To the Editor:
The hereditary stomatocytosis syndromes (HSt) are a heterogeneous group of disorders characterized by mouth-shaped (stomatocytic) erythrocyte morphology on peripheral blood smear.1,2 The clinical severity of HSt patients is variable; some patients experience hemolysis and anemia, whereas others are asymptomatic. The red blood cell membranes of HSt patients usually exhibit abnormal permeability to the cations sodium and potassium, with consequent modification of intracellular water content.1 2
One variant of HSt, dehydrated stomatocytosis (also known as xerocytosis), is an autosomal dominant disorder characterized by a net loss of monovalent cations, resulting in shrunken, dehydrated red blood cells.1-3 The peripheral blood smear of patients with dehydrated HSt exhibits variable morphology with target cells, xerocytes, and usually, but not always, stomatocytes. Erythrocytes from these patients demonstrate increased mean corpuscular hemoglobin concentrations and decreased osmotic fragility after incubation. In general, erythrocyte dehydration is mediated by the loss of intracellular potassium with an accompanying loss of water.4 The underlying abnormality leading to abnormal cation permeability and red blood cell dehydration in these patients is unknown.
The two major pathways mediating potassium efflux in erythrocytes are via electroneutral K-Cl cotransport and via an intermediate conductance, voltage insensitive, calcium-activated potassium channel, the Gardos channel.4 The Gardos channel appears to play a major role in volume regulation of both normal and sickle erythrocytes. An intermediate conductance, calcium-activated potassium channel, hIK1, has recently been identified.5-7 Biophysical and pharmacological studies of hIK1 make it a candidate for the Gardos channel. The murine homologue, mIK1, is expressed in spleen, in MEL cells, and in ES cells induced to undergo erythroid differentiation.8
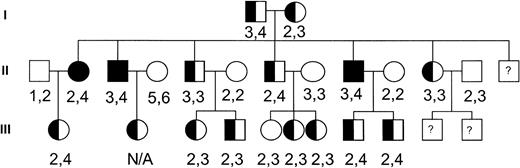
To investigate the potential role of this channel in the pathogenesis of dehydrated HSt, we performed genotyping of the hIK1 locus on genomic DNA samples from members of a large, well-characterized dehydrated HSt kindred with members spanning three generations.9 The HSt phenotype was assigned to family members as previously described.9 Genomic DNA isolated from peripheral blood lymphocytes was polymerase chain reaction (PCR) amplified and analyzed using standard techniques. The primers used in PCR were 5′-CTGGGGCAGGAGCACT-3′ and 5′-GCTTACCAAACCTAAAGGATGTC-3′. These primers correspond to D19S420 (GDB Accession ID: 199991), a locus located in the 3′ untranslated region of the hIK1 cDNA that has been localized to chromosome 19q13.2. They amplify a fragment approximately 262 bp in length. Amplified alleles were assigned and the genotyping results were assessed (Fig 1). There were no hIK1 gene alleles that cosegregated with the disease phenotype. Based on these observations, the hIK1 gene was excluded as a candidate disease gene in this kindred. Because there is great heterogenity in the clinical and biochemical characteristics of the dehydrated HSt syndromes, it will be important to perform genetic linkage to include or exclude this locus in other HSt kindreds.
Pedigree of the dehydrated HSt kindred. Solid symbols indicate affected individuals. Numbers correspond to hIK1 gene alleles amplified from genomic DNA of each individual.
Pedigree of the dehydrated HSt kindred. Solid symbols indicate affected individuals. Numbers correspond to hIK1 gene alleles amplified from genomic DNA of each individual.
The precise genetic basis(es) of the dehydrated HSt syndromes is unknown. Recently, linkage of dehydrated HSt to chromosome 16 has been identified in a three-generation Irish family.10
ACKNOWLEDGMENT
This work was supported in part by grants from the Yale Children’s Clinical Research Center, the National Institutes of Health, and the NHLBI Mammalian Genotyping Service.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal