Abstract
An unbiased genetic approach was used to identify a specific amino acid residue in the IIb subunit important for the ligand binding function of the integrin IIbβ. Chemically mutagenized cells were selected by flow cytometry based on their inability to bind the ligand mimetic antibody PAC1 and a cell line containing a single amino acid substitution in IIb at position 224 (D→V) was identified. Although well expressed on the surface of transfected cells, IIbD224Vβ3 as well as IIbD224Aβ3 did not bind IIbβ3-specific ligands or a RGD peptide, a ligand shared in common with vβ3. Insertion of exon 5 of IIb, residues G193-W235, into the backbone of the v subunit did not enable the chimeric receptor to bind IIbβ3-specific ligands. However, the chimeric receptor was still capable of binding to a RGD affinity matrix. IIbD224 is not well conserved among other integrin subunits and is located in a region of significant variability. In addition, amino acid D224 lies within a predicted loop of the recently proposed β-propeller model for integrin subunits and is adjacent to a loop containing amino acid residues previously implicated in receptor function. These data support a role for this region in ligand binding function of the IIbβ3 receptor.
PLATELET ADHESIVE interactions mediated by the integrin αIIbβ3 are fundamental to the maintenance of normal hemostasis, as illustrated by the inherited bleeding disorder Glanzmann’s thrombasthenia. Platelets from patients with this disease either lack the αIIbβ3receptor, express reduced levels of the receptor, or express receptor variants that lack ligand binding function.1 In areas of vessel or tissue trauma, the αIIbβ3receptor initiates the formation of a platelet-based plug by binding to soluble fibrinogen, which subsequently forms a bridge to other platelets and facilitates platelet aggregation. Understanding the precise molecular mechanisms underlying the structural basis of ligand-receptor interaction is an important step towards the modulation of platelet function and can provide new insights into the development of novel antithrombotic therapies. Furthermore, understanding the molecular basis of this interaction by αIIbβ3, the prototypic integrin could provide insights into other integrin-ligand interactions in a diverse range of physiological processes.
Multiple ligand contact sites have been identified on αIIbβ3. Studies of natural receptor variants, cross-linking of ligand mimetic peptides, and site-directed mutagenesis studies have provided convincing evidence for the importance of two discontinuous regions in the β3subunit, D119-S123 and D217-E220, in the ligand binding function of αIIbβ3.2-8 The importance of this region of β3 appears to be due to its structural similarity to the integrin α subunit I-domain,9 which has been demonstrated to be critically involved in the ligand binding function of those integrins that contain this structure.10This region of β3 may also function as a ligand/cation-binding MIDAS-like domain.8 The homologous region of other integrin β subunits is similarly critical for ligand binding function, substantiating that the amino terminal region of integrin β subunits is either directly involved in or structurally important for ligand binding receptor function.11-13
Potential ligand contact sites on integrin α subunits have been identified by mapping epitopes of inhibitory monoclonal antibodies (MoAbs) and subsequent site-directed mutagenesis studies.14-16 Residues of αIIb implicated in ligand binding function have also been identified by peptide studies17-19 and by the characterization of mutations present in patients with Glanzmann’s thrombasthenia. However, in contrast to many of the mutations identified in β3, nearly all of the naturally occurring mutations identified in αIIb reduce αIIbβ3 receptor expression to very low or undetectable levels.1 This suggests that the processing or structural stability of αIIb is very sensitive to substitutions. The limited occurrence of well-expressed natural receptor variants containing αIIb mutations has hindered the identification of specific residues in αIIb that are critical to the ligand binding function of the receptor. Despite these difficulties, secondary structure predictions have been used to identify several residues in αIIb whose substitutions blocked ligand binding but did not significantly affect expression of αIIbβ3.20 To overcome the inefficiencies of mutagenesis approaches in the absence of structural data and the rarity of Glanzmann’s thrombasthenia, we recently described a novel method for the identification of induced mutations that affect αIIbβ3 ligand binding function.21 The advantage of this method is its capacity to identify mutations that abolish ligand binding function but do not affect receptor expression. The present study identified a region in αIIb that is important for ligand binding and is consistent with the predicted binding sites in the recently proposed structural model22 for the integrin α subunits.
MATERIALS AND METHODS
Antibodies and reagents.
The IgMκ murine antibody PAC1 was kindly provided by Dr Sanford Shattil (The Scripps Research Institute, La Jolla, CA). PAC1 is an activation-specific, fibrinogen-mimetic MoAb that has been extensively characterized elsewhere.23 The αIIbβ3 complex-specific MoAbs AP224 and OPG225 were provided by Dr Thomas Kunicki (The Scripps Research Institute). The αIIbβ3 complex-specific MoAbs 4F10 and 2G12 were kindly provided by Dr Virgil Woods (University of California, San Diego, CA). The αIIbβ3 complex-specific MoAb D57, the anti-β3 MoAb 15, and the activating anti-β3 MoAbs, anti-LIBS1, anti-LIBS2, and anti-LIBS6, have been described elsewhere.26-28 The anti-αv specific MoAb 14229 was purchased from Chemicon (Temecula, CA). The anti-αIIb MoAb 98DF6 was obtained from Dr Jari Ylänne (University of Helsinki, Helsinki, Finland). The MoAb D57 was biotinylated using biotin-N-hydroxysuccinimide (Pierce, Rockford, IL) according to the manufacturer’s instructions. Fluorescein isothiocyanate-conjugated goat antimouse IgM and IgG were obtained from Biosource (Camarillo, CA). The αIIbβ3peptidomimetic Ro43-505430 was provided by Beat Steiner (Hoffmann-La Roche, Basel, Switzerland). The GRGDSPK-sepharose column was prepared as described.31
DNA constructs.
The expression constructs pc3a and CD2b encoding wild-type β3 and wild-type αIIb, respectively, have been previously described.8,32 A 3.3-kb fragment of αIIb containing the entire coding sequence and the 3′ untranslated region was excised from CD2b by digestion withXba I and ligated to the expression vector pcDNA3 (Invitrogen, San Diego, CA). The resulting construct was designated pc2b. The pCDM8 expression vector encoding wild-type αvhas been previously described.4 An insert containing the entire coding sequence of αv was removed from pCDM8 byXba I digestion and ligated to Xba I-digested pcDNA3. This construct was designated pcαv. Construction of the expression plasmids encoding the chimeric subunits αIIbα6A and β3β1 has been described elsewhere.28 These chimeric plasmids contain the extracellular and transmembrane domains of human αIIb or β3 fused to the cytoplasmic domains of α6Aand β1, respectively. CD3hyg is a derivative of CD3a32 containing the hygromycin resistance gene. Amino acid residues of αIIb are numbered, with the leucine residue at the amino terminus of the mature protein being residue number 1.
The chimeric α subunit, designated pcαvαIIb, consists of the backbone of αv from which amino acids 181-223 were removed and replaced with the corresponding amino acids of exon 5 of αIIb (amino acids G193-W235).33 It was constructed using the megaprimer method34 and three consecutive rounds of amplification. Chimeric oligonucleotide primers were constructed in which the 5′ ends contained αvsequences (30 bp for each primer) and the 3′ ends contained the sequences either of the 5′ or the 3′ end of exon 5 of αIIb (27 and 29 bp, 5′ and 3′ primer, respectively). The first round of polymerase chain reaction (PCR) used pc2b as the template and generated the 129-bp exon 5 of αIIb containing 30 bp of αv sequence on both the 5′ and 3′ ends. This fragment was gel-purified and used as a 5′ megaprimer together with a 3′ αvprimer corresponding to the sequence 5′-GAATAGCCAAAGCTTGGTGGCATGC-3′ in a second round of PCR using pcαv as the template to generate a 712-bp fragment containing exon 5 of αIIb fused to 3′ αv sequences. The 712-bp fragment was used as a 3′ megaprimer along with a 5′ αv primer corresponding to the sequence 5′-CCGaGtaagCTTCGGCGATGGCTTTTCCGC-3′ in a final round of PCR with pcαv as a template. The resulting 1,369-bp fragment contained exon 5 of αIIb flanked by 5′ and 3′ αv sequences. This PCR fragment was digested with HindIII and ligated to HindIII-digested pcαv. The authenticity of the final construct was confirmed by DNA sequencing of the entire subcloned fragment.
Mutagenesis.
Site-directed mutagenesis of selected αIIb residues was performed using splice overlap extension as previously described.35 PCR-generated fragments containing αIIb point mutations were digested with EcoRI andCla I, gel-purified, and ligated to EcoRI-ClaI–digested pc2b. All amplified fragments were sequenced in their entirety to verify the introduction of the mutation and the absence of any other substitutions.
The αβPy stable cell line was generated by transfecting CHO cells with three plasmids: αIIbα6A, β3β1, and a plasmid encoding the neomycin resistance gene.21 Chemical mutagenesis of the αβPy stable cell line was performed by treating 2 × 106cells with 300 μg/mL ethyl methane sulfonate (EMS; Sigma, St Louis, MO) for 15 to 19 hours. After 1 week, approximately 1 × 108 cells were harvested and ligand binding mutants were selected by flow cytometry.
Flow cytometry.
Chemically mutagenized cells were individually sorted on a FAC-STAR Plus (Becton Dickinson, Mountain View, CA) using two-color flow cytometry with MoAbs D57 and PAC1 as described in detail.36 Cells that exhibited positive staining for receptor expression (D57 positive) but weak or absent PAC1 staining were individually sorted into 96-well plates and cultured for further analysis.
Surface expression of transfected integrins was analyzed by flow cytometry as described.27 PAC1 binding to transfected cells was analyzed by two-color flow cytometry as described.36PAC1 binding was analyzed only on the subset of cells positive for αIIbβ3 receptor expression that were gated using biotinylated D57. Some samples also contained 4 μmol/L of one of the anti-β3 MoAbs anti-LIBS1, anti-LIBS2, or anti-LIBS6. These antibodies bind to distinct epitopes on β3 and directly induce PAC1 binding to αIIbβ3.37 Other samples contained 2 μmol/L Ro43-5054.
Reverse transcriptase-PCR (RT-PCR).
Total cellular RNA was isolated from the selected cell lines using TRIzol (GIBCO/BRL, Grand Island, NY). cDNA was synthesized using oligo(dT) primers and the cDNA Cycle kit (Invitrogen). Resulting cDNA was amplified for 30 cycles with αIIb-specific primers. The PCR products were directly sequenced using fluorescent automated sequencing (Applied Biosystems, Foster City, CA).
Cell culture and transfections.
Cell lines were routinely passaged in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 1% nonessential amino acids, 2 mmol/L L-glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin. cDNAs were transfected into CHO cells with Lipofectamine (Life Technologies, Gaithersburg, MD) as previously described.36 cDNAs encoding αvαIIb, αIIbD224V, or selected αIIb mutants were transfected into CHO cells that had been previously transfected with the expression plasmid CD3hyg. Stably transfected cells were established by selection with 700 μg/mL G418. Clonal cell lines expressing αvαIIbβ3 were selected by flow cytometry using the antihuman αv-specific MoAb 142. Clonal lines expressing αIIbD224Vβ3 were sorted with the anti-αIIbβ3complex-specific antibody, D57.
Affinity chromatography.
Lysates of 3 × 107 cells expressing either wild-type αIIbβ3, αIIbD224Vβ3, wild-type αvβ3, or αvαIIbβ3 were prepared and applied to a GRGDSPK-Sepharose 4B column (1 mL bed volume) as described.31 The column was washed with 10 mL of lysis buffer and then eluted with 3 mL of buffer containing 1 mg/mL GRGDSPK. One-milliliter fractions were collected and resolved on 8% sodium dodecyl sulfate (SDS) polyacrylamide gels under nonreducing conditions. Proteins were transferred to nitrocellulose and blotted with the αIIb-specific MoAb, 98DF6 (3.5 μg/mL), for the αIIbβ3 and αIIbD224Vβ3 receptors; with the αv-specific MoAb, 142 (1:200 dilution of ascites), for the αvβ3 and αvαIIbβ3 receptors; or with the β3-specific MoAb, MoAb15 (3.5 μg/mL).
RESULTS
Selection and identification of ligand binding defective mutants.
We have previously demonstrated that fusing the cytoplasmic domains of α6A and β1 to the extracellular and transmembrane domains of αIIb and β3, respectively, caused αIIbβ3 to assume the high-affinity state as defined by constitutive binding of the ligand mimetic antibody PAC1.28 Therefore, to isolate cell lines with disrupted integrin ligand binding function, CHO cells stably transfected with the active receptor variant αIIbα6Aβ3β1were exposed to the chemical mutagen, EMS. Mutagenized cells were then sorted by two-color flow cytometry to isolate those cells that failed to bind PAC1 but continued to express αIIbβ3 based on the binding of the MoAb D57 as described.21 Selected cells were individually sorted and subsequent analysis of the clonal cell lines grouped the mutants into three phenotypic classes: (1) ligand binding mutants, (2) integrin activation mutants, and (3) cellular activation mutants.21Individually sorted clonal lines that failed to bind PAC1 in the presence of the activating MoAb anti-LIBS6 were defined as ligand binding defective mutants. To identify which subunit contained a mutation resulting in the loss of ligand binding function, isolated cell lines with a ligand binding defect were transfected individually with the original αIIbα6A or β3β1 subunit and then reanalyzed for restoration of PAC1 binding. For most of the ligand binding defective mutants, PAC1 binding was restored after retransfection with the parental β subunit β3β1, indicating the causative mutation was present in the β subunit. In contrast, PAC1 binding was reconstituted in one cell line only after retransfection with αIIbα6A, indicating that the defect was likely contained in the αIIb subunit. This cell line was subjected to further analysis.
Total RNA was isolated from this cell line and subjected to RT-PCR. The entire αIIbα6A subunit was amplified in three separate fragments and the resulting PCR products were sequenced directly. Analysis of the resulting sequence identified a single A→T mutation resulting in substitution of D224 by valine.
Characterization of ligand binding defective mutant.
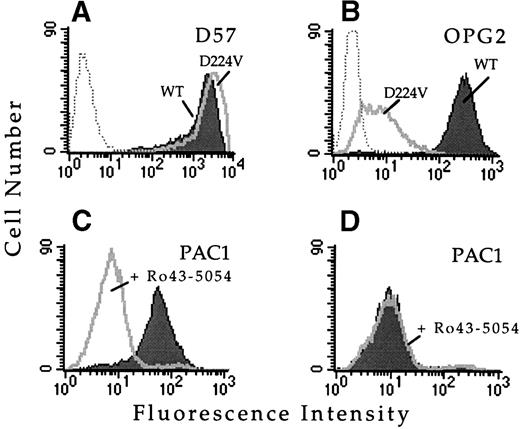
To confirm that this single amino acid substitution was responsible for the observed loss of ligand binding function, this mutation was introduced into wild-type αIIb cDNA and transfected into cells stably expressing the wild-type β3 subunit. Whether transiently or stably transfected, the αIIbD224Vβ3 receptor was expressed on the cell surface and bound a panel of complex-specific anti-αIIbβ3 MoAbs, including D57 (Fig 1A), 2G12, 4F10, and AP2, suggesting the mutation exerted minimal structural effects. However, cells expressing αIIbD224Vβ3 did not bind MoAb PAC1 in the presence or absence of several activating antibodies, including anti-LIBS6 (Fig 1D), anti-LIBS1, anti-LIBS2, and AP5. The inability of these MoAbs to activate PAC1 binding was not due to a loss of antibody epitopes, because all of these MoAbs bound to αIIbD224Vβ3, as assayed by flow cytometry (data not shown). Furthermore, cells expressing αIIbD224Vβ3 exhibited minimal binding of the activation-independent ligand mimetic MoAb OPG2 compared with cells expressing wild-type αIIbβ3 (Fig 1B). These results confirm that substitution of αIIbD224 alone was sufficient for the functional defect.
Substitution of IIb amino acid residue D224 results in a loss of ligand binding function. FACS histograms depicting the binding of the IIbβ3complex-specific MoAb D57, the activation-independent ligand mimetic MoAb OPG2, or the ligand mimetic MoAb PAC1 to CHO cells transfected with wild-type IIbβ3 or IIbD224Vβ3. (A) MoAb D57 staining of cells transfected with IIbβ3 (shaded histogram) or IIbD224Vβ3 (open histogram). Untransfected CHO cells are depicted by the dotted histogram. (B) MoAb OPG2 staining of the IIbβ3 (shaded histogram) or IIbD224Vβ3 (open histogram) transfected cells. Untransfected CHO cells are depicted by the dotted histogram. Cells expressing wild-type IIbβ3 (C) or IIbD224Vβ3 (D) were activated by incubation with 4 μmol/L anti-LIBS6 for 30 minutes followed by the addition of MoAb PAC1 (IgM). Cells were washed, stained with fluorescein-conjugated goat antimouse IgM for 30 minutes, and analyzed by FACS. The binding of PAC1was analyzed in the presence (open histogram) and absence (shaded histogram) of the competitve inhibitor Ro43-5054.
Substitution of IIb amino acid residue D224 results in a loss of ligand binding function. FACS histograms depicting the binding of the IIbβ3complex-specific MoAb D57, the activation-independent ligand mimetic MoAb OPG2, or the ligand mimetic MoAb PAC1 to CHO cells transfected with wild-type IIbβ3 or IIbD224Vβ3. (A) MoAb D57 staining of cells transfected with IIbβ3 (shaded histogram) or IIbD224Vβ3 (open histogram). Untransfected CHO cells are depicted by the dotted histogram. (B) MoAb OPG2 staining of the IIbβ3 (shaded histogram) or IIbD224Vβ3 (open histogram) transfected cells. Untransfected CHO cells are depicted by the dotted histogram. Cells expressing wild-type IIbβ3 (C) or IIbD224Vβ3 (D) were activated by incubation with 4 μmol/L anti-LIBS6 for 30 minutes followed by the addition of MoAb PAC1 (IgM). Cells were washed, stained with fluorescein-conjugated goat antimouse IgM for 30 minutes, and analyzed by FACS. The binding of PAC1was analyzed in the presence (open histogram) and absence (shaded histogram) of the competitve inhibitor Ro43-5054.
The failure of αIIbD224Vβ3 to bind the αIIbβ3-specific ligands PAC1 and OPG2 could represent the loss of αIIbβ3-specific ligand recognition or the loss of ligand binding function in general. To discriminate between these two possibilities, the RGD recognition function of αIIbD224Vβ3 was examined by affinity chromatography. Lysates of αIIbD224Vβ3-expressing or wild-type αIIbβ3-expressing cells were applied to a GRGDSPK Sepharose column, washed, and then eluted with GRGDSPK peptide. Eluted fractions were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and then immunoblotted with anti-αIIb or anti-β3 MoAbs (Fig 2). Wild-type αIIbβ3 bound to the matrix and was specifically eluted by GRGDSPK peptide. In contrast, the mutant αIIbD224Vβ3 showed only minimal binding to the affinity matrix. The failure of αIIbD224Vβ3 to bind to the RGD peptide indicates that this substitution results not only in the loss of binding of αIIbβ3-specific ligands such as PAC1 and OPG2, but also the loss of binding to ligands shared in common with other integrins.
Binding of IIbβ3 and IIbD224Vβ3 to a GRGDSPK-Sepharose 4B column. Lysates of CHO cells stably transfected with the indicated receptors were loaded onto a 1 mL GRGDSPK-Sepharose column. The column was washed then eluted with 1 mmol/L GRGDS peptide (3 mL). One-milliliter column fractions were collected, resolved on a 8% nonreducing polyacrylamide gel, transfered onto nitrocelluose, and immunoblotted with the IIb-specific MoAb 98DF6 (3.5 μg/mL). L, lysate; FT, flow through; CE, RGD eluted fraction.
Binding of IIbβ3 and IIbD224Vβ3 to a GRGDSPK-Sepharose 4B column. Lysates of CHO cells stably transfected with the indicated receptors were loaded onto a 1 mL GRGDSPK-Sepharose column. The column was washed then eluted with 1 mmol/L GRGDS peptide (3 mL). One-milliliter column fractions were collected, resolved on a 8% nonreducing polyacrylamide gel, transfered onto nitrocelluose, and immunoblotted with the IIb-specific MoAb 98DF6 (3.5 μg/mL). L, lysate; FT, flow through; CE, RGD eluted fraction.
Site-directed mutagenesis and analysis.
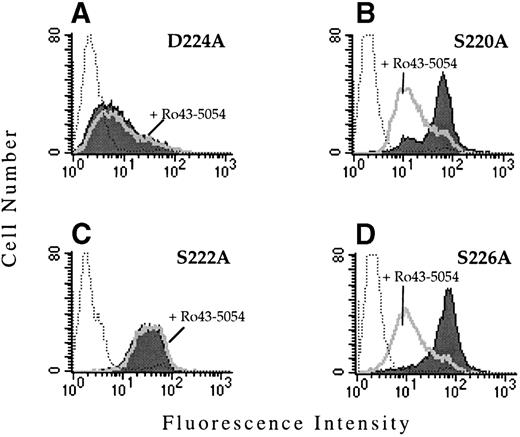
To determine whether the observed functional defect was due to the substitution with valine, site-directed mutagenesis was used to replace D224 by alanine. When transiently transfected into CD3a cells, αIIbD224A formed a complex with β3 and was expressed on the cell surface as assayed by flow cytometry. However, similar to cells expressing the αIIbD224Vβ3mutant, cells expressing αIIbD224Aβ3 did not bind PAC1 in the presence or absence of the activating antibody, anti-LIBS6 (Fig 3A).
PAC1 binding to the IIbβ3-mutant receptors. The binding of the activation-dependent ligand mimetic antibody PAC1 to CHO cells transfected with the indicated IIb mutant and the β3 subunit was examined by flow cytometry. To determine PAC1 binding, transfected cells were incubated (activated) with 4 μmol/L anti-LIBS6, followed by the addition of PAC1. The binding of PAC1 was analyzed in the presence (open histogram) and absence (shaded histogram) of the competitve inhibitor Ro43-5054. Untransfected CHO cells stained with PAC1 in the presence of anti-LIBS6 are depicted by the dotted histogram.
PAC1 binding to the IIbβ3-mutant receptors. The binding of the activation-dependent ligand mimetic antibody PAC1 to CHO cells transfected with the indicated IIb mutant and the β3 subunit was examined by flow cytometry. To determine PAC1 binding, transfected cells were incubated (activated) with 4 μmol/L anti-LIBS6, followed by the addition of PAC1. The binding of PAC1 was analyzed in the presence (open histogram) and absence (shaded histogram) of the competitve inhibitor Ro43-5054. Untransfected CHO cells stained with PAC1 in the presence of anti-LIBS6 are depicted by the dotted histogram.
The region surrounding D224 consists of a number of amino acids containing oxygenated side chains, particularly serine. This grouping of serine residues resembles the alignment of Asp and Ser residues that constitute a portion of the MIDAS motif found in the I domains of certain integrin α subunits and in the related structure present in all integrin β subunits. A number of these serine residues (S220, S222, and S226) were replaced by alanine and the effect on ligand binding was examined after transient and stable transfection of CD3a cells. The receptors αIIbS220Aβ3, αIIbS222Aβ3, and αIIbS226Aβ3 were all expressed on the cell surface and retained their ability to bind MoAb PAC1 (Fig 3B through D). Interestingly, Ro43-5054 did not inhibit the binding of PAC-1 to αIIbS222Aβ3. The mechanism for this loss of inhibition is unknown.
Ligand binding specificity of αIIbβ3.
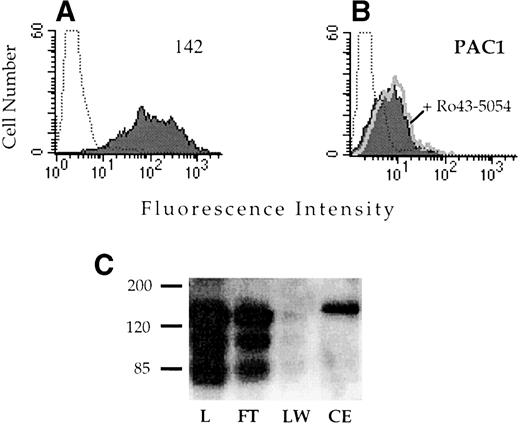
Alignment of the integrin α subunits shows that the region corresponding to αIIbD224 is not well conserved (Fig 4). Because other known ligand binding regions of the integrin receptors often exhibit a high degree of similarity, this lack of conservation suggested that this region might be important in determining the ligand recognition specificity of αIIbβ3. To test this hypothesis, a chimeric α subunit was constructed by inserting exon 5 of αIIb, which contains D224, into the corresponding location in the related αv subunit. The αvαIIb chimeric subunit was stably expressed in CD3a cells and assayed for the capacity to bind the αIIbβ3-specific ligand PAC1. The chimeric receptor was well expressed on the cell surface (Fig 5A); however, these cells did not exhibit binding to PAC1 in the presence or absence of the activating antibody anti-LIBS6 (Fig 5B). Similarly, binding to the activation-independent ligand mimetic antibody OPG2 was not observed (data not shown). Introduction of the αIIb sequence into αv did not disrupt ligand binding in general, because the chimeric receptor retained the capacity to bind to a RGD affinity matrix (Fig 5C).
Alignment of integrin subunits between the type III and IV homologous repeats. The alignment was created using the UWGCG program “pretty.” The amino acid sequences of the integrin subunits are shown using the single letter code. Alanine substitutions are indicated (*). The boundaries of exon 5 of IIb are indicated by vertical lines. Bold letters indicate residues with oxygen containing side chains. CN, consensus sequence.
Alignment of integrin subunits between the type III and IV homologous repeats. The alignment was created using the UWGCG program “pretty.” The amino acid sequences of the integrin subunits are shown using the single letter code. Alanine substitutions are indicated (*). The boundaries of exon 5 of IIb are indicated by vertical lines. Bold letters indicate residues with oxygen containing side chains. CN, consensus sequence.
Analysis of the ligand binding function of the vIIbβ3 chimeric receptor. CHO cells stably transfected with vIIbβ3 were stained with the anti-v MoAb 142 (A) or the IIbβ3-specific ligand mimetic antibody PAC1 (B). To analyze PAC1 binding, cells were incubated with the activating antibody anti-LIBS6 and washed and then PAC1 binding was analyzed in the presence (open histogram) and absence (shaded histogram) of the inhibitor Ro43-5054. Stained, untransfected CHO cells are depicted by the dotted histogram. (C) Lysates of CHO cells stably transfected with vIIbβ3were loaded onto a 1-mL GRGDSPK-Sepharose column. The column was washed and then eluted with 1 mmol/L GRGDS peptide (3 mL). Fractions were resolved on a 8% nonreducing polyacrylamide gel, transferred to nitrocellulose, and immunoblotted with MoAb 142. L, lysate; FT, flow through; LW, last wash fraction; CE, eluted fraction.
Analysis of the ligand binding function of the vIIbβ3 chimeric receptor. CHO cells stably transfected with vIIbβ3 were stained with the anti-v MoAb 142 (A) or the IIbβ3-specific ligand mimetic antibody PAC1 (B). To analyze PAC1 binding, cells were incubated with the activating antibody anti-LIBS6 and washed and then PAC1 binding was analyzed in the presence (open histogram) and absence (shaded histogram) of the inhibitor Ro43-5054. Stained, untransfected CHO cells are depicted by the dotted histogram. (C) Lysates of CHO cells stably transfected with vIIbβ3were loaded onto a 1-mL GRGDSPK-Sepharose column. The column was washed and then eluted with 1 mmol/L GRGDS peptide (3 mL). Fractions were resolved on a 8% nonreducing polyacrylamide gel, transferred to nitrocellulose, and immunoblotted with MoAb 142. L, lysate; FT, flow through; LW, last wash fraction; CE, eluted fraction.
DISCUSSION
We report the identification of a novel mutation in the integrin αIIb subunit (D224→V) that results in loss of αIIbβ3 adhesion receptor function. Unlike most of the mutations identified in the αIIb subunit, this mutation did not significantly affect expression of αIIbβ3 on the cell surface. However, αIIbD224Vβ3 did not bind the activation-dependent ligand mimetic antibody, PAC1, and bound only minimally to the activation-independent ligand mimetic antibody, OPG2. Substitution of D224 by alanine also resulted in a loss of receptor function. Insertion of exon 5 of αIIb containing D224 into the backbone of the αv subunit did not enable the resulting chimera to bind αIIbβ3-specific ligands; however, it did not interfere with ligand binding to αvβ3. These data suggest that αIIbD224 may play an important role in ligand binding specific to αIIbβ3.
An unbiased genetic approach was used to identify chemically induced mutations that result in the loss of αIIbβ3ligand binding activity. A key feature of this approach is the ability to isolate cells that expressed αIIbβ3 on the cell surface but failed to bind the activation-dependent, ligand mimetic antibody PAC1.21 Whereas the majority of the isolated clones contained substitutions in the β3subunit, PAC1 binding was restored in one clone only after retransfection with the parental α subunit. This mutant receptor contained a single basepair change resulting in substitution of D224 by valine. This receptor was expressed on the cell surface as assayed with a panel of anti αIIbβ3 MoAbs, indicating that the substitution likely had minimal effects on the processing or structure of this subunit. This finding is in sharp contrast to most of the identified naturally occurring mutations in αIIb that significantly reduce or abolish receptor expression.1The effect of the mutation at D224 on ligand binding to αIIbβ3 was not restricted to αIIbβ3-specific ligands such as PAC1 and OPG2. This mutation also significantly decreased the binding of a RGD peptide. Thus, this mutation affects the binding of ligands specific to αIIbβ3 and the binding of ligands shared in common with other integrins.
D224 is located in a group of serine residues distributed in a pattern that resembles a portion of the MIDAS motif known to be important for cation and ligand binding in the I domain of integrin α subunits and in a related motif in the integrin β subunits.8,9,11-13,38 39 However, substitution of several of these serine residues (S220, S222, and S226) with alanine did not result in the loss of PAC1 binding, suggesting that they do not directly play a critical role in the ligand binding process. Surprisingly, the substitution of S222 did appear to inhibit the capacity of the peptidomimetic Ro43-5054 to block PAC1 binding. The mechanism of the effect of substitution of D224 on ligand binding is unclear. It is unlikely to be the result of a gross structural alteration, because the mutant receptor bound a panel of complex-specific anti-αIIbβ3 antibodies. Other possibilities include direct interaction of D224 with ligand or, alternatively, through an indirect method by interacting cooperatively with other regions present in αIIb or in the β3 subunit. The precise functional assignment of this residue cannot be defined without high resolution structural data for the αIIbβ3 receptor.
The ligand recognition specificity of the integrins is determined in large part by the α subunits. In the case of αIIbβ3, the specificity of ligand recognition maps to the first 334 residues of the αIIbsubunit.40 The location of D224 is consistent with this data. Alignment between the integrin α subunits demonstrates that the region surrounding D224 is not well conserved, suggesting that this region of αIIb may be involved in a specific rather than general role of ligand recognition. Previous work from this laboratory had demonstrated that a larger region of αIIb (ie, amino acids R140-P334) did not confer αIIbβ3ligand binding specificity to an αvαIIbchimeric α subunit.40 In the present study, a smaller chimera (αIIb amino acids 193-235) was constructed consistent with αIIb intron/exon boundaries. Although this chimeric integrin consisting of the αv subunit and exon 5 of αIIb was well expressed, it did not bind the αIIbβ3-specific ligand PAC1. Moreover, disruption of the native αv region with the αIIb sequences did not abolish the binding of RGD peptides to this chimera. Thus, the region of αIIb that contains D224 may fully substitute for the analogous αvsequences or the analogous αv region is not similarly important for ligand binding to αvβ3. Although the lack of conservation between the α subunits in this region makes it difficult to accurately align residues with D224, these data do indicate that D224 is important for ligand binding function of αIIbβ3.
A structural model for the N-terminal approximately 440 amino acids of the integrin α subunits has recently been proposed.22This model predicts that the seven homologous sequence repeats found in the α subunits adopt the fold of a β-propeller domain. Enzymes with known β-propeller folds have their active sites at the top of the β-propeller, typically where adjacent loops run in opposite directions, such as seen between strands 2 and 3 within a β sheet and strands 4 and 1 between two sheets. The residue identified in this study, αIIbD224, lies between repeats III and IV in the αIIb subunit and is located in the large 4-1 loop region located at the top of the β propeller between the W3 and W4 β sheets. In a previous study, the αIIb region, G184-193, was implicated as important for ligand binding and is located in the adjacent 2-3 loop found in the W3 sheet at the top of the β-propeller.20 Thus, these two loop regions could act synergistically to form a portion of the ligand binding pocket between the αIIb and β3 subunits. The recent identification of an αIIb L183→P mutation in a Glanzmann’s thrombasthenia patient41 that produces quantitative and qualitative abnormalities in the receptor also substantiates the importance of this region in ligand binding. The identification of residues critical for α4β1-ligand interactions14 and for the binding of fibronectin to the integrin α5β116 further supports a role for the loop structures of the α subunits on the upper face of the predicted β-propeller model in ligand binding specificity.
The region identified in this study is located between the homologous repeats III and IV found in all integrin α subunits. Interestingly, this region is known to be alternatively spliced in α6and α7 and has been postulated to be alternatively spliced in α3.42,43 In α6 and α7, alternative splicing produces two variants, one of which (the α7X2 variant) contains a Asp residue homologous to D224 in αIIb (see Fig 4). The α7 splice variants are both developmentally regulated and tissue specific. Although no functional significance has yet been determined for these different variants, it has been speculated that this region may be involved in ligand specificity and/or different ligand binding affinities. Alternative splicing between the third and fourth repeats has also been reported for the Drosophila integrin PS2α subunit.44 45 Together, the differences among such variants could suggest a general function for this region in ligand recognition specificity consistent with the low conservation of this region among different α subunits.
In summary, this study has identified a novel region in αIIb that is important for ligand binding. This location of this region is within a predicted ligand binding site in the β propeller model proposed for the α subunits. This region is not highly conserved among the α subunits, and substitutions in the analogous regions of αv did not affect ligand binding function of αvβ3. These results indicate that this region of αIIb may play a role in ligand binding that is specific to αIIbβ3.
Supported in part by National Heart, Lung and Blood Institute Grant No. HL42977 (J.C.L.) and a fellowship from the National Institute of Health (1F32HL09321; to E.C.T.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Joseph C. Loftus, PhD, Mayo Clinic Scottsdale, 13400 E Shea Blvd, Scottsdale, AZ 85259; e-mail:loftus.joseph@mayo.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal