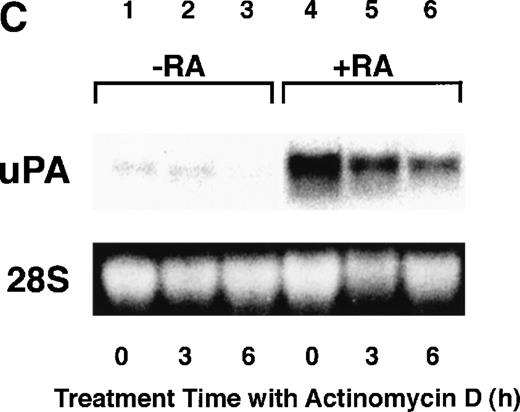
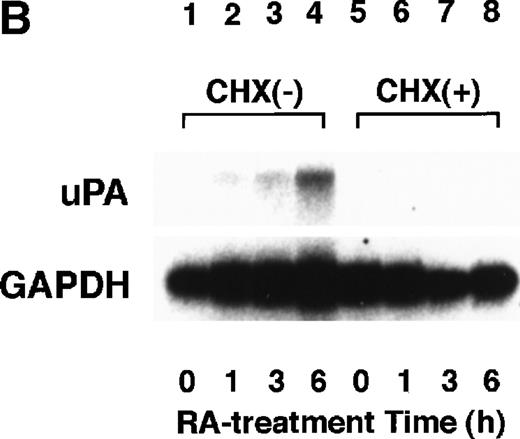
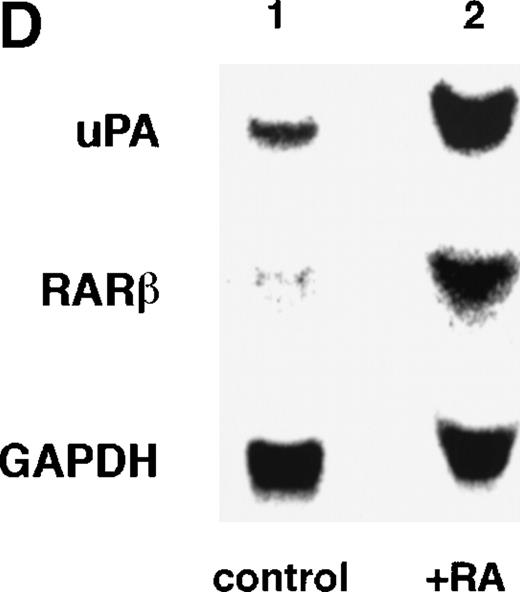
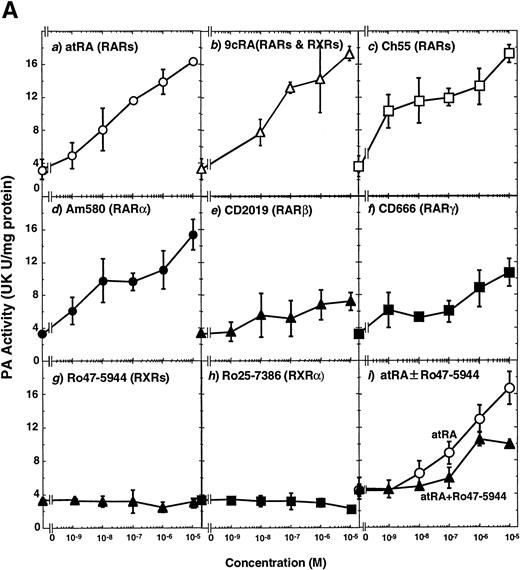
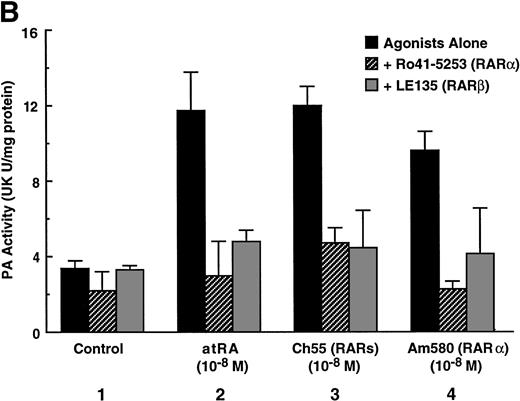
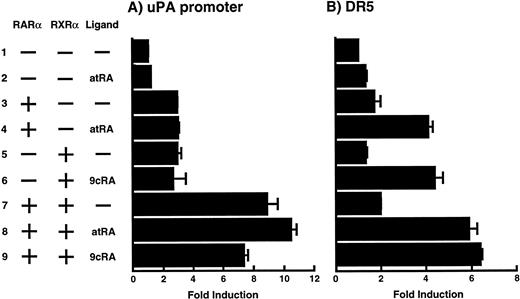
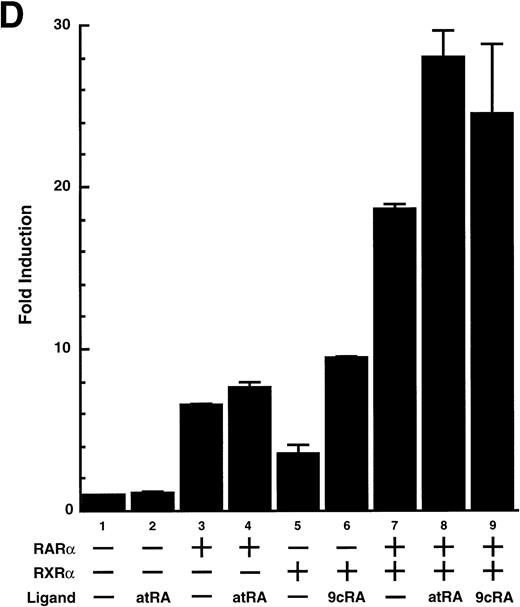
To analyze the molecular mechanism underlying the RA regulation of uPA transcription in endothelial cells, we first investigated whether RARs and/or RXRs might be involved. As seen in Fig 1A, upon stimulation with atRA, the expression of RARα increased significantly along with a strong and transient induction of RARβ, consistent with previous findings in other cell types.1,3,10,35 Although RARγ decreased until 9 hours, the sum of the three RAR isoforms increased. In contrast, such a distinctive increase was not observed for RXRs, of which RXRα was the major species. The induction of uPA concurred with the increase of RARα and RARβ, whereas this induction did not occur when protein synthesis was inhibited by cycloheximide (Fig 1B). RA stimulation did not alter the degradation rate of uPA mRNA after actinomycin D treatment, suggesting that RA did not significantly stabilize uPA mRNA (Fig 1C). A similar induction of uPA and RAR was observed in human aortic endothelial cells (Fig 1D). These results imply that the induction of uPA mRNA by RA is due to upregulation of uPA gene expression, which is dependent on new protein synthesis, and might be promoted by RARs, initially in particular by RARα. To address the role of RARs, we examined the effect of subtype-specific agonists and antagonists (Fig 2). The agonists and antagonists used here have different receptor selectivities and can bind to and transactivate either RARs or RXRs or one of receptor subtypes (eg, α, β, or γ).16-23 Agonists capable of binding to and activating RARα enhanced cellular PA levels almost identically (Fig 2A, panels a through d), whereas RARβ- or RARγ-selective agonists showed a much weaker effect (Fig 2A, panels e and f, respectively). RXR-selective agonists administered alone were not effective (Fig 2A, panels g and h) and weakly inhibited uPA induction when combined with atRA (Fig 2A, panel i). The enhancement of cellular PA levels by atRA, Ch55, and Am580 was abolished by both the RARα- and RARβ-selective antagonists (Fig 2B). The specificities of the agonists and antagonists to discriminate RARs and RXRs from each other are strict, whereas those to discriminate each receptor subtype are relatively low (ie, 10-to 100-fold specific). So, these data suggest the involvement of RARs in uPA induction. To confirm the involvement of RARs in uPA transcription, transfection studies were performed with the uPA promoter-luciferase and RARs and/or RXRs expressing vectors (Fig 3A). Luciferase activity was enhanced 1.2-fold after treatment with atRA (sample 2). A significant increase (∼3-fold) was induced when cells were cotransfected with either RARα or RXRα (samples 3 and 5, respectively). Their effects were dose-dependent, and a similar effect was obtained with other isoforms (data not shown). Cotransfection with RARα plus RXRα gave the largest increase (∼9-fold; sample 7). Interestingly, these enhancing effects were not dependent on the presence of ligands (samples 4, 6, 8, and 9), although ligand dependence was observed in a parallel experiment in which DR5-luciferase was used as reporter (Fig 3B). Collectively, the above-noted results suggest that RA causes induction of RARα as well as RARβ in endothelial cells, which leads to enhanced uPA transcription with the help of RXRs.
Induction of uPA and RARs by RA in endothelial cells. (A) Cell lysates were prepared from confluent BAEC cultures after the cultures had been incubated in MEM-BSA with 1 μmol/L atRA for varying lengths of time. Total RNA was isolated from cell lysates, and the changes in uPA mRNA levels as well as RARs and RXRs mRNA levels were assessed by Northern blotting as described in Materials and Methods. (B) After BAECs were treated for the indicated times with 1 μmol/L atRA in the absence or presence of 6 μg/mL cycloheximide (CHX), changes in uPA mRNA levels were determined by Northern blotting. (C) After exposure to either vehicle or 1 μmol/L atRA for 12 hours, BAECs were treated for the indicated times with 1 μg/mL actinomycin D. The rate of disappearance of uPA mRNA was determined by Northern blotting. Because mRNA for GAPDH also decreased along with incubation with actinomycin D, we presented ethidium bromide-labeled 28S RNA as internal. (D) Northern analyses were performed using total RNA isolated from human aortic endothelial cells treated with 1 μmol/L atRA for 12 hours.
Induction of uPA and RARs by RA in endothelial cells. (A) Cell lysates were prepared from confluent BAEC cultures after the cultures had been incubated in MEM-BSA with 1 μmol/L atRA for varying lengths of time. Total RNA was isolated from cell lysates, and the changes in uPA mRNA levels as well as RARs and RXRs mRNA levels were assessed by Northern blotting as described in Materials and Methods. (B) After BAECs were treated for the indicated times with 1 μmol/L atRA in the absence or presence of 6 μg/mL cycloheximide (CHX), changes in uPA mRNA levels were determined by Northern blotting. (C) After exposure to either vehicle or 1 μmol/L atRA for 12 hours, BAECs were treated for the indicated times with 1 μg/mL actinomycin D. The rate of disappearance of uPA mRNA was determined by Northern blotting. Because mRNA for GAPDH also decreased along with incubation with actinomycin D, we presented ethidium bromide-labeled 28S RNA as internal. (D) Northern analyses were performed using total RNA isolated from human aortic endothelial cells treated with 1 μmol/L atRA for 12 hours.
Induction of uPA in BAECs by RAR-activating retinoids. Cellular PA levels were determined using a chromogenic substrate, S2403, after treatment of BAECs for 12 hours in MEM-BSA either with (A) various concentrations of atRA (panel a), 9cRA (panel b), Ch55 (panel c), Am580 (panel d), CD2019 (panel e), CD666 (panel f), Ro47-5944 (panel g), and Ro25-7386 (panel h) or with a combination of various concentrations of atRA and 0.1 μmol/L Ro47-5944 (panel i) or (B) with 10−8 mol/L atRA (sample 2), Ch55 (sample 3), and Am580 (sample 4) in the absence or presence of 10−5 mol/L Ro41-5253 (▨) or LE135 (▩). The specificity and characterization of each compound are described under Materials and Methods. Data are expressed as urokinase (UK) units (U) per milligram of protein in the sample. Each point represents the average ± SD (n = 3).
Induction of uPA in BAECs by RAR-activating retinoids. Cellular PA levels were determined using a chromogenic substrate, S2403, after treatment of BAECs for 12 hours in MEM-BSA either with (A) various concentrations of atRA (panel a), 9cRA (panel b), Ch55 (panel c), Am580 (panel d), CD2019 (panel e), CD666 (panel f), Ro47-5944 (panel g), and Ro25-7386 (panel h) or with a combination of various concentrations of atRA and 0.1 μmol/L Ro47-5944 (panel i) or (B) with 10−8 mol/L atRA (sample 2), Ch55 (sample 3), and Am580 (sample 4) in the absence or presence of 10−5 mol/L Ro41-5253 (▨) or LE135 (▩). The specificity and characterization of each compound are described under Materials and Methods. Data are expressed as urokinase (UK) units (U) per milligram of protein in the sample. Each point represents the average ± SD (n = 3).
Enhancement of uPA promoter transactivation activity by RAR and RXR. BAECs were cotransfected with UK promoter-luciferase (0.4 μg/dish; A) or DR5-luciferase (0.4 μg/dish; B) plus RAR and/or RXR expressing vectors (250 ng each/dish), along with pRL-CMV (Renilla luciferase, 100 ng/dish) as described in Materials and Methods. Total DNA transfected was adjusted to 1.225 μg/dish with pSG5. The next day of transfection, the cells were treated or untreated with 1 μmol/L atRA or 9cRA for 12 hours. Luciferase activity of each cell was measured using the Dual-Luciferase Reporter Assay System, and changes in firefly luciferase activity were calculated and plotted after normalization to Renilla luciferase activity. Sample 1, reporter only; sample 2, atRA; sample 3, RAR; sample 4, RAR + atRA; sample 5, RXR; sample 6, RXR + 9cRA; sample 7, RAR and RXR; sample 8, RAR and RXR + atRA; sample 9, RAR and RXR + 9cRA. Each number represents the average ± SD (n = 3). Each similar experiment was repeated three times and representative results are shown here.
Enhancement of uPA promoter transactivation activity by RAR and RXR. BAECs were cotransfected with UK promoter-luciferase (0.4 μg/dish; A) or DR5-luciferase (0.4 μg/dish; B) plus RAR and/or RXR expressing vectors (250 ng each/dish), along with pRL-CMV (Renilla luciferase, 100 ng/dish) as described in Materials and Methods. Total DNA transfected was adjusted to 1.225 μg/dish with pSG5. The next day of transfection, the cells were treated or untreated with 1 μmol/L atRA or 9cRA for 12 hours. Luciferase activity of each cell was measured using the Dual-Luciferase Reporter Assay System, and changes in firefly luciferase activity were calculated and plotted after normalization to Renilla luciferase activity. Sample 1, reporter only; sample 2, atRA; sample 3, RAR; sample 4, RAR + atRA; sample 5, RXR; sample 6, RXR + 9cRA; sample 7, RAR and RXR; sample 8, RAR and RXR + atRA; sample 9, RAR and RXR + 9cRA. Each number represents the average ± SD (n = 3). Each similar experiment was repeated three times and representative results are shown here.
Involvement of Sp1 in RA induction of uPA.
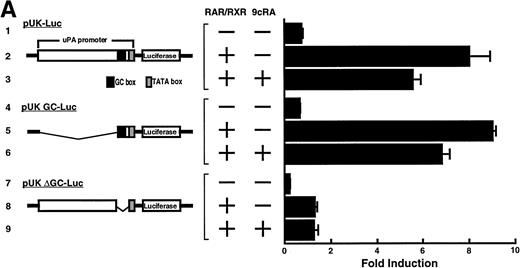
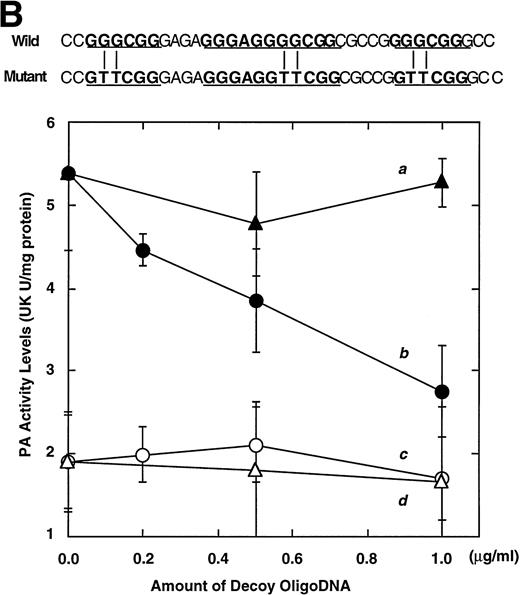
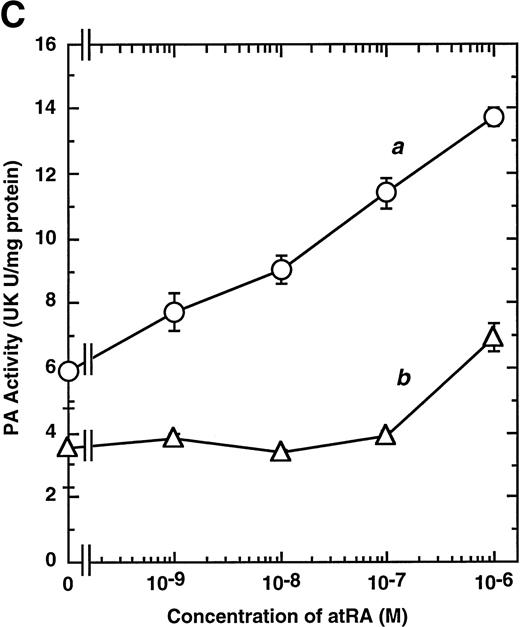
Computer analyses have shown that the uPA promoter used for the reporter assays does not contain any hitherto known RAREs, suggesting that RARs and or RXRs must transactivate the uPA promoter by a unique mechanism such as the interaction between RARs/RXRs and other transcription factors. Recently, Safe et al have reported that estrogen receptor, another member of the nuclear hormone receptor super family, physically interacts with the ubiquitous transcription factor Sp1 and regulates the transactivation of its target genes.36,37 We noticed that promoters of RA-inducible genes, including uPA, uPA receptor, and transglutaminase, contain GC boxes,30,38,39the cognate binding motif for Sp1.40 Therefore, we hypothesized that RARs/RXRs might also physically interact with Sp1 and regulate transactivation of uPA promoter. To explore this idea, we first examined whether Sp1 might be involved in the enhancing effect of RAR/RXR on uPA promoter transactivation. We used three different uPA promoter-luciferase constructs, including wild-type uPA promoter (pUK-Luc) or its mutants, either containing only GC and TATA boxes (pUK GC-Luc) or deficient in GC boxes (pUK ΔGC-Luc). As shown in Fig 4A, the basal activities of pUK GC-Luc (sample 4) and pUK ΔGC-Luc (sample 7) were, respectively, 93% and 27% of that of the wild-type pUK-Luc (sample 1), indicating the importance of GC box region for driving basal transactivation of uPA gene, as already reported previously.30 Transfection with RARα/RXRα uniformly enhanced the transactivation of these three constructs, suggesting a potential existence of RAR/RXR responsible element(s) in both GC box and out-of-GC box regions within the uPA promoter. However, the majority of enhancing effects by RAR/RXR on pUK-Luc was reproduced in pUK GC-Luc (samples 5 and 6), implying that Sp1 might be involved in RA induction of uPA. We, therefore, examined this possibility by modifying Sp1 function in two ways. First, we found that the Sp1 decoy, a 32-mer oligonucleotide, the sequence of which was derived from the uPA GC box, dose-dependently reduced the induction of uPA levels (Fig 4B, curve b), whereas an oligonucleotide containing the mutated Sp1 site had no effect (Fig 4B, curve a). The action of the Sp1 decoy was specific. It did not block the binding of other transcription factors, including RAR, and did not influence cell viability (data not shown). A similar suppression was obtained when cells were treated with mithramycin (Fig 4C, curve b), a specific inhibitor of the binding of Sp1 to the GC box (Fig 4D).41-43 The concentration of mithramycin used under the current experimental condition was not toxic to the cells, and the specificity of mithramycin has been ensured in the previous report.41 43 Collectively, these results suggest that RA induction of uPA is dependent on Sp1 in addition to RARs/RXRs.
Involvement of Sp1 in RA induction of uPA. (A) Promoter assay. Transfection study was performed as described before using wild-type pUK-luciferase (pUK-Luc, samples 1 through 3), pUK GC box-luciferase (pUK GC-Luc, samples 4 through 6), and pUK-luciferase deficient in GC box (pUK ▵GC-Luc, samples 7 through 9). Data are expressed as the relative luciferase activity compared with the activity of pUK-Luc cotransfected with empty pSG5 and untreated with 9cRA (sample 1). Samples 1, 4, and 7, reporter only; samples 2, 5, and 8, RAR and RXR; samples 3, 6, and 9, RAR and RXR + 9cRA. (B) Transcription factor decoy experiments. After BAECs were transfected with various amounts of Sp1 decoy or its mutant, whose sequences are presented above the illustration, the cultures were incubated either with vehicle or with 1 μmol/L atRA for 12 hours, cell lysates were prepared, and cellular PA levels were determined. Curves a and b, RA-treated cells; curves c and d, unstimulated cells. Curves a and d, mutant oligo; curves b and c, Sp1 decoy. (C) Cellular PA levels were determined after treatment of BAEC cultures with various concentrations of atRA for 12 hours in the absence (curve a) or presence (curve b) of 10 nmol/L mithramycin. For (A) through (C), the results are presented by the average ± SD (n = 3). Each similar experiment was repeated three times and representative results are shown here. (D) After treatment of BAECs with 1 μmol/L atRA for 12 hours in the absence (lane 1) or presence (lane 2) of 10 nmol/L mithramycin, nuclear extracts were prepared and Sp1 binding to uPA GC box was determined by gel shift assays as described in Materials and Methods.
Involvement of Sp1 in RA induction of uPA. (A) Promoter assay. Transfection study was performed as described before using wild-type pUK-luciferase (pUK-Luc, samples 1 through 3), pUK GC box-luciferase (pUK GC-Luc, samples 4 through 6), and pUK-luciferase deficient in GC box (pUK ▵GC-Luc, samples 7 through 9). Data are expressed as the relative luciferase activity compared with the activity of pUK-Luc cotransfected with empty pSG5 and untreated with 9cRA (sample 1). Samples 1, 4, and 7, reporter only; samples 2, 5, and 8, RAR and RXR; samples 3, 6, and 9, RAR and RXR + 9cRA. (B) Transcription factor decoy experiments. After BAECs were transfected with various amounts of Sp1 decoy or its mutant, whose sequences are presented above the illustration, the cultures were incubated either with vehicle or with 1 μmol/L atRA for 12 hours, cell lysates were prepared, and cellular PA levels were determined. Curves a and b, RA-treated cells; curves c and d, unstimulated cells. Curves a and d, mutant oligo; curves b and c, Sp1 decoy. (C) Cellular PA levels were determined after treatment of BAEC cultures with various concentrations of atRA for 12 hours in the absence (curve a) or presence (curve b) of 10 nmol/L mithramycin. For (A) through (C), the results are presented by the average ± SD (n = 3). Each similar experiment was repeated three times and representative results are shown here. (D) After treatment of BAECs with 1 μmol/L atRA for 12 hours in the absence (lane 1) or presence (lane 2) of 10 nmol/L mithramycin, nuclear extracts were prepared and Sp1 binding to uPA GC box was determined by gel shift assays as described in Materials and Methods.
Physical interaction between RARs/RXRs and Sp1.
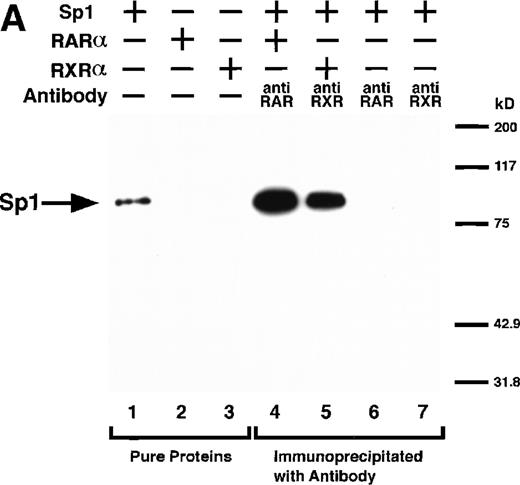
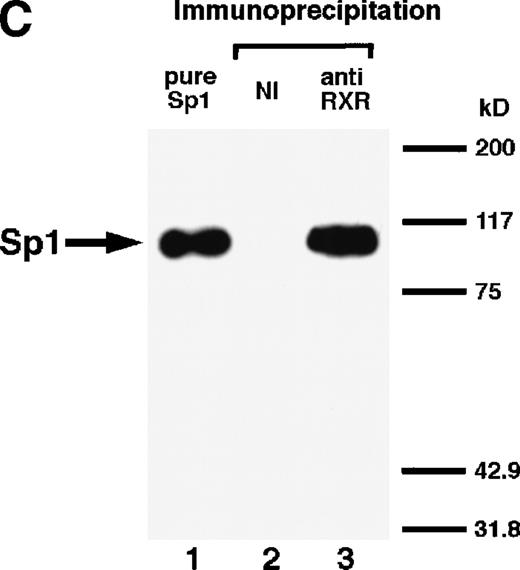
We next explored whether RARs/RXRs and Sp1 would interact physically. Figure 5A shows the results of immunoprecipitation studies. Sp1 was coprecipitated with either anti-RAR or RXR antibody, only in the presence of RARα (lane 4) or RXRα (lane 5), but not in their absence (lanes 6 and 7), suggesting a physical interaction between RARα/RXRα and Sp1. The physical interaction was also observed for 35S-labeled Sp1 in reticulocyte lysates incubated with RARα-GST (Fig 5B) as well as for Sp1 incubated with RAR/RXR present in BAEC nuclear extracts (Fig 5C). The physical interaction between RAR/RXR and Sp1 was further supported by the results of the mammalian two-hybrid assays (Fig 5D). Transactivation by Sp1 of Gal4-UAS-luciferase was potentiated by cotransfection with RARα and/or RXRα (samples 3, 5, and 7), suggesting a direct interaction between RARα/RXRα with Sp1 in vivo. Again, in this assay, obvious ligand dependence was not observed for RARα (sample 4). However, RXRα alone or RARα/RXRα showed some ligand dependence (samples 6, 8, and 9). Similar results were obtained for other subtypes of RARs/RXRs (data not shown).
Physical interaction between RAR/RXR and Sp1. (A) After the mixture of Sp1 plus either RAR or RXR was incubated, respectively, with anti-RAR or RXR antibody at 4°C for 2 hours, proteins were precipitated with protein A-Sepharose, eluted, and analyzed by Western blotting with anti-Sp1 antibody. Lanes 1 through 3, pure proteins; lane 1, Sp1; lane 2, RAR; lane 3, RXR. Lanes 4 through 7, proteins precipitated with anti-RAR or RXR antibody; lane 4, Sp1 preincubated together with RAR and precipitated with anti-RAR antibody; lane 5, Sp1 preincubated together with RXR and precipitated with anti-RXR antibody; lane 6, Sp1 incubated with anti-RAR antibody without RAR; lane 7, Sp1 incubated with anti-RXR antibody without RXR. (B) 35S-labeled Sp1 protein generated by in vitro translation reaction was incubated with GST or RAR-GST immobilized on glutathione-Sepharose beads. After extensive washings, the bound proteins were eluted and subjected to SDS-PAGE followed by autoradiography. Lane 1 input (0.5 μL of the total reaction mixture); lane 2, proteins bound to GST; lane 3, proteins bound to RAR-GST. (C) After the mixture of Sp1 and RAR/RXR was incubated with nonimmune antibody or anti-RXR antibody in BAEC nuclear extracts at 4°C overnight, proteins were precipitated with protein A-Sepharose, eluted, and analyzed by Western blotting with anti-Sp1 antibody. Lane 1, pure Sp1; lane 2, proteins precipitated with nonimmune (NI) antibody; lane 3, proteins precipitated with anti-RXR antibody. (D) BAECs were cotransfected with a combination of Sp1-Gal4 expressing vector (0.125 μg/dish) and Gal4-UAS-luciferase (0.5 μg/dish), plus RAR and/or RXR expressing vectors (250 ng each/dish), along with pRL-CMV (100 ng/dish; Promega) as described before. The day after transfection, the cells were treated or untreated with 1 μmol/L atRA or 9cRA for 12 hours. Luciferase activity of each cell was measured, and relative changes in firefly luciferase activity were plotted after normalization to Renilla luciferase activity. Sample 1, reporter only; sample 2, atRA; sample 3, RAR; sample 4, RAR + atRA; sample 5, RXR; sample 6, RXR + 9cRA; sample 7, RAR and RXR; sample 8, RAR and RXR + atRA; sample 9, RAR and RXR + 9cRA. Each number represents the average ± SD (n = 3). A similar experiment was repeated three times and representative results are shown here.
Physical interaction between RAR/RXR and Sp1. (A) After the mixture of Sp1 plus either RAR or RXR was incubated, respectively, with anti-RAR or RXR antibody at 4°C for 2 hours, proteins were precipitated with protein A-Sepharose, eluted, and analyzed by Western blotting with anti-Sp1 antibody. Lanes 1 through 3, pure proteins; lane 1, Sp1; lane 2, RAR; lane 3, RXR. Lanes 4 through 7, proteins precipitated with anti-RAR or RXR antibody; lane 4, Sp1 preincubated together with RAR and precipitated with anti-RAR antibody; lane 5, Sp1 preincubated together with RXR and precipitated with anti-RXR antibody; lane 6, Sp1 incubated with anti-RAR antibody without RAR; lane 7, Sp1 incubated with anti-RXR antibody without RXR. (B) 35S-labeled Sp1 protein generated by in vitro translation reaction was incubated with GST or RAR-GST immobilized on glutathione-Sepharose beads. After extensive washings, the bound proteins were eluted and subjected to SDS-PAGE followed by autoradiography. Lane 1 input (0.5 μL of the total reaction mixture); lane 2, proteins bound to GST; lane 3, proteins bound to RAR-GST. (C) After the mixture of Sp1 and RAR/RXR was incubated with nonimmune antibody or anti-RXR antibody in BAEC nuclear extracts at 4°C overnight, proteins were precipitated with protein A-Sepharose, eluted, and analyzed by Western blotting with anti-Sp1 antibody. Lane 1, pure Sp1; lane 2, proteins precipitated with nonimmune (NI) antibody; lane 3, proteins precipitated with anti-RXR antibody. (D) BAECs were cotransfected with a combination of Sp1-Gal4 expressing vector (0.125 μg/dish) and Gal4-UAS-luciferase (0.5 μg/dish), plus RAR and/or RXR expressing vectors (250 ng each/dish), along with pRL-CMV (100 ng/dish; Promega) as described before. The day after transfection, the cells were treated or untreated with 1 μmol/L atRA or 9cRA for 12 hours. Luciferase activity of each cell was measured, and relative changes in firefly luciferase activity were plotted after normalization to Renilla luciferase activity. Sample 1, reporter only; sample 2, atRA; sample 3, RAR; sample 4, RAR + atRA; sample 5, RXR; sample 6, RXR + 9cRA; sample 7, RAR and RXR; sample 8, RAR and RXR + atRA; sample 9, RAR and RXR + 9cRA. Each number represents the average ± SD (n = 3). A similar experiment was repeated three times and representative results are shown here.
Using gel shift assays, we next examined whether the direct interaction between RAR/RXR and Sp1 affected the ability of Sp1 to bind to DNA. We tested the effect of RARα on the binding of various amounts of Sp1 to both the uPA GC box and consensus GC box. As shown in Fig 6A, RARα potentiated the binding of Sp1 to both GC box sequences. The binding of Sp1 to these GC boxes was dose-dependent and drew a sigmoid curve, and RARα functioned to shift this curve toward lower concentration range. Namely, when the basal Sp1 (DNA binding) activity was low, the potentiating effect became large, whereas when the basal activity was high, the potentiating effect was small. A similar potentiating effect was obtained with RXRα administered either alone or in combination with RARα (Fig 6B, lanes 7 and 9). RARα and RXRα either alone or in combination did not bind to the uPA GC box (Fig 6B, lanes 4, 6, and 8), suggesting that RAR/RXR binding to DNA is not required for exertion of synergism. The potentiating effect of RARα/RXRα on Sp1 binding was not influenced by adding atRA or 9cRA (Fig 6C, lanes 5 or 6). No RAR-Sp1 or RXR-Sp1 complex was detected, as judged from supershift experiments (Fig 6C, lanes 8 through 11). The detected band was supershifted with anti-Sp1 antibody (lane 8) but not with anti-RAR antibody and/or anti-RXR antibody (lanes 9 through 11), suggesting that the increased band detected does not contain RARα/RXRα. We cannot explain this reason, but predict that probably affinity between RAR/RXR and Sp1 might be relatively weak compared with the interaction between RAR and RXR,1,2,44,45 as observed for association between RAR/RXR and coactivator or corepressor.4-7 Hence, although Sp1 bound to increased numbers of radiolabeled uPA GC box after physical interaction with RARα and/or RXRα, RAR-Sp1 and RXR-Sp1 associations could be disrupted during electrophoresis. In fact, when we performed the immunoprecipitation from the mixture containing RAR, Sp1, and radiolabeled uPA GC box oligonucleotide, we succeeded in detecting specific radioactivity in anti-RAR antibody-precipitated fraction. This radioactivity represents a specific binding of Sp1 to uPA GC box, suggesting that RAR, Sp1, and radiolabeled uPA GC box may form complexes in solution. The potentiating effect on Sp1 binding was not observed when BSA was used instead of RARs/RXRs (data not shown). We observed a similar result when comparing the binding of endogenous Sp1 present in the nuclear extract of control versus RARs transfected cells (data not shown). Collectively, these results suggest that physical interaction between RARs/RXRs and Sp1 may potentiate Sp1 binding to the uPA GC box and enhance uPA transcription. Finally, we confirmed this potentiation by a cotransfection experiment. When transcriptional activity of the uPA promoter was enhanced about fivefold by transfection of either Sp1 or RARα/RXRα alone, cotransfection of these resulted in a 17-fold increase in activity.
Potentiation of Sp1 binding to uPA GC box by RAR and/or RXR. (A) After Sp1 (10 to 20 ng) was preincubated in the absence or presence of RAR-GST (200 ng), the reaction mixture was incubated with 32P-labeled uPA GC box or consensus GC box, and thereafter protein-DNA complexes were separated by a 4% polyacrylamide gel electrophoresis and visualized on an image analyzer. Odd numbers, Sp1 alone; even numbers, Sp1 plus RAR. (B) Gel shift analyses were performed as before using both RAR-GST and RXR-GST. Lane 1, Sp1 (10 ng) alone; lane 2, GST alone; lane 3, Sp1 plus GST; lane 4, RAR alone; lane 5, Sp1 plus RAR; lane 6, RXR alone; lane 7, Sp1 plus RXR; lane 8, RAR and RXR; lane 9, Sp1 plus RAR/RXR. (C) Control experiments. Lane 1, control (Sp1 [10 ng] plus RAR/RXR); lane 2, + 20-fold excess of unlabeled oligonucleotide (cold); lane 3, + 50-fold excess of unlabeled oligonucleotide (cold); lane 4, + 0.5% ethanol (vehicle); lane 5, + 1 μmol/L atRA; lane 6, + 1 μmol/L 9cRA; lane 7, + nonimmune (NI) antibody (IgG); lane 8, + anti-Sp1 IgG; lane 9, + anti-RAR IgG; lane 10, + anti-RXR IgG; lane 11, + both anti-RAR IgG and anti-RXR IgG. The final concentration of each antibody was 80 μg/mL.
Potentiation of Sp1 binding to uPA GC box by RAR and/or RXR. (A) After Sp1 (10 to 20 ng) was preincubated in the absence or presence of RAR-GST (200 ng), the reaction mixture was incubated with 32P-labeled uPA GC box or consensus GC box, and thereafter protein-DNA complexes were separated by a 4% polyacrylamide gel electrophoresis and visualized on an image analyzer. Odd numbers, Sp1 alone; even numbers, Sp1 plus RAR. (B) Gel shift analyses were performed as before using both RAR-GST and RXR-GST. Lane 1, Sp1 (10 ng) alone; lane 2, GST alone; lane 3, Sp1 plus GST; lane 4, RAR alone; lane 5, Sp1 plus RAR; lane 6, RXR alone; lane 7, Sp1 plus RXR; lane 8, RAR and RXR; lane 9, Sp1 plus RAR/RXR. (C) Control experiments. Lane 1, control (Sp1 [10 ng] plus RAR/RXR); lane 2, + 20-fold excess of unlabeled oligonucleotide (cold); lane 3, + 50-fold excess of unlabeled oligonucleotide (cold); lane 4, + 0.5% ethanol (vehicle); lane 5, + 1 μmol/L atRA; lane 6, + 1 μmol/L 9cRA; lane 7, + nonimmune (NI) antibody (IgG); lane 8, + anti-Sp1 IgG; lane 9, + anti-RAR IgG; lane 10, + anti-RXR IgG; lane 11, + both anti-RAR IgG and anti-RXR IgG. The final concentration of each antibody was 80 μg/mL.
DISCUSSION
First, we demonstrated that RARs potentiate transactivation of the uPA gene, which lacks a canonical RARE, with help of RXRs. Interestingly, this effect appeared to be ligand-independent, suggesting a novel underlying mechanism. In support of this idea, we next demonstrated that RARs/RXRs directly interact with Sp1 and enhance transactivation of the uPA promoter, at least in part, by potentiating the binding of Sp1 to the uPA GC box. The result shown in Fig 6A suggests that the effect of RAR/RXR we found is a more general effect on Sp1-mediated transcription. The result shown in Fig 3A is thought to reflect the synergism between endogenous Sp1 and transfected RARs/RXRs. We cannot explain the discrepancy in ligand dependency among different experimental settings, eg, reporter assays (Figs 3A and 4A; apparent independence), two-hybrid assays (Fig 5D; partial dependence), and gel shift assays (Fig 6C; apparent independence). If the ligand-independence of the RAR/RXR-Sp1 effect is genuine, then the question arises as to which factor is responsible for the RA-dependency in uPA induction by RA. One likely answer is that at the very least RA is needed to increase RARs in the cells, although there are many other possibilities. RAR/RXR may act predominantly to strengthen the weak Sp1 binding. Alternatively, RAR/RXR may act both increasing Sp1 binding and enhancing the transcription of Sp1. We predict that effects on DNA binding may be ligand independent, whereas effects on transactivation may be ligand dependent.
We demonstrated that RARα/RXRα enhanced the transactivation of the wild-type uPA promoter-luciferase construct approximately sixfold (Fig4A) and that of Gal4-UAS-luciferase construct about 25- to 30-fold in the mammalian two-hybrid system (Fig 5D). This difference can be explained as follows. As can be seen in Fig 6A, the binding of Sp1 to the GC box is dose-dependent and draws a sigmoid curve, and RAR/RXR function to shift this curve toward lower concentration range. Namely, when the basal Sp1 (DNA binding) activity is low, the potentiating effect becomes large, whereas when the basal activity is high, the potentiating effect is small. A similar theory can be said for reporter assays. The basal activity of the Gal4-UAS-luciferase is low, because, theoretically, only Gal4 fusion protein is able to transactivate this reporter gene. In contrast, the full-length uPA promoter-luciferase is transactivated to some extent by endogenous Sp1 as well as other transcription factors. Therefore, the potentiating effect of RAR/RXR is more dominant in the former assays compared with the latter assays.
The three subtypes of RARs seem to share equivalent ability in terms of interacting with and potentiating Sp1. Therefore, the dominant function of each subtype may be determined by the nature of the subtype expressed at a particular time. Under the present conditions, endothelial cells expressed RARα and RARγ initially, whereas the expression of RARα and RARβ was dramatically increased upon stimulation. The results showing that the expression of RARα and RARβ increased and that specific antagonists of these factors blocked the induction of uPA indicate the participation of RARα and RARβ, but fail to elucidate a role for RARγ. Although the contribution of RARγ appeared to be minimal, judging by the results obtained using the RARγ-selective agonist (Fig 2A, panel f), our data do not rule out the involvement of RARγ in this system, especially after 12 hours, when the expression of RARα and RARβ started to decrease, in contrast to the expression of RARγ, which started to increase (Fig1A). Complementary expression of RARβ and RARγ has been reported in the developing mouse embryo.46-48 A similar argument can be raised about the role of RXRs. Although the expression of RXRα/β did not change as observed for RARα/β (Fig 1A) and RXR-selective retinoids were ineffective in enhancing uPA expression by BAECs (Fig 2A, panels g through i), RXRs themselves were capable of interacting with (Fig 5) and potentiating Sp1 binding (Fig 6) and stimulating uPA transactivation (Fig 3), both alone and in combination with RARs. This discrepancy could be explained partly because RXR-selective retinoids were unable to induce RARs nor RXRs in BAEC cultures (data not shown). The involvement of RARγ and RXRα/β in our system could be verified by examining the effect of an RARγ or RXRα/β-specific antagonist, if such molecules could be obtained. Recently, Piedrafita and Pfahl49 reported that specific degradation of Sp1 occurs in apoptic T cells after treatment with a relatively high concentration of RARγ-specific retinoids, whereas RA showed no significant effect. Because we used RA on endothelial cells, our system is different.
The functional interaction between RARs/RXRs and Sp1 has previously been described for the RA induction of the retinol-binding protein gene.50 However, in this case, RARs and Sp1 bind to the RARE and GC box, respectively, and function synergistically. On the other hand, a physical interaction between Sp1 and other members of the nuclear receptor family has been reported.36,37,51 52 The current study provides an initial evidence that RARs/RXRs also physically and functionally interact with Sp1 and enhance its binding affinity for target DNA without any need for binding of RARs/RXRs to RARE. Although we found that RAR/RXR-Sp1 interaction occurs in a tube in the absence of ligands, we cannot exclude the possibility that physiological interaction was dependent on a low concentration of RA that we may not have been able to eliminate from the serum (<10−10 mol/L). It is also possible that endogenous RARs/RXRs require ligand to keep corepressors away and to associate with Sp1, whereas large numbers of exogenously transfected RARs/RXRs might be free from limited numbers of corepressors and therefore might not require a ligand.
Several important issues remain unresolved: (1) It might be possible that RAR/RXR-Sp1 interaction not only potentiates Sp1-mediated transcription, but also allows RAR/RXR to drive transcription by enabling binding to the GC box via Sp1. Figure 5D demonstrates RARα and RXRα enhancement of the transcriptional activity of a chimeric transcription factor, Sp1-Gal4, suggesting that RAR/RXR alter the transactivation domain of Sp1 and/or bind to Sp1-activation domain and confer their transactivation activity. (2) We predict that RARs may act as allosteric effectors that change the conformation of Sp1. Thus, it will be important to determine the nature of the change in Sp1 structure induced by binding to RARs/RXRs that is responsible for increased binding to GC box. (3) It will also be important to elucidate whether other nuclear factors, including corepressors and coactivators, modulate the RAR/RXR-Sp1 interaction. (4) Finally, because functional repression of gene expression through interaction between RAR and AP-1, NF-IL6, or Myb has been reported,53-55 one may ask whether the RAR/RXR-Sp1 interaction affects other transcription factors shown to be important for basal as well as induced transcription from the uPA gene, such as Egr-1, PEA3, NF-κB, CREB, UEF1-4, and AP-1.56-59 We are now trying to map the binding site(s) in both RAR and Sp1 molecules as the first step towards answering these questions.
ACKNOWLEDGMENT
The authors thank F. Blasi and S.L. Friedman for helpful advice and critical reading of the manuscript; S. Michel, M. Klaus, W.-D. Schleuning, J.T. Kadonaga, S. Kato, and P. Chambon for reagents and constructs; and C. Iijima, M. Yoshizawa, and S. Hayashi for technical assistance.
Supported partly by Grant-in-Aids from the Ministry of Education, Science, Sports and Culture (10780395), Grant-in-Aids from Tokyo Biochemical Research Foundation, Grants for “Biodesign Research Program” and “Multibioprobe Research Program” from RIKEN, and The Sumitomo Foundation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Soichi Kojima, PhD, Laboratory of Molecular Cell Sciences, Tsukuba Life Science Center, The Institute of Physical and Chemical Research (RIKEN), Koyadai, Tsukuba, Ibaraki 305-0074, Japan; e-mail: kojima@rtc.riken.go.jp.


![Fig. 6. Potentiation of Sp1 binding to uPA GC box by RAR and/or RXR. (A) After Sp1 (10 to 20 ng) was preincubated in the absence or presence of RAR-GST (200 ng), the reaction mixture was incubated with 32P-labeled uPA GC box or consensus GC box, and thereafter protein-DNA complexes were separated by a 4% polyacrylamide gel electrophoresis and visualized on an image analyzer. Odd numbers, Sp1 alone; even numbers, Sp1 plus RAR. (B) Gel shift analyses were performed as before using both RAR-GST and RXR-GST. Lane 1, Sp1 (10 ng) alone; lane 2, GST alone; lane 3, Sp1 plus GST; lane 4, RAR alone; lane 5, Sp1 plus RAR; lane 6, RXR alone; lane 7, Sp1 plus RXR; lane 8, RAR and RXR; lane 9, Sp1 plus RAR/RXR. (C) Control experiments. Lane 1, control (Sp1 [10 ng] plus RAR/RXR); lane 2, + 20-fold excess of unlabeled oligonucleotide (cold); lane 3, + 50-fold excess of unlabeled oligonucleotide (cold); lane 4, + 0.5% ethanol (vehicle); lane 5, + 1 μmol/L atRA; lane 6, + 1 μmol/L 9cRA; lane 7, + nonimmune (NI) antibody (IgG); lane 8, + anti-Sp1 IgG; lane 9, + anti-RAR IgG; lane 10, + anti-RXR IgG; lane 11, + both anti-RAR IgG and anti-RXR IgG. The final concentration of each antibody was 80 μg/mL.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/12/10.1182_blood.v93.12.4264/4/m_blod41227006aw.jpeg?Expires=1769090200&Signature=ZqGeaXcwQNPnNRonptPjgwP56NwGpSrwQefcpSZjwF2oRk2aZ1LhXYoEzjWXKb5CIJECtkJUsAvOVF5hNbPfRfwoHnqplL4PsyIzv69LfayDDSzBY2sJygFQCOSvDgW8GAuyZvYr1CETxlVmltBPEiTeT7ZY1zCz66Ql8I7c18M9KuBcHl-M~cHJJRPioVFU-MsQrTOSh4SIZwlv2swJX26dSS181vlshgxT6qdzAKouSQ~WmgaD27Aa30G7DBEIsF917x0~wvMLDQAyy3tMqQnGsUPJamjkgTLX05oFhUlVdnoybpqyUGXJIIQYUPmAH1rPmI50ZEaglXrjvb-iDA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Potentiation of Sp1 binding to uPA GC box by RAR and/or RXR. (A) After Sp1 (10 to 20 ng) was preincubated in the absence or presence of RAR-GST (200 ng), the reaction mixture was incubated with 32P-labeled uPA GC box or consensus GC box, and thereafter protein-DNA complexes were separated by a 4% polyacrylamide gel electrophoresis and visualized on an image analyzer. Odd numbers, Sp1 alone; even numbers, Sp1 plus RAR. (B) Gel shift analyses were performed as before using both RAR-GST and RXR-GST. Lane 1, Sp1 (10 ng) alone; lane 2, GST alone; lane 3, Sp1 plus GST; lane 4, RAR alone; lane 5, Sp1 plus RAR; lane 6, RXR alone; lane 7, Sp1 plus RXR; lane 8, RAR and RXR; lane 9, Sp1 plus RAR/RXR. (C) Control experiments. Lane 1, control (Sp1 [10 ng] plus RAR/RXR); lane 2, + 20-fold excess of unlabeled oligonucleotide (cold); lane 3, + 50-fold excess of unlabeled oligonucleotide (cold); lane 4, + 0.5% ethanol (vehicle); lane 5, + 1 μmol/L atRA; lane 6, + 1 μmol/L 9cRA; lane 7, + nonimmune (NI) antibody (IgG); lane 8, + anti-Sp1 IgG; lane 9, + anti-RAR IgG; lane 10, + anti-RXR IgG; lane 11, + both anti-RAR IgG and anti-RXR IgG. The final concentration of each antibody was 80 μg/mL.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/12/10.1182_blood.v93.12.4264/4/m_blod41227006bw.jpeg?Expires=1769090200&Signature=H2hzC8K21HHFwHpTIjJDvrqnuNycxcZn1hq1vGtGW0KUUEKy~cMk6PjGFuVSGH0zuUIpRD6F2MTQ5LVTX3c6X8Bec40JBfCMwgK~p2SJjW3zNmpxh1LiABmbQSB2MsPnnFiDjR7VgjBk9nBkh7dSckHfGUA3E1xpVPiUuQnFLIh8JX-oDMYMyMcPdcpW~~PyDT3s69fpbJMpQjvjRjoEF3V7WrVgJ2z~kxxL4AKOgnnPNs8Tk3kdMtZspSruPDHCuSE1Se9P6zGVvauutzn6M1Er6akmYgSfn~h2fhyYX8pNqHwQ8PjMl2FFI~aDk-K18DPEuqqbenO1yOJ5e7bz5A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Potentiation of Sp1 binding to uPA GC box by RAR and/or RXR. (A) After Sp1 (10 to 20 ng) was preincubated in the absence or presence of RAR-GST (200 ng), the reaction mixture was incubated with 32P-labeled uPA GC box or consensus GC box, and thereafter protein-DNA complexes were separated by a 4% polyacrylamide gel electrophoresis and visualized on an image analyzer. Odd numbers, Sp1 alone; even numbers, Sp1 plus RAR. (B) Gel shift analyses were performed as before using both RAR-GST and RXR-GST. Lane 1, Sp1 (10 ng) alone; lane 2, GST alone; lane 3, Sp1 plus GST; lane 4, RAR alone; lane 5, Sp1 plus RAR; lane 6, RXR alone; lane 7, Sp1 plus RXR; lane 8, RAR and RXR; lane 9, Sp1 plus RAR/RXR. (C) Control experiments. Lane 1, control (Sp1 [10 ng] plus RAR/RXR); lane 2, + 20-fold excess of unlabeled oligonucleotide (cold); lane 3, + 50-fold excess of unlabeled oligonucleotide (cold); lane 4, + 0.5% ethanol (vehicle); lane 5, + 1 μmol/L atRA; lane 6, + 1 μmol/L 9cRA; lane 7, + nonimmune (NI) antibody (IgG); lane 8, + anti-Sp1 IgG; lane 9, + anti-RAR IgG; lane 10, + anti-RXR IgG; lane 11, + both anti-RAR IgG and anti-RXR IgG. The final concentration of each antibody was 80 μg/mL.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/12/10.1182_blood.v93.12.4264/4/m_blod41227006cw.jpeg?Expires=1769090200&Signature=0R73eGC7zVPkH8v0a2oOK~wkguMNtTDMimir8h2zBu3DJQ5C1k31Yc9toG~pQD7DoTKUCZOwEHIrt3rW0WzDsojVwIZ511hMPzxeZUpxf2PqYHW66V9S-C5NjoZqqkj0RMSAy4EzOOWABSEwlNO-ShxI35KHC5RAZjxf-lJZDzmTvyZQyVZFc1e0lXNIlLHJ6onKg3twGlkKtqfMeeAP1iD6qg9KJzOs0k-WdvwKNQzJuAejqM9Ih1hBFWsYdkxEY~tNqZgCRbhu0JucMiqla1eLmOqEtjg5UZNKGyXOSaviqFU~XNBPj0-WFoPHClEqejJqX0NfryW7~9RR1aJaCQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal