Abstract
Combined factors V and VIII deficiency is an autosomal recessive bleeding disorder associated with plasma levels of coagulation factors V and VIII approximately 5% to 30% of normal. The disease gene was recently identified as the endoplasmic reticulum-Golgi intermediate compartment protein ERGIC-53 by positional cloning, with the detection of two founder mutations in 10 Jewish families. To identify mutations in additional families, the structure of the ERGIC-53 gene was determined by genomic polymerase chain reaction (PCR) and sequence analysis of bacterial artificial chromosome clones containing the ERGIC-53 gene. Nineteen additional families were analyzed by direct sequence analysis of the entire coding region and the intron/exon junctions. Seven novel mutations were identified in 10 families, with one additional family found to harbor one of the two previously described mutations. All of the identified mutations would be predicted to result in complete absence of functional ERGIC-53 protein. In 8 of 19 families, no mutation was identified. Genotyping data indicate that at least two of these families are not linked to the ERGIC-53 locus. Taken together, these results suggest that a significant subset of combined factors V and VIII deficiency is due to mutation in one or more additional genes.
COMBINED DEFICIENCY of coagulation factors V and VIII is an autosomal recessive bleeding disorder with greater than 89 cases reported since the original description by Oeri et al in 1954.1-5 Affected individuals present with a moderate bleeding tendency and plasma levels of factors V and VIII (both antigen and activity) in the range of 5% to 30% of normal. Combined factors V and VIII deficiency was initially mapped to the long arm of chromosome 18.6-8 Recent positional cloning efforts identified the disease gene as ERGIC-53, a component of the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) of previously unknown function.9 This latter report identified two different founder mutations: (1) a G insertion in codon 30 resulting in a frameshift of the 510 amino acid protein product in all affected individuals from five Middle Eastern Jewish families and (2) a splice donor mutation in intron 9 in all affected individuals from five Sephardic Jewish families. Immunofluorescence studies and Western analysis using Epstein-Barr virus (EBV)-immortalized B lymphocytes from affected individuals confirmed the absence of detectable ERGIC-53 antigen.
Although the function of ERGIC-53 is unknown, it is a type 1 transmembrane protein with homology to leguminous lectins that exhibits mannose-selective and calcium-dependent binding and has been hypothesized to play a role in the transport of glycoproteins through the secretory pathway.10-14 The demonstration that its deficiency results in decreased plasma levels of coagulation factors V and VIII suggests an important role for ERGIC-53 in ER to Golgi transport and implies the existence of cargo-specific pathways within the secretory system. Vollenweider et al15have recently shown that mistargeting of ERGIC-53 to the ER impairs the secretion of the lysosomal enzyme cathepsin C, although transport of two other lysosomal enzymes and three post-Golgi membrane glycoproteins was unaffected. These results suggested that the recycling of ERGIC-53 between the ERGIC and ER is required for the intracellular transport of only a small subset of glycoproteins. The common features shared between coagulation factors V and VIII and other potential ERGIC-53–dependent proteins remain to be determined.
We now report the gene structure for ERGIC-53, as well as mutation analysis for an additional 19 combined factors V and VIII deficiency families. Seven new mutations were identified in 10 of 19 families. In all except one of these families, the affected individual(s) was homozygous for the mutation. One affected individual from a nonconsanguineous family is a compound heterozygote, although the mutation has been identified on only one allele. All of the mutations would predict complete deficiency of a normal ERGIC-53 product. In addition, one of the families was shown to carry the Middle Eastern Jewish founder mutation.9 Failure to identify a mutation in eight families, taken together with clear evidence against linkage to the ERGIC-53 locus in two of these families, suggests that a significant subset of combined factors V and VIII deficiency is due to mutations in one or more additional genes.
MATERIALS AND METHODS
ERGIC-53 Gene Structure
Polymerase chain reaction (PCR) and sequence analysis of bacterial artificial chromosomes and total human genomic DNA.
Either 2 nanograms of bacterial artificial chromosome (BAC) clones 293H21 or 65B23 (Research Genetics, Huntsville, AL) purified as previously described9 or 20 ng of total human genomic DNA was amplified in a 50-μL reaction containing 200 μmol/L each dNTP, 50 to 100 ng each primer, 1X PCR buffer (50 mmol/L KCl, 10 mmol/L Tris, pH 9.0, 1.5 mmol/L MgCl2, 0.01% gelatin, 0.1% Triton X-100), and Taq DNA polymerase. PCR was performed for 35 cycles using an MJ Research PTC-100 96V thermocycler (MJ Research, Watertown, MA). Forward and reverse primers were synthesized from the ERGIC-53 cDNA sequence (GenBank X71661) and used in various combinations to determine the location of introns. PCR products were separated by electrophoresis through agarose and visualized by ethidium bromide staining. Any PCR product whose length was longer than that expected for the cDNA was assumed to contain an intron and was subjected to DNA sequence analysis using the Thermo Sequenase radiolabelled terminator cycle sequencing kit (Amersham Life Science Products, Arlington Heights, IL).
Direct sequence analysis of BACs 293H21 and 65B23.
One microgram purified BAC DNA was directly sequenced using the Thermo Sequenase radiolabeled kit as above (Amersham Life Science Products) in a reaction containing the dITP nucleotide master mix and 20 ng of primer specific for the ERGIC-53 coding sequence. Numerous reactions were performed, each with a different specific primer. Cycling conditions were 35 cycles of denaturation at 94°C for 30 seconds, annealing at 50°C for 30 seconds, and extension at 60°C for 6 minutes. Sequencing products were electrophoresed through standard 8% sequencing gels at 100 W. After electrophoresis, the gels were fixed, dried, and exposed to Kodak Biomax film (Eastman Kodak, Rochester, NY).
Mutation analysis in combined factors V and VIII deficiency patients.
Twenty-forty nanograms of genomic DNA, prepared as previously described,6 from individuals affected with combined factors V and VIII deficiency (factors V and VIII antigen and activity levels <30% of normal) and their unaffected parents and siblings (when available) from 19 unrelated families were amplified by PCR. The entire coding region of ERGIC-53 was amplified in multiple PCR reactions using 25 ng (37.5 ng for exon 8) of the appropriate primers as listed in Table 1. Exons 2 and 3 were amplified in a single PCR product as were exons 9 and 10 using the forward primer for exon 2 or 9 and the reverse primer for exon 3 or 10. Only a portion of exon 13 through the stop codon was included in the amplification product. Cycling conditions were 1 minute each of denaturation, annealing, and elongation for 35 cycles using an MJ Research PTC-100 96V thermocycler (MJ Research). PCR products were separated by electrophoresis through 4% agarose (3% Nusieve; FMC Bioproducts, Rockland, ME, 1% BRL) to ensure presence of sufficient quantities for sequence analysis. PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Santa Clarita, CA) and eluted in 30 μL dH20. PCR products were sequenced on both strands, as described above, using the same primers as for the PCR reactions.
Haplotype analysis.
Haplotype analysis for short tandem repeat markers flanking the ERGIC-53 gene was performed as previously described.6Haplotype analysis using ERGIC-53 intragenic polymorphisms was performed by PCR and sequence analysis as described above.
RESULTS
ERGIC-53 Gene Structure
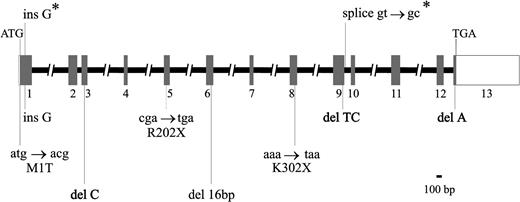
To determine the intron/exon boundaries of the ERGIC-53 gene, a combination of human genomic and BAC DNA PCR and direct sequence analysis of BAC DNA was used. In previous studies to clone the combined factors V and VIII deficiency gene, BAC clones 293H21 and 65B23, obtained by screening a human genomic DNA BAC library (Research Genetics), had been shown by Southern analysis to contain the largest portions of the ERGIC-53 gene (data not shown) and were therefore used in the PCR and direct sequencing studies.9 As seen in Fig 1 and Table 1, the ERGIC-53 gene contains 13 exons ranging in size from 59 bp (exon 7) to greater than 1,244 bp (exon 13). Exon 13 encodes only amino acids 499-510 with the remaining greater than 1,200 bp corresponding to the 3′ untranslated sequence (UTR). The full size of the 3′ UTR is unknown, as no polyadenylation signal is present in the available cDNA sequence.16 Exon 1 contains at least 21 bp of 5′ UTR, although the transcription start site has not been determined and the existence of an additional upstream noncoding exon cannot be excluded.
Structure of the ERGIC-53 gene and mutations identified in combined factors V and VIII deficiency. Exons, indicated by rectangles, are numbered from 1 to 13. The coding portion of the gene is shaded, with the open portions of exons 1 and 13 representing the 5′ UTR and 3′ UTR, respectively. The exons, as well as introns 2, 9, and 12, are shown to scale. The // in the remaining introns indicates that the size was not determined. The locations of the two founder mutations identified in 10 Jewish families9are shown above (indicated by *), while the eight mutations detected by sequence analysis in the 19 families of this study (see Table 2) are shown below the gene.
Structure of the ERGIC-53 gene and mutations identified in combined factors V and VIII deficiency. Exons, indicated by rectangles, are numbered from 1 to 13. The coding portion of the gene is shaded, with the open portions of exons 1 and 13 representing the 5′ UTR and 3′ UTR, respectively. The exons, as well as introns 2, 9, and 12, are shown to scale. The // in the remaining introns indicates that the size was not determined. The locations of the two founder mutations identified in 10 Jewish families9are shown above (indicated by *), while the eight mutations detected by sequence analysis in the 19 families of this study (see Table 2) are shown below the gene.
Location and Length of ERGIC-53 Exons and Sequences of Primers Used to PCR Complete Coding Region of ERGIC-53 From Genomic DNA
| Exon . | Nucleotides* . | Exon Length . | Primers . |
|---|---|---|---|
| 1 | ?-235 | 235+ | 5′-TCGCGTTCCAGAATCCAAG-3′ |
| 5′-TCAGCACACCAGGGTAGC-3′ | |||
| 2 | 236-390 | 155 | 5′-CAGTTTGGAAATGTACATTGAG-3′ |
| 5′-GGGAACAGTTAGAGGCTAG-3′ | |||
| 3 | 391-498 | 108 | 5′-CATGCCTCTAACTGTTCCC-3′ |
| 5′-CTCACAGCCTAACTCTGTTG-3′ | |||
| 4 | 499-560 | 62 | 5′-TGTAAGTCACTTCATAGTAC-3′ |
| 5′-CAATGTATTTCATAAGGATTCC-3′ | |||
| 5 | 561-660 | 100 | 5′-TGAAAAGCTGAGTGTCTTGT-3′ |
| 5′-GAAAAGTGATACTGTAACATTG-3′ | |||
| 6 | 661-784 | 124 | 5′-GAAACAAAACTGAATAGTAGTC-3′ |
| 5′-ACAAGTCTACATATCCCTAAv | |||
| 7 | 785-843 | 59 | 5′-AGAGTGCCATTGCCTTTACC-3′ |
| 5′-CAAACCTAAGTTAGTCTTCC-3′ | |||
| 8 | 844-976 | 132 | 5′-CATGTATAGAGATATCTTAATG-3′ |
| 5′-GTCCATGATCAACAGCCTC-3′ | |||
| 9 | 977-1170 | 194 | 5′-CACTTTGGTCACTTACGTTA-3′ |
| 5′-TCTATGAGCACATAGTACAG-3′ | |||
| 10 | 1171-1241 | 71 | 5′-GGGAAGTAAAGAAGAAGGGC-3′ |
| 5′-AATCACATAACACACAAACG-3′ | |||
| 11 | 1242-1395 | 154 | 5′-GTGATTTTATTGTATCAAGAG-3′ |
| 5′-AGTATGAGTTCTTCCTTTCC-3′ | |||
| 12 | 1396-1517 | 122 | 5′-GGGGATAGAAGGTTTTCTGG-3′ |
| 5′-GAACATAGATAACTTAGTTG-3′ | |||
| 13 | 1518-? | 1244+ | 5′-CTGTTCATTTCAGTTCACAT-3′ |
| 5′-AATTCCCTCAAAACGACATC-3′ |
| Exon . | Nucleotides* . | Exon Length . | Primers . |
|---|---|---|---|
| 1 | ?-235 | 235+ | 5′-TCGCGTTCCAGAATCCAAG-3′ |
| 5′-TCAGCACACCAGGGTAGC-3′ | |||
| 2 | 236-390 | 155 | 5′-CAGTTTGGAAATGTACATTGAG-3′ |
| 5′-GGGAACAGTTAGAGGCTAG-3′ | |||
| 3 | 391-498 | 108 | 5′-CATGCCTCTAACTGTTCCC-3′ |
| 5′-CTCACAGCCTAACTCTGTTG-3′ | |||
| 4 | 499-560 | 62 | 5′-TGTAAGTCACTTCATAGTAC-3′ |
| 5′-CAATGTATTTCATAAGGATTCC-3′ | |||
| 5 | 561-660 | 100 | 5′-TGAAAAGCTGAGTGTCTTGT-3′ |
| 5′-GAAAAGTGATACTGTAACATTG-3′ | |||
| 6 | 661-784 | 124 | 5′-GAAACAAAACTGAATAGTAGTC-3′ |
| 5′-ACAAGTCTACATATCCCTAAv | |||
| 7 | 785-843 | 59 | 5′-AGAGTGCCATTGCCTTTACC-3′ |
| 5′-CAAACCTAAGTTAGTCTTCC-3′ | |||
| 8 | 844-976 | 132 | 5′-CATGTATAGAGATATCTTAATG-3′ |
| 5′-GTCCATGATCAACAGCCTC-3′ | |||
| 9 | 977-1170 | 194 | 5′-CACTTTGGTCACTTACGTTA-3′ |
| 5′-TCTATGAGCACATAGTACAG-3′ | |||
| 10 | 1171-1241 | 71 | 5′-GGGAAGTAAAGAAGAAGGGC-3′ |
| 5′-AATCACATAACACACAAACG-3′ | |||
| 11 | 1242-1395 | 154 | 5′-GTGATTTTATTGTATCAAGAG-3′ |
| 5′-AGTATGAGTTCTTCCTTTCC-3′ | |||
| 12 | 1396-1517 | 122 | 5′-GGGGATAGAAGGTTTTCTGG-3′ |
| 5′-GAACATAGATAACTTAGTTG-3′ | |||
| 13 | 1518-? | 1244+ | 5′-CTGTTCATTTCAGTTCACAT-3′ |
| 5′-AATTCCCTCAAAACGACATC-3′ |
Nucleotide numbers are based on the Genbank file X71661.
Direct sequence analysis enabled the determination of the size and full sequence for three introns. Introns 2, 9, and 12 were completely sequenced (GenBank Accession numbers AF081867, AF081880, and AF081885) and are 83, 132, 290 bp, respectively. PCR analysis indicates that intron 1 is approximately 4 kb in length. The sizes of the remaining introns were not determined. All sequence data obtained to date have been deposited in GenBank under accession numbers AF081865, AF081866,AF081867, AF081868, AF081869, AF081870, AF081871, AF081872, AF081873,AF081874, AF081875, AF081876, AF081877, AF081878, AF081879, AF081880,AF081881, AF081882, AF081883, AF081884, and AF081885.
Mutation Analysis in Combined Factors V and VIII Deficiency Patients
We previously reported the identification of two founder mutations in the ERGIC-53 gene in 10 Israeli families with combined factors V and VIII deficiency.9 In the current study, we have analyzed DNA samples from 19 additional unrelated families/individuals with the disorder, including 7 families from Italy, 4 from Venezuela, 3 from Japan,17 3 from the United States, and 1 each from France and Turkey. The entire coding region of the ERGIC-53 gene and all intron/exon junctions were sequenced in all affected individuals, as well as in selected obligate carrier parents and unaffected siblings of the patients when available (a total of 39 individuals).
Results of the sequence analysis are summarized in Table 2. A total of eight different mutations were identified, accounting for 11 of 19 families. Seven of the eight mutations are either nonsense or frameshift mutations resulting in a truncated protein that would be predicted to lack normal ERGIC-53 function. All but one of the affected individuals is homozygous for their respective mutations, consistent with a high frequency of consanguinity in these families. The affected individual in family 2 is heterozygous for the exon 3 frameshift mutation with no candidate mutation detected on the other allele, either in the ERGIC-53 coding region or the sequences flanking the intron/exon junctions. The mutation detected in family 11, ins G85-89, is the same mutation previously reported in five Middle Eastern Jewish families.9 Haplotype analysis of DNA markers in this region demonstrated that the mutation in the affected individual in this family, also of Middle Eastern Jewish origin (Iranian-Jewish) is carried on the same haplotype,6 consistent with a common origin (data not shown).
ERGIC-53 Mutation Analysis in 19 Combined Factors V and VIII Deficiency Families
| Family . | Country of Origin . | ERGIC-53 Gene Mutation* . | Amino Acid Substitution . |
|---|---|---|---|
| 1 | Japan | C604 → T, exon 5 | Arg202 → stop |
| 2 | Japan | del C422, exon 3† | Frameshift at aa 141 |
| 3 | Japan | Unknown | |
| 4 | Italy | T2 → C, exon 1 | Met1 → Thr |
| 5 | Italy | Unknown | |
| 6 | Italy | Unknown | |
| 7 | Italy | T2 → C, exon 1 | Met1 → Thr |
| 8 | Italy | Unknown | |
| 9 | Italy | T2 → C, exon 1 | Met1 → Thr |
| 10 | Italy | T2 → C, exon 1 | Met1 → Thr |
| 11 | Venezuela (Iranian Jewish) | ins G85-89, exon 1 | Frameshift at aa 31 |
| 12 | Venezuela | del A720-A735 (16 bp), exon 6 | Frameshift at aa 240 |
| 13 | Venezuela | Unknown | |
| 14 | Venezuela | Unknown | |
| 15 | France | A904 → T, exon 8 | Lys302 → stop |
| 16 | Turkey | Unknown | |
| 17 | United States (Armenian) | del A1519-1524, exon 13 | Frameshift at aa 508 |
| 18 | United States | del TC1109-1112, exon 9 | Frameshift at aa 371 |
| 19 | United States | Unknown |
| Family . | Country of Origin . | ERGIC-53 Gene Mutation* . | Amino Acid Substitution . |
|---|---|---|---|
| 1 | Japan | C604 → T, exon 5 | Arg202 → stop |
| 2 | Japan | del C422, exon 3† | Frameshift at aa 141 |
| 3 | Japan | Unknown | |
| 4 | Italy | T2 → C, exon 1 | Met1 → Thr |
| 5 | Italy | Unknown | |
| 6 | Italy | Unknown | |
| 7 | Italy | T2 → C, exon 1 | Met1 → Thr |
| 8 | Italy | Unknown | |
| 9 | Italy | T2 → C, exon 1 | Met1 → Thr |
| 10 | Italy | T2 → C, exon 1 | Met1 → Thr |
| 11 | Venezuela (Iranian Jewish) | ins G85-89, exon 1 | Frameshift at aa 31 |
| 12 | Venezuela | del A720-A735 (16 bp), exon 6 | Frameshift at aa 240 |
| 13 | Venezuela | Unknown | |
| 14 | Venezuela | Unknown | |
| 15 | France | A904 → T, exon 8 | Lys302 → stop |
| 16 | Turkey | Unknown | |
| 17 | United States (Armenian) | del A1519-1524, exon 13 | Frameshift at aa 508 |
| 18 | United States | del TC1109-1112, exon 9 | Frameshift at aa 371 |
| 19 | United States | Unknown |
Nucleotide numbers are based on the Genbank file X71661 using the A (nucleotide 22) of the ATG initiatior methionine as +1.
Patient is heterozygous for this mutation with no mutation detected on the other allele.
A single, common mutation was identified in four of the seven Italian families in our patient cohort (Table 2). The affected individuals in these four families were homozygous for the only missense mutation observed in our study. This results in the substitution of threonine for the initiator methionine. The next methionine codon is not encountered until exon 2 and is out of frame. This mutation should thus result in the complete absence of ERGIC-53 expression in these patients. In all four of the families, the mutation is carried on a shared haplotype, again suggesting a founder effect.
A number of additional sequence differences also detected in the ERGIC-53 gene were identified as likely polymorphisms or rare sequence variants (Table 3). Most of these changes were observed in numerous unrelated families and are likely to represent common polymorphisms. The intron 6 polymorphism appears to be in complete linkage disequilibrium with the intron 7 polymorphism; that is, the two polymorphisms are always inherited together with no individual identified to carry only one.
Polymorphisms of the ERGIC-53 Gene
| Location . | Polymorphism . |
|---|---|
| Exon 1-A | G31 → A, Arg14 → Gln |
| Exon 1-B | C116 → T, Val39 → Arg |
| Exon 2 | G351 → A, silent |
| Intron 4 | del GT(+5 → +12) |
| Intron 63-150 | ins T in pyrimidine tract T12 at 3′ of intron (−10 → −21) |
| Intron 73-150 | ins TGGT(+40) |
| Exon 8 | C936 → T, silent |
| Exon 11 | A1228 → T, Met410 → Leu |
| Intron 11 | C(+14) → T |
| Location . | Polymorphism . |
|---|---|
| Exon 1-A | G31 → A, Arg14 → Gln |
| Exon 1-B | C116 → T, Val39 → Arg |
| Exon 2 | G351 → A, silent |
| Intron 4 | del GT(+5 → +12) |
| Intron 63-150 | ins T in pyrimidine tract T12 at 3′ of intron (−10 → −21) |
| Intron 73-150 | ins TGGT(+40) |
| Exon 8 | C936 → T, silent |
| Exon 11 | A1228 → T, Met410 → Leu |
| Intron 11 | C(+14) → T |
These two intron polymorphisms are in complete linkage disequilibrium.
Of note, all chromosomes sequenced to date contain a thymine at nucleotide 457, not the adenine reported in the published cDNA16, resulting in a serine residue at position 153 in place of the reported threonine. An independently reported sequence for a mannose-specific lectin termed MR6012 is identical to ERGIC-53 except for the same serine for threonine substitution at position 153. This suggests a sequencing or cloning artifact in the original report, although a rare sequence variant in the source of the cDNA library cannot be excluded.
Haplotype Analysis in ERGIC-53 Mutation–Negative Families
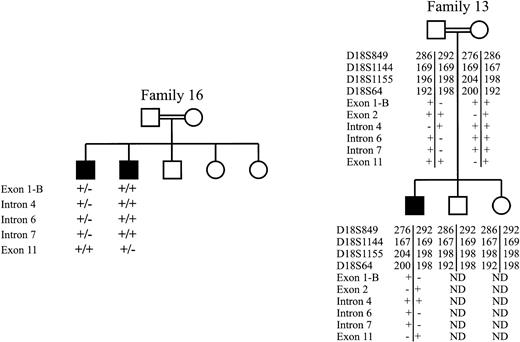
Shown in Fig 2 is haplotype analysis for two of the eight families in our study for which no mutation was detected in the entire ERGIC-53 coding sequence or intron sequences flanking the intron/exon junctions. The affected siblings in family 16 inherited different ERGIC-53 alleles from their parents, indicating lack of linkage to the ERGIC-53 gene. The parents of the affected individual in family 13 are first cousins, making it likely that the patient is homozygous for the same mutant allele inherited from the carrier parents. However, the patient is heterozygous for four short tandem repeat markers flanking the ERGIC-53 gene, as well as five of six intragenic polymorphisms. These results indicate that the mutations responsible for combined factors V and VIII deficiency in these two families are unlikely to lie within or near the ERGIC-53 gene.
Haplotype analysis for two families in which no ERGIC-53 mutation was detected. Affected individuals (shaded) in family 16, shown at the left, were determined to differ at five ERGIC-53 intragenic polymorphisms (Table 3). The + indicates the presence of the more common sequence, while the − indicates the presence of the polymorphism or rare sequence variant. Individuals in family 13, shown at the right, were genotyped for four short tandem repeat polymorphisms flanking the ERGIC-53 gene.6 9 Numbers represent basepair length of the PCR products generated with primers flanking the repeat sequence. Both parents and the affected individuals were also typed for six intragenic polymorphisms (Table 3).
Haplotype analysis for two families in which no ERGIC-53 mutation was detected. Affected individuals (shaded) in family 16, shown at the left, were determined to differ at five ERGIC-53 intragenic polymorphisms (Table 3). The + indicates the presence of the more common sequence, while the − indicates the presence of the polymorphism or rare sequence variant. Individuals in family 13, shown at the right, were genotyped for four short tandem repeat polymorphisms flanking the ERGIC-53 gene.6 9 Numbers represent basepair length of the PCR products generated with primers flanking the repeat sequence. Both parents and the affected individuals were also typed for six intragenic polymorphisms (Table 3).
DISCUSSION
We report the identification of seven novel mutations in the ERGIC-53 gene in patients with combined factors V and VIII deficiency. Mutations were identified in 11 of 19 families studied. One founder mutation, by virtue of haplotype analysis, was identified in four of seven Italian families. All patients, except one, are homozygous for the respective mutation, consistent with the high frequency of consanguinity in these families. The large number of families reported here and in the accompanying report18 suggests that combined factors V and VIII deficiency may be considerably more frequent than originally suspected. However, consistent with previous reports,2-5the majority of families in both studies are of Mediterranean/Middle Eastern origin. Taken together with the evidence for a founder effect among Middle Eastern Jews9 (and this study), Sephardic Jews,9 Italians (this study), Iranians,18 and Pakistanis,18 these data suggest the possibility of a specific positive selection for heterozygotes among individuals in these geographic areas, perhaps resistance to a regional infectious pathogen, as has been elegantly demonstrated for the hemoglobinopathies.19
ERGIC-53 is a type 1 transmembrane protein that constitutively recycles between the ER, the ERGIC, and cis Golgi.10,14,16The recent identification of ERGIC-53 as the gene responsible for combined factors V and VIII deficiency provided the first direct evidence that this protein might function as a sorting receptor in the ER for a specific subset of secretory proteins.9 The C-terminal sequence of ERGIC-53 (507LysLysPhePhe510) contains a dilysine ER retrieval signal.20 Kappeler et al21 have demonstrated by site directed mutagenesis that the two terminal phenylalanine residues are required for exit of ERGIC-53 from the ER. These signals also cooperate with lumenal and transmembrane domains in the normal intracellular trafficking of ERGIC-53. Vollenweider et al15 recently studied a recombinant mutant ERGIC-53 in which AlaAla had replaced the diphenylalanine motif. This mutant was prevented from recycling and accumulated in the ER, exerting a dominant-negative effect on the endogenous normal ERGIC-53. A selective delay in secretion of the lysosomal protein cathepsin C was observed, consistent with the proposed role of ERGIC-53 as a specific sorting receptor. The absence of any other apparent disease phenotype in combined factors V and VIII deficiency patients, outside of the bleeding disorder, suggests that the defect in the targeting of cathepsin C or other similar proteins may be too subtle to produce a significant functional abnormality.
All eight mutations reported here result in premature termination or truncation removing the LysLysPhePhe motif. The most distal of these mutations, a one base deletion in exon 13, results in a frameshift at amino acid 508 (Table 2, family 17). This mutation substitutes LysAsnSerPhe for the LysLysPhePhe motif, followed by 14 additional amino acids before the next stop codon. Despite the presence of greater than 99% (507 of 510 amino acids) of the ERGIC-53 protein, the predicted protein, if stably folded and able to exit the ER, lacks the critical dilysine ER retention signal20 and would likely be constitutively secreted from the cell. In addition to deleting the KKFF motif, all of the other identified mutations also remove the transmembrane domain and cytoplasmic domain.22
Only one missense mutation has been identified to date in the ERGIC-53 gene in patients with combined factors V and VIII deficiency. Detected in 4 of 7 Italian families included in the study, this single nucleotide change results in the substitution of the initiator methionine with threonine. As the next methionine codon is not encountered until exon 2 and it is out of frame, no functional ERGIC-53 would be predicted in these patients. This same mutation was identified by Neerman-Arbez et al18 in 2 of 8 Italian families in their study of 35 families. The finding of only mutations that are completely inactivating suggests that even minimal residual ERGIC-53 function might be sufficient for normal factor V and factor VIII secretion.
Although all 10 Middle Eastern and Sephardic Jewish families in our original report had an identifiable mutation, this result was biased by the finding of two founder mutations accounting for all patients in this restricted population.9 In contrast, no ERGIC-53 mutation was detected in 8 of 19 families in the current study. Taken together with our previous study of 10 Israeli families, we have failed to find a mutation in approximately 30% (8 of 29) of the families studied. These latter results are similar to those of Neerman-Arbez et al,18 who were unable to detect a mutation in nine of 35 families. Mutations in portions of the ERGIC-53 gene that were not sequenced, such as upstream regulatory regions, 3′ UTR, or intron sequences could account for these patients, although mutations of this type are generally infrequent in other genetic disorders. However, a hot spot for recurrent mutation, as results in the common FVIII gene inversion in hemophilia A, could account for this observation.23 The affected individual in family two, for whom the mutation on only one allele was identified, is most likely explained as a compound heterozygote with an undetected mutation in the other allele of ERGIC-53. This patient’s plasma levels of factors V and VIII (12% FV, 14% FVIII)17 are similar to those of other combined factors V and VIII deficiency patients, offering no particular insight into the nature of the second, undetected mutation.
Our data for lack of linkage to ERGIC-53 in two mutation-negative families (Fig 2) provide compelling evidence that the combined factors V and VIII deficiency in at least a subset of these patients is due to mutations in another gene. Consistent with this hypothesis, Neerman-Arbez et al18 observed apparently normal levels of ERGIC-53 by Western analysis of cell extracts from EBV-immortalized B lymphocytes in two of three families for which no ERGIC-53 mutation was detected by single-strand conformation polymorphism and sequence analysis. Taken together with our linkage findings, these data strongly suggest the presence of at least one additional locus for combined factors V and VIII deficiency. No phenotypic differences are discernible between affected individuals for whom an ERGIC-53 mutation has been detected and mutation-negative families or the specific subset demonstrating lack of linkage to the ERGIC-53 locus. It is likely that the gene(s) encoded by this alternative locus also functions in the secretory pathway for coagulation factors V and VIII, perhaps acting directly upstream or downstream of ERGIC-53. The future identification of this alternative gene(s) is likely to shed important light on the biological function of this unique cargo-specific ER to Golgi transport pathway.
ACKNOWLEDGMENT
We are indebted to the families who donated samples for this study and to Dr Tadashi Kamiya for assistance with the Japanese families.
Supported by Grants No. HL39693 and HL57346 from the National Institutes of Health, Bethesda, MD. D.G. and R.J.K. are investigators of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to David Ginsburg, MD, 4520 MSRB I, 1150 W Medical Center Dr, Ann Arbor, MI 48109-0650; e-mail:ginsburg@umich.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal