Abstract
It has been reported that reactivation of human herpesvirus-6 (HHV-6) causes a failure of hematopoiesis. To clarify the mechanisms of bone marrow suppression induced by HHV-6 infection, it is necessary to establish an in vitro model of HHV-6 infection in hematopoietic progenitor cells. We have established two novel Philadelphia chromosome–positive myeloid cell lines, SAS413 and SAS527, which possess different hematologic characteristics and show distinct susceptibility to infection by HHV-6, from a patient with blast crisis of chronic myelogenous leukemia (CML). HHV-6 subgroup A (HHV-6A) showed marked replication in SAS413, forming syncytia and inducing cell lysis in short-term culture. On the other hand, HHV-6A–inoculated SAS527 continued to proliferate without cell lysis and only a few cells showed HHV-6 antigen expression. In contrast to HHV-6A infection, inoculation with HHV-6 subgroup B (HHV-6B) did not induce any cytopathic effect (CPE) or viral antigen expression in either of the cell lines. Although HHV-6B replication was undetectable, the presence of the HHV-6 genome in both cell lines was shown by polymerase chain reaction (PCR) during culture for more than 10 months, suggesting that HHV-6B latently infected SAS413 and SAS527. Phorbol ester treatment of SAS527 latently infected with HHV-6B resulted in reactivation of HHV-6, as shown by the appearance of a CPE, positive reactivity for the HHV-6 antigen, and isolation of infectious HHV-6. These novel cell lines should be useful for studying the mechanisms of HHV-6–induced hematopoietic failure and HHV-6 latency and reactivation, as well as differentiation, of the myeloid cell lineage.
HUMAN HERPESVIRUS-6 (HHV-6) was first isolated from immunocompromised patients with lymphoproliferative disorders.1 HHV-6 is divided into two subgroups, HHV-6A and HHV-6B, on the basis of reactivity with monoclonal antibodies and HHV-6–specific T-cell clones and restriction enzyme cleavage patterns.2-4 Recent studies have shown that HHV-6 induces biologic alterations of T lymphocytes5,6 and transactivation of the human immunodeficiency virus type 1 (HIV-1) long terminal repeat.7,8 HHV-6 is thought to infect most individuals at an early age and to persistently infect throughout life, as with other HHVs. It has been established that HHV-6 is a causative agent of exanthem subitum,9 and various disorders are caused by reactivation of HHV-6. It has been reported recently that reactivation of HHV-6 occurs frequently in patients with immunodeficient conditions such as acquired immune deficiency syndrome and recipients of organ transplantation, and it causes bone marrow suppression in some patients.10-15 To clarify the mechanisms of hematopoietic failure mediated by HHV-6, it is necessary to establish an in vitro experimental model of HHV-6 infection of hematopoietic precursor cells.
We have established two novel myeloid cell lines, designated SAS413 and SAS527, from a patient with blast crisis of chronic myelogenous leukemia (CML). Although their bcr gene rearrangement patterns are identical, their growth patterns, cytochemistry, and surface phenotypes are different. In the present study, to establish an in vitro experimental system for HHV-6 infection of myeloid cells, we examined the characteristics of these cell lines with a focus on their susceptibility to infection with HHV-6A and HHV-6B. The results showed that SAS413 and SAS527 possess distinct characteristics of susceptibility to HHV-6A, and both support latent infection with HHV-6B. In addition, phorbol ester treatment of SAS527 that was latently infected with HHV-6B resulted in reactivation of HHV-6. These cell lines should therefore be useful for studying the mechanisms of HHV-6–induced hematopoietic failure and HHV-6 latency and reactivation in myeloid cells.
MATERIALS AND METHODS
Case history.
A 41-year-old man was admitted to Uwajima City Hospital on May 20, 1989 because of leukocytosis. A cytogenetic study of his bone marrow cells demonstrated the karyotype 46, XY, t(9;22)(q34;q11). The patient was diagnosed as being in the chronic phase of CML and was treated with interferon alfa, busulfan, and 6-mercaptopurine. He remained in stable condition until January 1994, when the number of blasts in peripheral blood and the size of the spleen increased rapidly. Karyotype analysis of the bone marrow cells demonstrated karyotype 46, XY, t(9;22)(q34;q11), −5, 17p+. The patient was diagnosed as having CML blast crisis and was treated with combined chemotherapy. Despite intensive chemotherapy, he died on May 31, 1994 due to intracranial hemorrhage.
Cell culture.
Peripheral blood mononuclear cells, of which greater than 90% were leukemic blasts, were isolated by Ficoll-Conray gradient centrifugation on April 13 and May 27, 1994. The cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) in 16-mm wells at 37°C in a 5% CO2 incubator. Half of the culture medium was exchanged with fresh medium every 3 or 4 days. Rapid cell growth was observed about 1 month after initiation of cell culture. The growing cells were maintained in RPMI 1640 supplemented with 10% FCS for more than 3 years, and were designated SAS413 and SAS527, respectively.
Cytochemical staining.
Cytocentrifuged cell preparations were stained with May-Grünwald-Giemsa solution, myeloperoxidase, naphthol AS-D chloroacetate esterase, and α-naphthyl butyrate esterase. Expression of terminal deoxynucleotidyl transferase (TdT) was examined by immunofluorescence.
Chromosome analysis.
Chromosome analysis was performed on bone marrow cells and established cell lines as described previously.16 Chromosomes were banded by the trypsin-Giemsa method.
Southern blot analysis.
DNA samples prepared from peripheral blood mononuclear cells, of which greater than 90% were leukemic cells, and established cell lines were digested with restriction enzymes and size-fractionated on 0.8% agarose gel. The DNA was then transferred to nylon filters, and hybridized with a 32P-labeled 1.2-kbHindIII-BglII bcr gene fragment.
Cell-surface molecule expression.
Expression of surface molecules on peripheral blood leukemic blasts and cell lines was examined by direct and indirect immunofluorescence using a flow cytometer. Control cells for background fluorescence were stained with fluorescein isothiocyanate (FITC)-conjugated mouse Ig or FITC-conjugated goat anti-mouse Ig. The analysis gate was set to include 99% of the control cells, and the percentage of positive cells was recorded.
Inoculation of cells with viruses.
The U1102 strain of HHV-6A and the Z29 strain of HHV-6B were grown in cord blood mononuclear cells prestimulated with phytohemagglutinin (PHA).9 The cells were inoculated with HHV-6A or HHV-6B at a multiplicity of infection of approximately 1 50% tissue culture infective dose. Virus-inoculated cells were cultured continuously in RPMI 1640 supplemented with 10% FCS for more than 10 months, and virus replication and the presence of the virus genome were monitored repeatedly during the culture. Effects of HHV-6 inoculation on cell growth were examined by calculating viable cells periodically using the trypan blue exclusion test.
Detection of virus replication.
The appearance of a cytopathic effect (CPE) of HHV-6 was examined with an inverted microscope. The indirect immunofluorescence assay for detection of HHV-6 antigen was performed using HHV-6–seropositive human serum as described previously.17 Briefly, virus-infected cells were mounted on glass slides and fixed in cold acetone. HHV-6–seropositive human serum diluted 20-fold was applied to the slide, followed by incubation for 30 minutes at 37°C. After washing, FITC-conjugated goat anti-human IgG (Organon Teknika, Durham, NC) was added and incubated for 30 minutes at 37°C. After washing, the slides were examined using a fluorescence microscope.
Transmission electron microscopy.
Transmission electron microscopy of SAS413 and SAS527 was performed as described previously.18 Briefly, the cells were fixed with 2.0% glutaraldehyde in 0.1 mol/L phosphate buffer (pH 7.4), postfixed with 1% osmium tetroxide, and gradually dehydrated. Samples were embedded in Epon 812, sectioned, stained with uranyl acetate and lead citrate, and examined with an H-800 electron microscope (Hitachi Co, Ibaragi, Japan).
Detection of HHV-6 genome.
A polymerase chain reaction (PCR) assay for the HHV-6 genome was performed as described previously.19 The primers used were as follows: 5′-GTGTTTCCATTGTACTGAAACCGGT-3′ and 5′-TAAACATCAATGCGTTGCATACAGT-3′. The samples were amplified through 35 cycles. Annealing was performed at 60°C for 90 seconds, extension at 72°C for 120 seconds, and denaturation at 95°C for 60 seconds. The expected product of HHV-6 DNA from the use of these primers is 776 bp. We also used the following primers to examine the presence of the HHV-7 genome in the cell lines: 5′-TATCCCAGCTGTTTTCATATAGTAAC-3′ and 5′-GCCTTGCGGTAGCACTAGATTTTTTG-3′. The expected product of HHV-7 DNA from the use of these primers is 186 bp.
Detection of HHV-6 mRNA expression.
Expression of mRNA for the HHV-6 immediate-early gene was investigated by reverse transcriptase (RT)-PCR. Total RNAs were extracted from HHV-6–inoculated cell lines, and cDNA was synthesized by reverse transcription with Moloney murine leukemia virus RT. cDNA amplification by PCR was performed using the following primers: 5′-TTCTCCAGATGTGCCAGGGAAATCC-3′ and 5′-CATCATTGTTATCGCTTTCACTCTC-3′. The expected length of the amplified cDNA sequences for HHV-6A and HHV-6B was 325 and 553 bp, respectively.20 21 Amplification of cDNA for the β-actin gene was also performed as the control for RT-PCR using the following primers: 5′-TCCTGTGGCATCCACGAAACT-3′ and 5′-GAAGCATTTGCGGTGGACGAT-3′.
Treatment of cell lines with phorbol ester.
It is well known that some virus types can be reactivated from latency by cell stimulation with phorbol ester.22-25 Accordingly, we attempted to isolate HHV-6 from the cell lines that were infected latently with HHV-6B. The cell lines were cultured in RPMI 1640 medium containing 10% FCS with or without the phorbol ester 12-0-tetradecanoylphorbol-13-acetate ([TPA] Sigma Chemical Co, St Louis, MO) at a concentration of 1.6 × 10−7 mol/L for 7 days.
Isolation of HHV-6.
Isolation of HHV-6 from cell lines was performed as described previously.9 Briefly, the virus-infected cell lines were frozen and thawed and then sonicated. The cell lysates were then added to cord blood lymphocytes prestimulated with PHA for 3 days. The cells were cultured in RPMI 1640 medium supplemented with 10% FCS, and HHV-6 replication was monitored as already described.
RESULTS
Morphology and cytochemical studies.
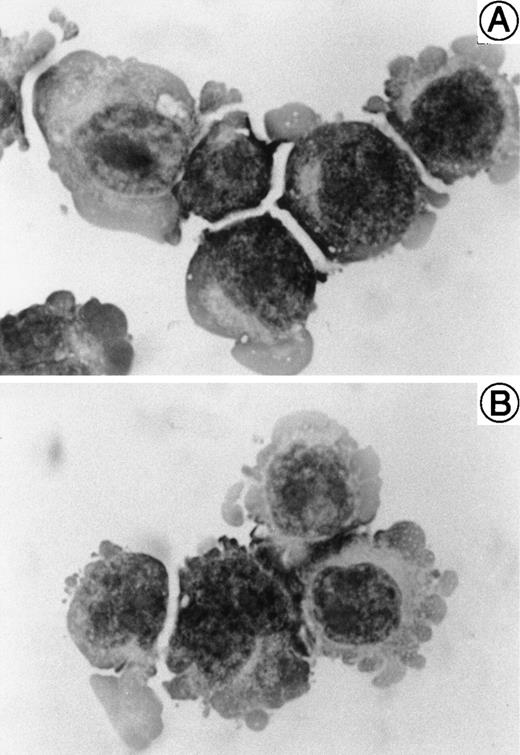
The morphology of SAS413 and SAS527 is shown in Fig 1. SAS413 grew as a single-cell suspension with slight aggregation, and most SAS527 cells showed adherent growth on the surface of a plastic flask. May-Grünwald-Giemsa staining of SAS413 showed a myeloblastic appearance, with basophilic cytoplasm and a round nucleus without any cytoplasmic granules. SAS527 was monoblastic in appearance, with irregular-shaped cytoplasm and a convoluted nucleus. Results for cytochemical studies of SAS413 were negative for myeloperoxidase, negative for naphthol AS-D chloroacetate esterase, weakly positive for α-naphthyl butyrate esterase, and negative for TdT, and results for SAS527 were negative for myeloperoxidase, negative for naphthol AS-D chloroacetate esterase, weakly positive for α-naphthyl butyrate esterase, and weakly positive for TdT.
Cell-surface molecule expression.
Surface molecule expression of the original leukemic blasts and HHV-6–infected and –uninfected SAS413 and SAS527 is shown in Table 1. The original leukemic blasts were positive for CD13, CD14, and CD33, suggesting that the lineage of the CML blast crisis was myelomonocytoid. SAS413 showed expression of a megakaryocyte-associated antigen, CD41a, and an erythroblast-associated antigen, glycophorin A, as well as the myelomonocytoid-associated antigens CD13, CD14, and CD33, suggesting that this cell line may have the characteristics of a multipotent hematopoietic precursor cell. In contrast, SAS527 was positive for CD9, CD13, CD14, CD33, CD34, and CD56. No apparent changes were observed in either SAS413 or SAS527 following infection with HHV-6A or HHV-6B, except for increased expression of CD21 and glycophorin A on SAS413 induced by HHV-6A and HHV-6B infection during short-term culture and downregulation of CD34 on HHV-6A– and HHV-6B–infected SAS527 during long-term culture.
Surface Phenotype of the Cell Lines
| Surface Molecule . | Leukemic Blasts . | SAS413 . | SAS527 . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Uninfected . | HHV-6A+ . | HHV-6B+ . | Uninfected . | HHV-6A+ . | HHV-6B+ . | |||||
| Day 12 . | Day 12 . | Day 210 . | Day 12 . | Day 210 . | Day 12 . | Day 210 . | ||||
| CD2 | 0 | 0.9 | 1.3 | 0.1 | 0 | 1.6 | 0.8 | 0.1 | 0.8 | 0.1 |
| CD3 | 0 | 0 | 0.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CD4 | 0 | 0 | 0.4 | 0.2 | 0 | 0.2 | 0 | 0 | 0 | 0 |
| CD5 | 0 | 0.4 | 0.7 | 0.2 | 0 | 1.0 | 0.7 | 0 | 0.6 | 0 |
| CD7 | 0 | 0.2 | 0.3 | 0.3 | 0.1 | 1.4 | 0.8 | 0.2 | 0.5 | 0.2 |
| CD8 | 0 | 0.5 | 0.9 | 0.1 | 0 | 1.1 | 0.4 | 0 | 0.7 | 0 |
| CD9 | 0.4 | 20.0 | 27.8 | 13.4 | 36.5 | 97.6 | 96.3 | 96.0 | 92.6 | 93.7 |
| CD10 | 9.1 | 2.0 | 0.1 | 0.3 | 0.1 | 5.4 | 7.3 | 0.7 | 4.5 | 1.2 |
| CD11b | 0 | 0 | 0 | 0.2 | 0 | 5.2 | 5.7 | 0 | 9.9 | 0 |
| CD13 | 86.8 | 80.0 | 70.7 | 86.9 | 77.1 | 94.7 | 91.9 | 99.5 | 91.2 | 99.8 |
| CD14 | 46.5 | 30.9 | 41.5 | 43.5 | 37.6 | 22.3 | 33.5 | 21.0 | 31.5 | 24.9 |
| CD16 | 0.2 | 0.3 | 0.3 | 3.2 | 0.1 | 0.1 | 1.4 | 0 | 1.3 | 0 |
| CD19 | 0 | 0.1 | 0.3 | 0.2 | 0 | 0.4 | 0.4 | 0 | 0.5 | 0 |
| CD20 | 0.2 | 0.2 | 0.3 | 2.9 | 0 | 1.8 | 1.8 | 0.4 | 2.2 | 0.4 |
| CD21 | 19.9 | 0.5 | 15.9 | 29.4 | 0.1 | 0.9 | 0.1 | 0 | 0.2 | 0.1 |
| CD25 | 0 | 0.4 | 0.2 | 0.1 | 0 | 7.6 | 2.3 | 0 | 2.6 | 0 |
| CD33 | 99.8 | 99.5 | 99.4 | 99.5 | 99.8 | 92.7 | 99.0 | 97.5 | 99.2 | 98.9 |
| CD34 | 0 | 0 | 0 | 0.2 | 0 | 93.4 | 97.8 | 61.8 | 97.8 | 63.1 |
| CD41a | 15.4 | 34.4 | 30.6 | 30.5 | 58.5 | 5.6 | 1.6 | 5.5 | 3.1 | 8.3 |
| CD56 | 0.4 | 0.1 | 0.5 | 0.2 | 0 | 94.9 | 99.8 | 98.5 | 99.9 | 99.3 |
| HLA-DR | 0.2 | 0.4 | 1.0 | 1.0 | 0.1 | 4.5 | 5.2 | 0 | 5.8 | 0 |
| Glycophorin A | ND | 76.3 | 95.0 | 99.0 | 73.5 | 4.1 | 8.1 | 1.8 | 8.3 | 3.3 |
| Surface Molecule . | Leukemic Blasts . | SAS413 . | SAS527 . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Uninfected . | HHV-6A+ . | HHV-6B+ . | Uninfected . | HHV-6A+ . | HHV-6B+ . | |||||
| Day 12 . | Day 12 . | Day 210 . | Day 12 . | Day 210 . | Day 12 . | Day 210 . | ||||
| CD2 | 0 | 0.9 | 1.3 | 0.1 | 0 | 1.6 | 0.8 | 0.1 | 0.8 | 0.1 |
| CD3 | 0 | 0 | 0.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CD4 | 0 | 0 | 0.4 | 0.2 | 0 | 0.2 | 0 | 0 | 0 | 0 |
| CD5 | 0 | 0.4 | 0.7 | 0.2 | 0 | 1.0 | 0.7 | 0 | 0.6 | 0 |
| CD7 | 0 | 0.2 | 0.3 | 0.3 | 0.1 | 1.4 | 0.8 | 0.2 | 0.5 | 0.2 |
| CD8 | 0 | 0.5 | 0.9 | 0.1 | 0 | 1.1 | 0.4 | 0 | 0.7 | 0 |
| CD9 | 0.4 | 20.0 | 27.8 | 13.4 | 36.5 | 97.6 | 96.3 | 96.0 | 92.6 | 93.7 |
| CD10 | 9.1 | 2.0 | 0.1 | 0.3 | 0.1 | 5.4 | 7.3 | 0.7 | 4.5 | 1.2 |
| CD11b | 0 | 0 | 0 | 0.2 | 0 | 5.2 | 5.7 | 0 | 9.9 | 0 |
| CD13 | 86.8 | 80.0 | 70.7 | 86.9 | 77.1 | 94.7 | 91.9 | 99.5 | 91.2 | 99.8 |
| CD14 | 46.5 | 30.9 | 41.5 | 43.5 | 37.6 | 22.3 | 33.5 | 21.0 | 31.5 | 24.9 |
| CD16 | 0.2 | 0.3 | 0.3 | 3.2 | 0.1 | 0.1 | 1.4 | 0 | 1.3 | 0 |
| CD19 | 0 | 0.1 | 0.3 | 0.2 | 0 | 0.4 | 0.4 | 0 | 0.5 | 0 |
| CD20 | 0.2 | 0.2 | 0.3 | 2.9 | 0 | 1.8 | 1.8 | 0.4 | 2.2 | 0.4 |
| CD21 | 19.9 | 0.5 | 15.9 | 29.4 | 0.1 | 0.9 | 0.1 | 0 | 0.2 | 0.1 |
| CD25 | 0 | 0.4 | 0.2 | 0.1 | 0 | 7.6 | 2.3 | 0 | 2.6 | 0 |
| CD33 | 99.8 | 99.5 | 99.4 | 99.5 | 99.8 | 92.7 | 99.0 | 97.5 | 99.2 | 98.9 |
| CD34 | 0 | 0 | 0 | 0.2 | 0 | 93.4 | 97.8 | 61.8 | 97.8 | 63.1 |
| CD41a | 15.4 | 34.4 | 30.6 | 30.5 | 58.5 | 5.6 | 1.6 | 5.5 | 3.1 | 8.3 |
| CD56 | 0.4 | 0.1 | 0.5 | 0.2 | 0 | 94.9 | 99.8 | 98.5 | 99.9 | 99.3 |
| HLA-DR | 0.2 | 0.4 | 1.0 | 1.0 | 0.1 | 4.5 | 5.2 | 0 | 5.8 | 0 |
| Glycophorin A | ND | 76.3 | 95.0 | 99.0 | 73.5 | 4.1 | 8.1 | 1.8 | 8.3 | 3.3 |
The expression of surface molecules on SAS413 and SAS527 was examined by flow cytometric analysis at day 12 or day 210 after inoculation with HHV-6A or HHV-6B. Results are expressed as the percentage of positive cells.
Abbreviation: ND, not determined.
Genetic analyses.
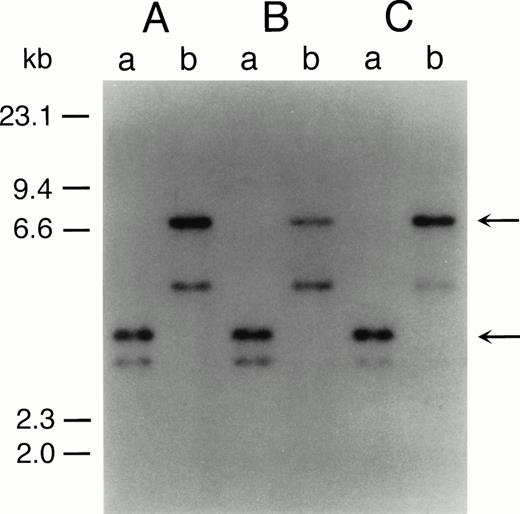
The representative karyotypes of SAS413 and SAS527 were 60, XY, +1, +6, +6, +8, +8, der(9)t(9;22)(q34;q11) × 2, +10, der(11)t(3;11)(p21;p13), +12, −18, +19, +19, +20, +20, +21, +21, −22, +2mar and 56, XY, +1, +6, i(8)(q10), +i(8), t(9;22)(q34;q11), +der(9)t(9;22), +add(10)(q22), +13, +add(17)(p11), add(18)(p11), +19, +19, add(19)(q13), +21, respectively. Identical rearranged bands were detected by Southern blot analysis of the bcr gene in all of the original leukemic cells and SAS413 and SAS527, indicating that both SAS413 and SAS527 originated from leukemic cells (Fig 2).
Southern blot analysis of DNA from original leukemic cells (A), SAS413 (B), and SAS527 (C). DNA was digested withBamHI (a) or BglII (b) and hybridized to a 3′bcr probe. Arrows indicate rearranged bands.
Southern blot analysis of DNA from original leukemic cells (A), SAS413 (B), and SAS527 (C). DNA was digested withBamHI (a) or BglII (b) and hybridized to a 3′bcr probe. Arrows indicate rearranged bands.
Replication of HHV-6A and HHV-6B in the cell lines.
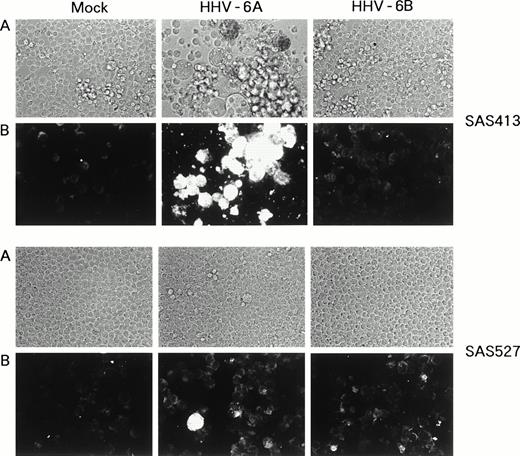
SAS413 appeared to show a typical CPE of HHV-6 after 4 days of inoculation with HHV-6A (Fig 3). At day 6 of virus inoculation, greater than 30% of the cells showed positive immunofluorescence for HHV-6 antigen expression. Inoculation of SAS413 with HHV-6A produced large ballooning cells and resulted in cell lysis 15 days after virus inoculation. On the other hand, a CPE was scarcely detected in HHV-6A–inoculated SAS527 during culture for more than 10 months. Immunofluorescence assays performed on days 6 and 240 after HHV-6A inoculation showed that only one in 500 to 1,000 SAS527 cells were positive for HHV-6 antigen expression. In contrast to HHV-6A, inoculation with HHV-6B did not induce any CPE in either SAS413 or SAS527, and no HHV-6 antigen expression was detected in HHV-6B–inoculated SAS413 or SAS527 during long-term culture.
Replication of HHV-6 in SAS413 and SAS527. HHV-6–inoculated SAS413 and SAS527 were observed with an inverted microscope to detect CPE (A) and analyzed by indirect immunofluorescence with HHV-6–seropositive human serum (B). Analyses were performed at day 6 of virus inoculation for HHV-6A–inoculated SAS413 and at day 240 for HHV-6B–inoculated SAS413, HHV-6A–inoculated SAS527, and HHV-6B–inoculated SAS527.
Replication of HHV-6 in SAS413 and SAS527. HHV-6–inoculated SAS413 and SAS527 were observed with an inverted microscope to detect CPE (A) and analyzed by indirect immunofluorescence with HHV-6–seropositive human serum (B). Analyses were performed at day 6 of virus inoculation for HHV-6A–inoculated SAS413 and at day 240 for HHV-6B–inoculated SAS413, HHV-6A–inoculated SAS527, and HHV-6B–inoculated SAS527.
Effects of HHV-6 inoculation on cell growth.
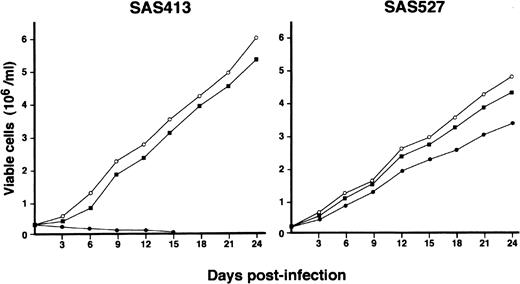
The growth of both SAS413 and SAS527 did not change following inoculation with HHV-6B (Fig 4). On the other hand, SAS527 growth decreased following inoculation with HHV-6A, although the cells were able to proliferate continuously. As detected using an inverted microscope, the growth of SAS413 was severely impaired by inoculation with HHV-6A. Two weeks after inoculation with HHV-6A, greater than half of the SAS413 cells became moribund, showing an apparent CPE.
Growth curve of SAS413 and SAS527. The growth of mock-infected (○), HHV-6A–inoculated (•), and HHV-6B–inoculated (▪) SAS413 and SAS527 was monitored by counting viable cells using the trypan blue exclusion test.
Growth curve of SAS413 and SAS527. The growth of mock-infected (○), HHV-6A–inoculated (•), and HHV-6B–inoculated (▪) SAS413 and SAS527 was monitored by counting viable cells using the trypan blue exclusion test.
Detection of HHV-6 by transmission electron microscopy.
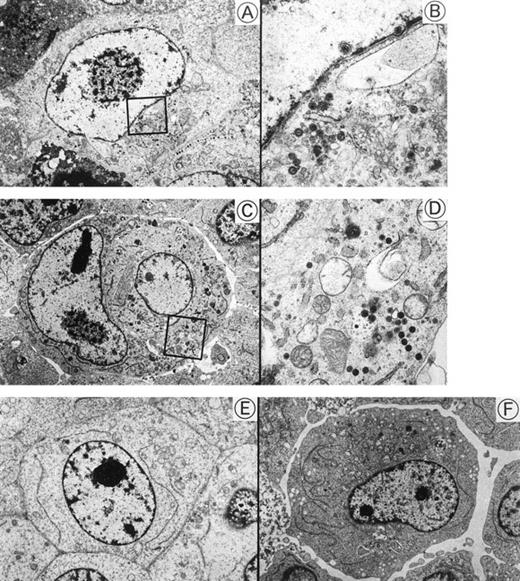
Transmission electron micrographs of HHV-6–inoculated SAS413 and SAS527 are shown in Fig 5. Many HHV-6 particles were observed in the cytoplasm and nuclei of SAS413 at day 6 of HHV-6A inoculation when a marked CPE was detected. HHV-6 particles were also detected in HHV-6A–inoculated SAS527 by electron microscopy on day 240 after virus inoculation, although the frequency of cells containing virus particles was markedly lower versus HHV-6A–inoculated SAS413. On the other hand, no virus structures were detected in either SAS413 or SAS527 after inoculation with HHV-6B. These results were identical to those obtained by the immunofluorescence assays.
Transmission electron microscopy of HHV-6–inoculated SAS413 and SAS527. Note that many HHV-6 particles are present in HHV-6A–inoculated SAS413 (A and B) and SAS527 (C and D), whereas no virus is detectable in HHV-6B–inoculated SAS413 (E) and SAS527 (F). The highly magnified boxed areas in (A) and (C) are shown in (B) and (D), respectively. HHV-6A–inoculated SAS413 cells were examined at day 6 of virus inoculation, and HHV-6B–inoculated SAS413, HHV-6A–inoculated SAS527, and HHV-6B–inoculated SAS527 cells were examined at day 240.
Transmission electron microscopy of HHV-6–inoculated SAS413 and SAS527. Note that many HHV-6 particles are present in HHV-6A–inoculated SAS413 (A and B) and SAS527 (C and D), whereas no virus is detectable in HHV-6B–inoculated SAS413 (E) and SAS527 (F). The highly magnified boxed areas in (A) and (C) are shown in (B) and (D), respectively. HHV-6A–inoculated SAS413 cells were examined at day 6 of virus inoculation, and HHV-6B–inoculated SAS413, HHV-6A–inoculated SAS527, and HHV-6B–inoculated SAS527 cells were examined at day 240.
Detection of HHV-6 genome in the cell lines.
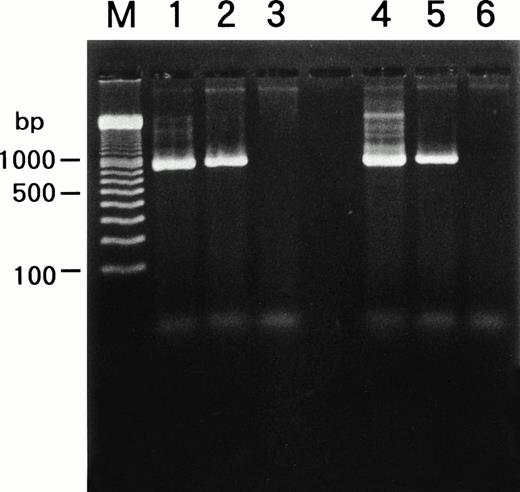
We examined whether HHV-6B infected SAS413 and SAS527 or persistently infected these cell lines using a PCR for the HHV-6 genome (Fig6). PCR was performed repeatedly during long-term culture, and the same results were obtained. HHV-6A and HHV-6B genomes were not detected in the original SAS413 or SAS527. Similarly, the genome for HHV-7, a virus closely related to HHV-6, was not detected in either of the cell lines (data not shown). As expected, HHV-6 DNA was detected in HHV-6A–infected SAS413. Similarly, the HHV-6 genome was detected in SAS527 cultured for more than 10 months after HHV-6A inoculation. Although apparent CPE and HHV-6 antigen expression were not detected in HHV-6B–inoculated SAS413 and SAS527, the HHV-6 genome was detected in both cell lines that were cultured for more than 10 months after HHV-6B inoculation. These data suggest that HHV-6A productively infected and caused cell death in SAS413 and persistently infected SAS527 during long-term culture. On the other hand, HHV-6B latently infected both SAS413 and SAS527 without viral expression.
Detection of HHV-6 genome in SAS413 and SAS527 by PCR. A PCR product of the expected size was detected in HHV-6A–inoculated SAS413 (lane 1), HHV-6B–inoculated SAS413 (lane 2), HHV-6A–inoculated SAS527 (lane 4), and HHV-6B–inoculated SAS527 (lane 5), but not in uninfected SAS413 (lane 3) or SAS527 (lane 6). DNA was extracted from HHV-6A–inoculated SAS413 at day 6 after virus inoculation, and from HHV-6B–inoculated SAS413, HHV-6A–inoculated SAS527, and HHV-6B–inoculated SAS527 at day 240. Lane M shows marker DNAs.
Detection of HHV-6 genome in SAS413 and SAS527 by PCR. A PCR product of the expected size was detected in HHV-6A–inoculated SAS413 (lane 1), HHV-6B–inoculated SAS413 (lane 2), HHV-6A–inoculated SAS527 (lane 4), and HHV-6B–inoculated SAS527 (lane 5), but not in uninfected SAS413 (lane 3) or SAS527 (lane 6). DNA was extracted from HHV-6A–inoculated SAS413 at day 6 after virus inoculation, and from HHV-6B–inoculated SAS413, HHV-6A–inoculated SAS527, and HHV-6B–inoculated SAS527 at day 240. Lane M shows marker DNAs.
Expression of HHV-6 mRNA.
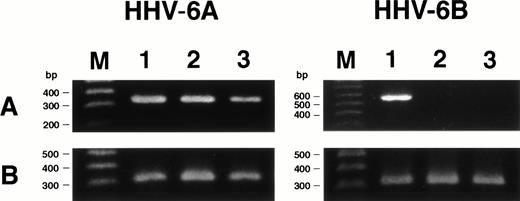
We examined whether HHV-6 infected SAS413 and SAS527 latently without HHV-6 gene expression or whether transcription of the HHV-6 gene occurred in virus-inoculated cell lines using RT-PCR for the immediate-early gene. As expected, mRNA for the immediate-early gene was detected in HHV-6A–inoculated SAS413 and SAS527, whereas it was not detectable in HHV-6B–inoculated SAS413 or SAS527 (Fig7). These results suggest that HHV-6A infected productively and HHV-6B infected latently in these cell lines.
Expression of HHV-6 mRNA in SAS413 and SAS527. (A) Expression of the HHV-6 immediate-early gene in the cell lines was examined by RT-PCR. cDNA synthesized from cord blood mononuclear cells (lane 1), SAS413 (lane 2), and SAS527 (lane 3) inoculated with HHV-6A or HHV-6B was amplified using primers corresponding to the immediate-early gene. (B) RT-PCR for β-actin gene. Lane M shows marker DNAs.
Expression of HHV-6 mRNA in SAS413 and SAS527. (A) Expression of the HHV-6 immediate-early gene in the cell lines was examined by RT-PCR. cDNA synthesized from cord blood mononuclear cells (lane 1), SAS413 (lane 2), and SAS527 (lane 3) inoculated with HHV-6A or HHV-6B was amplified using primers corresponding to the immediate-early gene. (B) RT-PCR for β-actin gene. Lane M shows marker DNAs.
Reactivation of HHV-6B by treatment with phorbol ester.
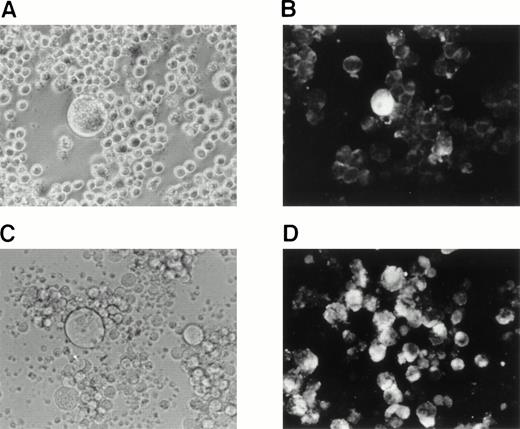
We sought to determine whether HHV-6B could be reactivated in SAS413 and SAS527 latently infected with HHV-6B. As it has been reported that phorbol ester treatment of cells latently infected with virus results in reactivation of the virus, we attempted to reactivate HHV-6B in SAS413 and SAS527 by treatment with the phorbol ester TPA. Experiments were performed 120 and 200 days after HHV-6B inoculation, and identical results were obtained. After 7 days of TPA treatment, a CPE was detected in SAS527 inoculated with HHV-6B (Fig8A). Immunofluorescence assays showed that about one in 300 cells were positive for HHV-6 antigen expression (Fig8B). Transmission electron microscopy also revealed HHV-6 particles in the cytoplasm of SAS527 (data not shown). In addition, HHV-6B was isolated from TPA-treated, but not from untreated, SAS527 by cocultivation with PHA-stimulated cord blood lymphocytes as demonstrated by the appearance of a marked CPE and positive reactivity with anti–HHV-6 antibody using indirect immunofluorescence (Fig 8C and D). As expected, although morphologic change was induced, no HHV-6 antigen was detected in mock-infected SAS527 after treatment with TPA. Treatment of HHV-6A–infected SAS527 with TPA resulted in the augmentation of HHV-6A replication as determined by the appearance of a CPE and positive reactivity to indirect immunofluorescence (data not shown). In contrast to SAS527, typical CPE or HHV-6 antigen expression was hardly detected in HHV-6B–inoculated SAS413 even after treatment with TPA (data not shown).
Reactivation of HHV-6B in SAS527. HHV-6B–inoculated SAS527 cells were cultured in the presence of TPA at a concentration of 1.6 × 10−7 mol/L for 7 days. The cells were then observed with an inverted microscope for detection of CPE (A) and analyzed by indirect immunofluorescence with HHV-6–seropositive human serum (B). The cells were also sonicated and added to PHA-stimulated cord blood lymphocytes. After 6 days, the cells were observed with an inverted microscope for detection of CPE (C) and analyzed by indirect immunofluorescence with HHV-6–seropositive human serum (D).
Reactivation of HHV-6B in SAS527. HHV-6B–inoculated SAS527 cells were cultured in the presence of TPA at a concentration of 1.6 × 10−7 mol/L for 7 days. The cells were then observed with an inverted microscope for detection of CPE (A) and analyzed by indirect immunofluorescence with HHV-6–seropositive human serum (B). The cells were also sonicated and added to PHA-stimulated cord blood lymphocytes. After 6 days, the cells were observed with an inverted microscope for detection of CPE (C) and analyzed by indirect immunofluorescence with HHV-6–seropositive human serum (D).
DISCUSSION
HHV-6 was originally defined as a B-lymphotropic virus,1but subsequent studies showed that it preferentially infects CD4+ T lymphocytes.26 The presence of HHV-6 DNA sequences has also been shown in several cases of lymphoid neoplasm.21,27-30 Although it has been reported that nonlymphoid cell lines also support the replication of HHV-6,31-35 HHV-6 infectivity against myeloid cell lines has not been extensively examined. The present study shows that both HHV-6A and HHV-6B can infect myeloid cells with different replication patterns. Recent studies have shown that the major sites for latent infection of HHV-6 in vivo are monocytes24 and salivary glands.19 The present data demonstrating that HHV-6 can infect myeloid cell lines suggest that myeloid precursor cells in bone marrow also support HHV-6 latency. The finding that HHV-6 is frequently isolated from bone marrow cells of posttransplant patients36 supports this hypothesis.
Although the HHV-6A and HHV-6B genomes were detected in both SAS413 and SAS527, their replication patterns in these cell lines were distinctly different. That is, HHV-6A markedly replicated in SAS413 with a remarkable CPE and persistently infected SAS527 with little CPE, whereas HHV-6B latently infected both cell lines without apparent viral replication. Although the infectivity of HHV-6A against various cell lines has been shown, it has been reported that lytic infection of HHV-6A occurs only in T lymphocytes. SAS413 is the first myeloid cell line reported to permit lytic infection of HHV-6A. Viral replication requires various factors derived from host cells. Thus, analysis of SAS413 may provide important information to clarify the intracellular factors essential for HHV-6 replication.
Activation of HHV-6 is frequently observed in patients who have received bone marrow transplantation, and this inhibits marrow engraftment.11,12,15,37,38 Although the precise mechanisms of bone marrow suppression mediated by HHV-6 are still obscure, frequent isolation of HHV-6 from bone marrow and the suppressive effect of HHV-6 on in vitro colony formation by bone marrow cells13,14 suggest that direct infection of hematopoietic precursor cells with HHV-6 may cause suppression of bone marrow progenitor cell differentiation. Since it has been reported that the vast majority of HHV-6 isolates from posttransplant patients are subgroup B,36,39 the SAS413 and SAS527 cell lines latently infected with HHV-6B seem to be a useful model to investigate the mechanisms of bone marrow suppression mediated by HHV-6B. It has been reported that phorbol ester treatment of cells latently infected with virus occasionally results in viral reactivation.22-25 In view of these previous findings, we attempted to induce HHV-6B reactivation in these cell lines by stimulation with the phorbol ester TPA. HHV-6B was indeed reactivated, and the infectious virus was isolated from TPA-treated SAS527 but not from SAS413. The unsuccessful isolation of HHV-6B from SAS413 may be due to the fact that SAS527, but not SAS413, has monocyte-lineage characteristics, since HHV-6 is known to show tropism to monocytes.24 This is the first experimental system using cell lines for HHV-6 reactivation, and it should provide important information for clarifying the mechanisms of HHV-6 reactivation and bone marrow suppression induced by HHV-6 reactivation.
It has been shown that the expression of various cell surface molecules is altered following infection with HHV-6. For example, de novo expression of CD4 is induced in CD4− lymphocytes by HHV-6A infection, rendering them susceptible to infection with HIV-1.40-42 Induction of CD4 mRNA synthesis and surface CD4 expression by HHV-6 infection has also been reported recently in the lymphomyeloid progenitor cell line KG-1.43 Although examination after long-term culture was not performed, KG-1 cells inoculated with HHV-6 showed HHV-6 antigen expression without a CPE or loss of cell viability, resembling HHV-6–inoculated SAS527. In light of these previous reports, we examined the expression levels of various surface molecules on SAS413 and SAS527, before and after HHV-6 infection. Although a slight increase of CD21 and glycophorin A expression and downregulation of CD34 were observed on SAS413 and SAS527, respectively, following infection with HHV-6A and HHV-6B, no apparent alteration of other surface molecules, including CD4 expression, was detected. Although the reason for the different effects of HHV-6 infection on CD4 expression in KG-1 and our present cell lines is unknown, it may be due to the dependency of de novo CD4 expression induced by HHV-6 infection on certain intracellular factor(s) produced by specific cells.
In summary, we have established the first myeloid cell line susceptible to lytic infection with HHV-6A and two myeloid cell lines that have contained the HHV-6B genome for more than 10 months without apparent CPE or HHV-6 antigen expression. We have also succeeded in reactivating HHV-6B from one of the cell lines by treatment with phorbol ester. These cell lines should be useful for two avenues of research. One is the mechanism of myeloid lineage differentiation, since these cell lines possess distinct hematologic characteristics with an identical genetic background. The other is the mechanism of HHV-6 latent infection and reactivation in myeloid precursor cells. Further investigations using these cell lines should provide important information to clarify these important issues in hematology and virology.
ACKNOWLEDGMENT
We gratefully acknowledge Drs J. Nicholas, J.B. Black, and C. Lopez for providing the viruses. We also thank M. Shudo for technical support.
Supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan, and by the Mochida Foundation for Medical and Pharmaceutical Research, the Inamori Foundation, and the Suzuken Memorial Foundation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Masaki Yasukawa, MD, PhD, First Department of Internal Medicine, Ehime University School of Medicine, Shigenobu, Ehime 791-0295, Japan; e-mail: yasukawa@m.ehime-u.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal