Abstract
Glycoprotein (GP) IX is a subunit of the von Willebrand receptor, GPIb-V-IX, which mediates adhesion of platelets to the subendothelium of damaged blood vessels. Previous characterization of the GPIX promoter identified a functional Ets site that, when disrupted, reduced promoter activity. However, the Ets protein(s) that regulated GPIX promoter expression was unknown. In this study, transient cotransfection of several GPIX promoter/reporter constructs into 293T kidney fibroblasts with a Fli-1 expression vector shows that the oncogenic protein Fli-1 can transactivate the GPIX promoter when an intact GPIX Ets site is present. In addition, Fli-1 binding of the GPIX Ets site was identified in antibody supershift experiments in nuclear extracts derived from hematopoietic human erythroleukemia cells. Comparative studies showed that Fli-1 was also able to transactivate the GPIb and, to a lesser extent, the GPIIb promoter. Immunoblot analysis identified Fli-1 protein in lysates derived from platelets. In addition, expression of Fli-1 was identified immunohistochemically in megakaryocytes derived from CD34+ cells treated with the megakaryocyte differentiation and proliferation factor, thrombopoietin. These results suggest that Fli-1 is likely to regulate lineage-specific genes during megakaryocytopoiesis.
HEMATOPOIETIC DEVELOPMENT is regulated by a complex interplay of external and internal cell signaling that leads to a decision process whereby primitive stem cells differentiate into specific lineages.1 An integral part of this process is the expression of lineage-restricted transcription factors that interact with the complex regulatory machinery that controls differential gene expression.2 Numerous laboratories have characterized hematopoietic transcription factors, which generally have been identified either on the basis of their binding to cis-acting promoter elements or on the basis of their dysregulated expression, often as fusion proteins, in leukemic cells.3
Megakaryocytes are the hematopoietic precursors of platelets, which play an essential role in thrombosis and hemostasis.4,5Several megakaryocyte-specific promoters, including platelet factor-4,6 glycoprotein (GP) IIb,7-10GPIbα,11 thrombopoietin receptor,12 and GPIX13 have been characterized. These promoters are often short, generally less than 600 base pairs, lack TATA boxes, and usually contain cis-acting regulatory elements regulated by the Sp1,14 GATA,15,16 and Ets17 18families of transcription factors.
The Ets factors comprise a family of transcription factors that regulate expression of several prominent hematopoietic genes. The prototypical Ets protein, Ets-1, was originally named because of its expression as part of an oncogenic protein by theE-twenty-six virus.19,20 Subsequent studies identified a number of Ets transcription factors that share an 82 amino acid motif, termed the Ets domain, that is required for binding to DNA sequences containing a consensus core of 5′-GGAA/T-3′ residues.21,22Numerous hematopoietic genes have been described that are regulated by Ets factors and several of these factors, including Tel, Erg, Ets-1, Ets-2, and Fli-1 are involved with leukemia18 suggesting a critical role in growth regulation of hematopoietic cells.
The Friend’s leukemia integration-1 (Fli-1) protein, also known as ErgB, is an Ets family member originally identified for its overexpression in erythroleukemia in mice infected with Friend’s leukemia virus.23 Fli-1 was independently discovered in Ewing’s sarcoma and peripheral neuroectodermal tumors with the Fli-1 carboxyterminus, including the DNA binding domain, linked to the aminoterminus of the Ewing’s sarcoma gene.24 Human Fli-1 is 97% homologous to the mouse gene.25 The mouse Fli-1 gene encodes two isoforms of 452 and 419 amino acids with molecular weights of 51 and 48 kD respectively, which use alternate initiation codons with the same reading frame.26 Both Fli-1 isoforms have been identified in vivo27 and generated in vitro.26 Fli-1 protein, like other Ets factors, has a helix-turn-helix domain (amino acids 121-196), believed to be involved with protein-protein interaction and an Ets domain (amino acids 277-360) that mediates DNA binding.28
The normal cellular target genes regulated by Fli-1 are unknown, but recent experiments suggest that Fli-1 might play a prominent role in the regulation of megakaryocytic genes. For example, Athanasiou et al29 showed that the human erythro-megakaryocytic cell line K562, which normally lacks Fli-1, shows an increase in expression of megakaryocytic features when transfected with a Fli-1 expression vector. Furthermore, promoters for the thrombopoietin receptor,12 von Willebrand factor,30 and GPIIb29 genes have been shown to be transactivated by Fli-1.
GPIX is a subunit of the platelet von Willebrand factor receptor, GPIb-IX-V, which is expressed in megakaryocytes.31Deficiency in GPIX is associated with the rare human disorder, Bernard-Soulier Syndrome.32 GPIX is expressed in and was originally cloned from, the erythro-megakaryocytic human erythroleukemia (HEL) cell line.33,34 Functional analysis of the GPIX promoter identified an Ets site located between -35 and -49 relative to the GPIX transcriptional start site that, when disrupted, reduced promoter activity as assessed by transient transfection into HEL cells.13 The GPIX Ets site also bound to HEL cell nuclear proteins in DNAse protection and gel mobility retardation experiments. However, the Ets factor(s) that regulate the GPIX promoter were unknown.13 Recent experiments35 showed that the Ets factor, Tel, could abrogate Fli-1 transactivation when reconstituted in the fibroblast-like 293T human embryonic kidney cell line. Experiments described herein extend these observations comparing Fli-1 transactivation of different GPIX constructs and showing Fli-1 binding in nuclear extracts derived from HEL cells. Additional experiments compare Fli-1 transactivation of the GPIX, GPIbα, and GPIIb megakaryocyte promoters. Finally, Fli-1 protein was identified in immunoblots of protein extracts derived from platelets and immunohistochemically in human megakaryocytes. In summary, these data provide evidence suggesting that Fli-1 is a likely candidate for regulation of genes in megakaryocytes.
MATERIALS AND METHODS
Plasmid constructs.
Construction of plasmids GPIX5′-686Luc, GPIX5′-203Luc, GPIX5′-69Luc, and pXP2 has been described.13 36Plasmid GPIX5′-37Luc was constructed using polymerase chain reaction (PCR) amplification of a GPIX promoter luciferase template using a 5′ sense primer that contained an Acc65 I restriction site adaptor that bound at -37, relative to the GPIX promoter, in conjunction with an antisense primer that bound in the luciferase gene. The resulting fragment was restriction digested with Acc65 I and Bgl II and inserted into the pXP2 luciferase expression plasmid that had been predigested with the same enzymes. A similar strategy was used in the construction of GPIbα5′-567Luc, GPIIb5′-596Luc, and GPIIb5′-75Luc, inserting promoter sequences extending to 567, 596, and 75 base pairs, upstream of their respective transcriptional start sites into pXP2. GPIX5′-203MutLuc was constructed using PCR amplification of a fragment of the GPIX promoter extending from -203 to -45 relative to the GPIX transcriptional start site, followed by restriction digestion and insertion into the HindIII and Acc 65 I sites of GPIX5′-37Luc. The resulting plasmid has GPIX promoter sequence identical to GPIX5′-203Luc except that the 7 base region that contains the Ets site between -37 and -45, ie, 5′-ACTTCCTTC-3′ was replaced with 5′-AAGGTACCC-3′. To screen for possible PCR errors the fidelity of the GPIX constructs was confirmed by sequencing. CMVFli-1 was constructed by insertion of a XhoI-NotI fragment containing the human Fli-1 gene into pCRTM3 (Invitrogen, Carlsbad, CA) predigested with the same enzymes.
Cell culture and transient transfections.
Human kidney 293T fibroblasts37 and K562 cells38 were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 0.584g/L glutamine, 4.5g/L glucose, 100 U/mL penicillin, 0.1 mg/mL streptomycin, 25 mmol/L HEPES and 10% heat-inactivated fetal bovine serum. The cells were cultured at 37° in 5% CO2 and passaged three times per week. HEL cells39 were cultured as described.13 Transient transfections on cell lines were performed using the SUPERFECT TM (Qiagen, Inc, Valencia, CA) according to the manufacturer’s instructions. Luciferase assays were performed as previously described.13 Each assay was performed in duplicate and adjusted for variations in transfection efficiency by normalizing for activity generated by a cotransfected CMVβgal internal control plasmid.
Generation and purification of peripheral mobilized hematopoietic CD34+ stem cells was performed on a service basis through the laboratory of Scott Rowley at the Fred Hutchinson Cancer Research Center. A male volunteer was injected with 5μg/kg/d with granulocyte colony-stimulating factor followed by apheresis and purification of the peripheral mobilized stem cells on an anti-CD34 antibody purification column (CellPro, Borhell, WA). The CD34+cells were cultured for 7 days in DMEM with Nutridoma (GIBCO-BRL, Gaithersburg, MD), 100 units/mL penicillin, 0.1 mg/mL streptomycin, and 10 ng/mL of thrombopoietin.
Gel mobility retardation assays.
Gel mobility retardation assays were performed as described.40 Briefly, oligonucleotides that include the GPIX Ets site, 5′-ATTTTCATCACTTCCTTCCGC-3′ and its complement were end-labeled with polynucleotide kinase and (γ-32) adenosine triphosphate (ATP), followed by annealing and purification on a nondenaturing polyacrylamide gel. To test for factor binding a mixture was made of 30 μg of HEL nuclear extracts, prepared as described,40 in 10 mmol/L Tris (pH 7.5), 50 mmol/L KCl, 5 mmol/L MgCl2, 1 mmol/L dithiothreitol, 1 mmol/L EDTA, 12.5% glycerol, 0.1% Triton X-100 with 2 μg poly (dI-dC) as a nonspecific competitor. The 15 μL reactions were preincubated for 20 minutess on ice before addition of probe (30,000 disintegrations/minute ∼.24 pmoles) and further incubation at room temperature for 30 minutes. In samples containing antibodies, 5 μg of each antibody was added to the binding mixture followed by incubation for an additional 30 minutes before loading on the gel. Anti–Fli-1 (ab 1) is a purified mouse immunoglobulin G (IgG) antihuman Fli-1 Ets domain monoclonal antibody (Pharmingen, San Diego, CA, catalog # 15491A); anti–Fli-1 (ab 2) is a rabbit polyclonal IgG directed against the carboxyterminal 19 amino acids of human Fli-1 (Santa Cruz Biotechnology, Santa Cruz, CA, catalog # sc-356) and the nonspecific control antibody is Mouse IgG anti-IgG. After incubation the samples were separated on a 5% polyacrylamide gel containing 50 mmol/L Tris, 380 mmol/L glycine, and 2 mmol/L EDTA running buffer. The gels were fixed in 10% methanol, 10% acetic acid, 5% glycerol, dried, and exposed to Kodak (Rochester, NY) X-OMAT AR film at −80°C with an intensifying screen.
Immunoblotting.
Immunoblotting was performed as described.41 Briefly, cell lysates derived from 5 × 105 cells or 1.6 × 107 purified platelets42 were resolved on 7.5% sodium dodecyl sulfate polyacrylamide gels followed by transfer to polyvinylidene difluoride membranes (Bio-Rad cat # 162-184; Bio-Rad, Hercules, CA). Primary antibodies ab 1 and ab 2 are described above. The anti-Fli1 antibody used in Fig 4 has been described.35 Secondary antibodies used were either the monoclonal antirabbit γ-chain–specific IgG (Sigma catalog # A-2556; Sigma, St Louis, MO) or the antirabbit Fab-specific IgG (Sigma catalog # A-2179). The bands were visualized using Western Blue stabilized substrate for alkaline phosphatase (Promega Catalog # S384B; Promega, Madison, WI).
Immunofluorescence.
Megakaryocytes derived from thrombopoietin-treated CD34+cells and 293T cells that were either mock transfected or transfected with CMVFli-1 expression vector were fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) (pH 7.2) for 10 minutes followed by washing in PBS containing 1% bovine serum albumin. The cells were incubated with rabbit anti–fli-1 antibody (Ab 2 described above) and anti-GPIIb mouse monoclonal antibody (Pharmingen) for 30 minutes followed by incubation with Goat antirabbit IgG linked to Cy3 fluorescent dye and Goat antimouse IgG linked to Cy2 fluorescent dye (Jackson ImmunoResearch, West Grove, PA). The samples were incubated for 45 minutes at room temperature in a humidified chamber in the dark followed by washing in PBS and mounting in the DNA stain bisbenzimide trihydrochloride (Sigma) (similar to Hoechst No. 33258, Kansas City, MO) dissolved in 15% polyvinyl alcohol, 10% glycerol, 0.01% Sodium azide, in 50 mmol/L Tris pH 9.0. Immunofluorescence was analyzed on a Microcomputer Imaging Device using the M2 software package (Imaging Research, Ontario, Canada).
RESULTS
Transactivation of the GPIX promoter by Fli-1 in 293T fibroblasts.
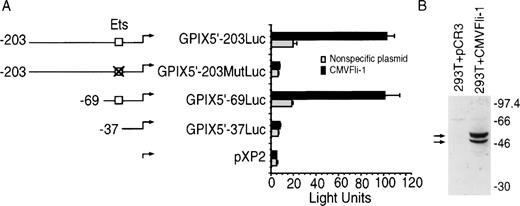
To test whether Fli-1 could regulate GPIX promoter activity, constructs were generated containing either intact, disrupted, or deleted GPIX Ets sites linked to a luciferase reporter gene. The promoter constructs were transiently cotransfected into fibroblast-like 293T cells in the presence of either a Fli-1–expression vector, CMVFli-1, or nonspecific plasmid, and the cells assayed for the presence of luciferase activity. The left side of Fig 1A diagrams the five luciferase constructs used in this experiment. GPIX5′-203Luc, GPIX5′-69Luc, and GPIX5′-37Luc contain promoter sequences extending to 203, 69, and 37 base pairs upstream of the GPIX transcriptional start site, respectively. GPIX5′-203Luc and GPIX5′-69Luc contain intact GPIX Ets sites. GPIX5′-37Luc contains GPIX promoter sequence that terminates just downstream of the GPIX Ets consensus sequence. GPIX5′-203MutLuc is identical to GPIX5′-203Luc, except the Ets site has been replaced with irrelevant sequence. Plasmid pXP2 is the promoterless parental luciferase construct from which the GPIX promoter constructs were derived. The right side of Fig 1A shows activity generated by the reporter vectors. Only constructs containing an intact Ets site, GPIX5′-203 and GPIX5′-69, showed a significant increase, approximately four- to fivefold, in luciferase activity in the presence of CMVFli-1. In contrast, GPIX5′-203MutLuc, GPIX-37Luc, and pXP2, which lack intact GPIX Ets sites, showed no significant increase in activity in the presence of CMVFli-1. This indicates that the Fli-1 protein transactivates the GPIX promoter and that this activity is mediated by the Ets site. Comparison of luciferase activities generated in the presence of nonspecific plasmid, ie, the gray bars, identifies a small but measurable Ets activity by both of the constructs containing intact Ets sites, GPIX5′-203Luc and GP5′-69Luc, that is absent in GPIX5′-203MutLuc, GPIX5′-37Luc, and pXP2, which lack Ets sites. This indicates that 293T cells contain weak endogenous Ets transactivation activity that is likely mediated by endogenous 293T Ets factor(s) that operate through the GPIX Ets sequence. The identity of this (these) factor(s) is unknown.
Fli-1 transactivation of the GPIX promoter in 293T kidney fibroblasts. (A) Left: Diagrams of the luciferase reporter constructs used in Fli-1 transactivation assays. The open boxes identify intact GPIX Ets sites. The crossed out box identifies the region of the GPIX promoter containing the Ets site that is replaced with irrelevant sequence (See Materials and Methods). Plasmid pXP2 is a promoterless construct that encodes the luciferase gene. (A) Right luciferase activity, indicated as light units, generated in transiently transfected 293T kidney fibroblasts. Each sample was transfected with 1.5 μg of luciferase reporter construct and 4.5 μg of the Fli-1 expression vector, CMVFli-1, or nonspecific plasmid, normalized using a CMVβgal control plasmid. Error bars represent deviations between duplicate samples. Each plasmid was tested in at least seven independent experiments. (B) shows an immunoblot analysis of lysates derived from 293T kidney fibroblasts transfected with either pPCR3, the empty expression vector lacking Fli-1, or CMVFli-1. Arrows identify the two isoforms of Fli-1 and numbers indicate the migration pattern of molecular weight markers.
Fli-1 transactivation of the GPIX promoter in 293T kidney fibroblasts. (A) Left: Diagrams of the luciferase reporter constructs used in Fli-1 transactivation assays. The open boxes identify intact GPIX Ets sites. The crossed out box identifies the region of the GPIX promoter containing the Ets site that is replaced with irrelevant sequence (See Materials and Methods). Plasmid pXP2 is a promoterless construct that encodes the luciferase gene. (A) Right luciferase activity, indicated as light units, generated in transiently transfected 293T kidney fibroblasts. Each sample was transfected with 1.5 μg of luciferase reporter construct and 4.5 μg of the Fli-1 expression vector, CMVFli-1, or nonspecific plasmid, normalized using a CMVβgal control plasmid. Error bars represent deviations between duplicate samples. Each plasmid was tested in at least seven independent experiments. (B) shows an immunoblot analysis of lysates derived from 293T kidney fibroblasts transfected with either pPCR3, the empty expression vector lacking Fli-1, or CMVFli-1. Arrows identify the two isoforms of Fli-1 and numbers indicate the migration pattern of molecular weight markers.
To test for authentic Fli-1 expression lysates derived 293T cells that were transfected with CMVFli-1 or plasmid pCR3, which is identical to CMVFli-1 except that it lacks a Fli-1 complementary DNA, were analyzed using immunoblot analysis using a Fli-1–specific antibody. Figure 1B shows that the CMVFli-1 expression construct expressed two detectable bands migrating with molecular weights of approximately 48 and 51 kD. The two Fli-1 translation products are from different translational start codons using the same reading frame, as has been previously reported.27 26 The data indicate that the CMVFli-1 vector encodes Fli-1 that is accurately translated into both isoforms of the immunoreactive Fli-1 protein.
Identification of Fli-1 binding to the GPIX Ets site in the erythro-megakaryocytic HEL cell line.
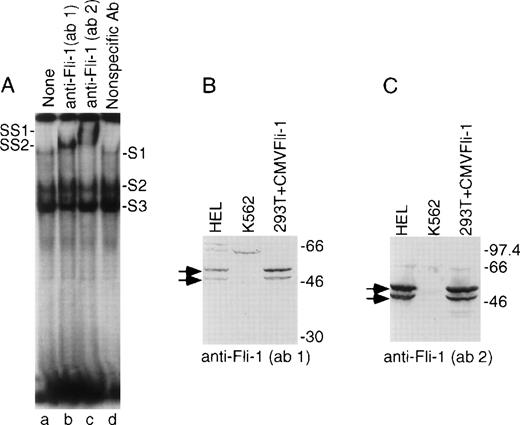
To test whether the GPIX Ets element can be regulated by Fli-1 in hematopoietic cells, gel retardation supershift experiments were performed using labeled, double-stranded oligonucleotide analogs of the GPIX Ets site, nuclear extracts from HEL cells, and anti–Fli-1 antibodies. Figure 2A identifies three DNA-protein complexes designated S1, S2, and S3. Previously published experiments identified two of the DNA-protein complexes, S2 and S3.13 Use of better quality nuclear extracts and improved binding assay and gel conditions (see Materials and Methods) allowed the identification of a third DNA-protein complex, S1, which migrates more slowly than the other two complexes. Lanes b and c show patterns of DNA-protein complex formation generated after incubation with a monoclonal antibody directed against the Ets domain of Fli-1 (Ab-1) or a polyclonal antibody directed against the Fli-1 carboxyterminus (Ab-2). Incubation with a nonspecific antibody shows a pattern of DNA-protein complexes that is similar to the absence of antibody (compare Fig 2A Lanes a and d). Comparison of lanes b and c with a and d indicate that in addition to formation of new retarded complexes, designated SS1 and SS2, there is a diminution of intensity in the S1 band.
Fli-1 in the hematopoietic HEL cell line binds to GPIX Ets sites. (A) Autoradiogram of a gel supershift experiment. Labeled double-stranded oligonucleotide corresponding to bases -54 to -34 within the GPIX promoter was mixed with nuclear extracts derived from HEL cells followed by addition of the indicated antibody. Shifted complexes are indicated as S1, S2, and S3. Supershifted complexes, observed only in lanes b and c, are indicated as SS1 and SS2. (B and C) Immunoblots of separated extracts from HEL cells (left lanes), K562 cells (center), and 293T cells (right) that were transiently transfected with the CMVFli-1 expression construct. (B) was probed with the anti–Fli-1 (ab 1) antibody used in (A) lane b. (C) was probed with the same anti–-Fli-1 (ab 2) antibody that was used in (A) lane c. Arrows identify bands of reactive protein that correspond to the sizes of the two isoforms of Fli-1. Numbers on right indicate molecular weight markers.
Fli-1 in the hematopoietic HEL cell line binds to GPIX Ets sites. (A) Autoradiogram of a gel supershift experiment. Labeled double-stranded oligonucleotide corresponding to bases -54 to -34 within the GPIX promoter was mixed with nuclear extracts derived from HEL cells followed by addition of the indicated antibody. Shifted complexes are indicated as S1, S2, and S3. Supershifted complexes, observed only in lanes b and c, are indicated as SS1 and SS2. (B and C) Immunoblots of separated extracts from HEL cells (left lanes), K562 cells (center), and 293T cells (right) that were transiently transfected with the CMVFli-1 expression construct. (B) was probed with the anti–Fli-1 (ab 1) antibody used in (A) lane b. (C) was probed with the same anti–-Fli-1 (ab 2) antibody that was used in (A) lane c. Arrows identify bands of reactive protein that correspond to the sizes of the two isoforms of Fli-1. Numbers on right indicate molecular weight markers.
To test the specificity of the anti–Fli-1 antibodies, and to test for the presence of authentic Fli-1 in HEL cells, immunoblots were performed on membranes containing proteins derived from HEL, K562, and 293T cells transfected with CMVFli-1 expression vector. The K562 cell line is a human erythro-megakaryocytic cell line that is known to not express detectable Fli-1.29 Figure 2B and C shows that both antibodies bound two prominent bands of approximately 48 and 51 kD, that comigrated at the same rate as Fli-1 protein expressed in 293T cells. There was no evident authentic Fli-1 expressed in K562 cells. These data indicate that HEL cells contain Fli-1 protein of the appropriate size and that the supershift complexes shown in Fig 2A are very likely to contain Fli-1 protein.
Fli-1 transactivation of megakaryocyte promoters GPIbα and GPIIb.
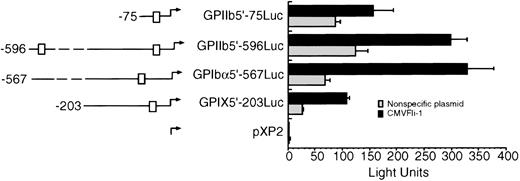
To test whether Fli-1 could transactivate other megakaryocyte promoters, 293T cells were transiently cotransfected with the CMVFli-1 expression vector and luciferase expression vectors containing sequences derived from the GPIX, GPIbα, and GPIIb promoters. The left side of Fig 3 diagrams the known Ets sites in each promoter construct, indicated as boxes. GPIIb has two Ets sites located at -515 and -40 and the GPIbα promoter contains one Ets site located at -144 relative to their respective transcription start sites. Both the GPIX GPIIb5′-596Luc and GPIIb5′-75Luc promoter constructs show an increase in promoter activity in the presence of Fli-1 of approximately four- to sixfold, whereas the GPIIb promoter constructs GPIIb5′-75Luc and GPIIb-596Luc appear to be less responsive to Fli-1 showing an induction of approximately twofold.
Fli-1 transactivation of GPIX, GPIb, and GPIIb promoter constructs. The left side of the figure diagrams the luciferase reporter constructs used in Fli-1 transactivation assays. The □ identifies Ets sites. The exact locations of the Ets boxes are identified in the text. The transfection was performed as described in Materials and Methods and the legend to Fig 1. Error bars represent deviations between duplicate samples. Each plasmid was tested in at least three independent experiments, all with essentially with the same results.
Fli-1 transactivation of GPIX, GPIb, and GPIIb promoter constructs. The left side of the figure diagrams the luciferase reporter constructs used in Fli-1 transactivation assays. The □ identifies Ets sites. The exact locations of the Ets boxes are identified in the text. The transfection was performed as described in Materials and Methods and the legend to Fig 1. Error bars represent deviations between duplicate samples. Each plasmid was tested in at least three independent experiments, all with essentially with the same results.
Identification of Fli-1 in platelets.
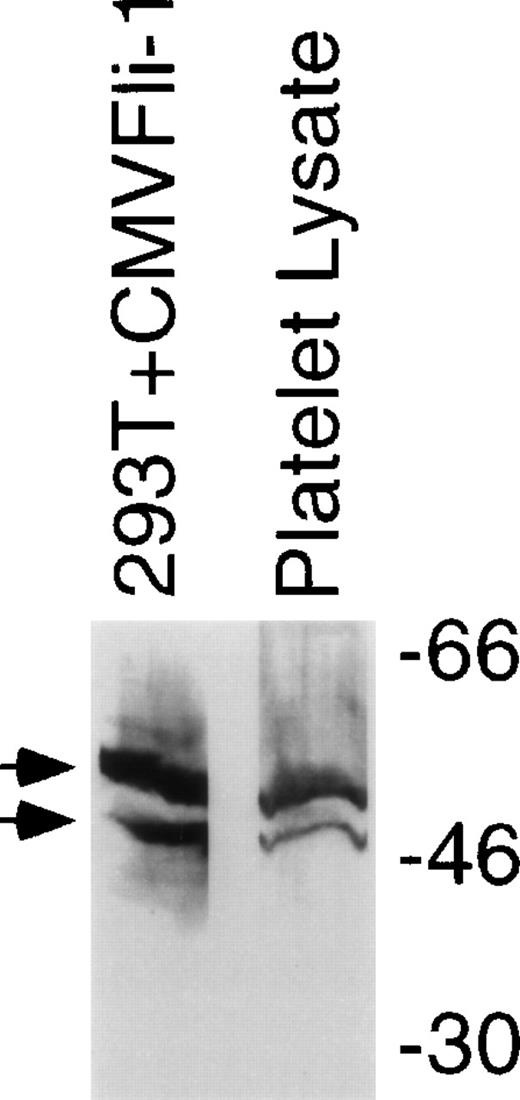
To investigate whether Fli-1 is a likely candidate for regulation of genes expressed in megakaryocytes immunoblot analysis was performed on lysates derived from human platelets. Figure 4 shows an immunoblot comparing Fli-1 protein in lysates derived from purified platelets with Fli-1 protein expressed in 293T cells transfected as described for Fig 1. Platelets contain protein that comigrates with Fli-1 expressed in transiently transfected 293T cells. Platelets do not have nuclei, and therefore lack transcription. Furthermore, platelets have few ribosomes and likely have very low levels of translational activity. This suggests that the Fli-1 contained in platelets is protein that was expressed in the megakaryocyte. Although platelets are a common source for platelet integrins and other surface proteins,42 they are not usually used as source for gene regulatory proteins. The presence of Fli-1 in platelets may be a reflection of the abundance and/or stability of the protein.
Fli-1 protein is contained in platelets. Immunoblot analysis using an anti–Fli-1 antibody was performed on lysates derived from platelets that were separated in parallel with molecular weight markers (shown on right) and lysate derived from 293T cells transiently transfected with the CMVFli-1 expression construct. Arrows identify the two isoforms of Fli-1.
Fli-1 protein is contained in platelets. Immunoblot analysis using an anti–Fli-1 antibody was performed on lysates derived from platelets that were separated in parallel with molecular weight markers (shown on right) and lysate derived from 293T cells transiently transfected with the CMVFli-1 expression construct. Arrows identify the two isoforms of Fli-1.
Identification of Fli-1 in megakaryocytes.
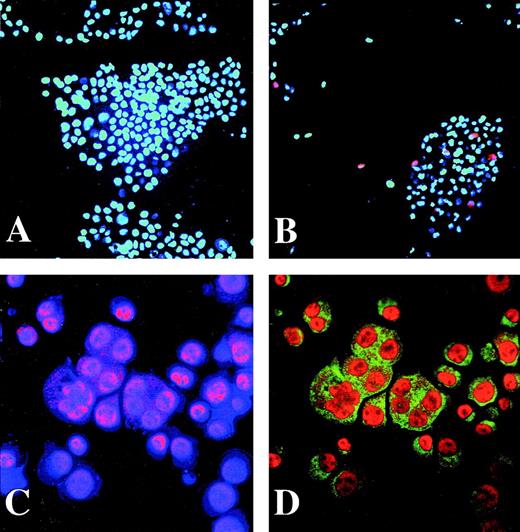
We next used indirect immunofluorescence to test whether Fli-1 is expressed in megakaryocytes. Figure 5 (A and B) shows 293T cells that were transfected with a nonspecific plasmid (A) or with the CMVFli-1 expression construct (B). The cyan color shows Hoechst staining in all cells, based on DNA content, and the magenta color identifies cells positive for expression of Fli-1. Only the population of 293T cells that that was transfected with the CMVFli-1 expression vector contained Fli-1–positive cells. (Compare 5A to 5B.) This established the specificity of our indirect immunofluorescence staining procedure. To test whether Fli-1 is expressed in megakaryocytes, indirect immunofluorescence was performed on human megakaryocytes derived from CD34+ cells treated for 7 days with thrombopoietin. Figure 5C and 5D shows primary megakaryocytes derived from peripheral mobilized CD34+cells treated with the megakaryocyte differentiation and proliferation agent, thrombopoietin. Figure 5C shows double staining with both the blue Hoechst DNA stain and red stain that identifies antibody directed against Fli-1. Colocalized DNA and Fli-1 signals appear as magenta. Figure 5D shows the same field of cells showing double staining with antibodies directed against the megakaryocyte differentiation marker GPIIb, green color, and Fli-1, which appears orange in 5D. The larger cells show multilobed nuclei that are typical of mature, polyploid megakaryocytes. The GPIIb antigen localizes to the cytoplasm and the Fli-1 antigen is expressed predominantly in the nucleus. However, there is detectable Fli-1 in the megakaryocyte cytoplasm (More easily visualized in 5C). This experiment shows that Fli-1 is expressed in megakaryocytes, consistent with a role for Fli-1 regulation of megakaryocyte genes.
Fli-1 protein is expressed in human megakaryocytes. A and B show immunohistochemical analysis for Fli-1 expression in 293T cells that were transiently transfected with either nonspecific plasmid (A) or the Fli-1 expression construct CMVFli-1 (B). Hoechst DNA staining is cyan and Fli-1 staining is red. Regions that colocalize for DNA and Fli-1 staining appear as magenta. Panels C and D show CD34+ cells that were treated with 10 ng/mL of thrombopoietin for 7 days before triple staining. Panel C shows Hoechst DNA stain (blue) and Fli-1 (red). Areas of colocalization appear as magenta. Panel D displays the same cells as panel C showing GPIIb antigen (green) and Fli-1 (orange). In panel D Fli-1 appears as orange. Original magnification of panels A and B was 40× and panels C and D was 200×.
Fli-1 protein is expressed in human megakaryocytes. A and B show immunohistochemical analysis for Fli-1 expression in 293T cells that were transiently transfected with either nonspecific plasmid (A) or the Fli-1 expression construct CMVFli-1 (B). Hoechst DNA staining is cyan and Fli-1 staining is red. Regions that colocalize for DNA and Fli-1 staining appear as magenta. Panels C and D show CD34+ cells that were treated with 10 ng/mL of thrombopoietin for 7 days before triple staining. Panel C shows Hoechst DNA stain (blue) and Fli-1 (red). Areas of colocalization appear as magenta. Panel D displays the same cells as panel C showing GPIIb antigen (green) and Fli-1 (orange). In panel D Fli-1 appears as orange. Original magnification of panels A and B was 40× and panels C and D was 200×.
DISCUSSION
Because of their relative rarity, fragility, and apoptotic developmental fate,43 gene expression in megakaryocytes is difficult to study compared with other hematopoietic lineages. Although several megakaryocyte promoters have been characterized, most analyses of these promoters have used hematopoietic cell lines to identify the factors and sequences that regulate megakaryocyte expression. In fact, a megakaryocytic-specific transcription factor has not yet been described. Thus, it is difficult to confirm precisely which factors govern megakaryocyte gene expression. Fortunately, comparisons of activities of transcriptional elements in hematopoietic cell lines and the few promoter regulatory studies in primary megakaryocytes have confirmed that sites that are active in hematopoietic cell lines are active in primary megakaryocytes.44 The recent cloning of the megakaryocyte inducer thrombopoietin45-48 has made it feasible to generate enough megakaryocytes to do biochemical-based experiments and analyze megakaryocyte developmental regulators49 giving rise to the possibility that megakaryocyte-specific factors, if such exist, may be identified.
Members of the Ets transcription factor family have been identified in numerous different tissues, but seem particularly important to hematopoietic development. There is little information available regarding the expression of Ets factors in megakaryocytes, although Pu.1 has been identified immunohistochemically.50 However, erythro-megakaryocytic cell lines are known to express several Ets factors, including Ets-1, Pu.1, Fli-1, Ets-2, Elf-1, Erm, and SAP-1.12,51 Several reports have described transactivation of megakaryocytic promoters using reconstitution assays in nonhematopoietic cells. For example, Ets-1 is known to transactivate the GPIIb promoter in HeLa cells that are transiently cotransfected with GPIIb promoter constructs.52 It has also been shown that the Ets factor Pu.1 can transactivate the promoter for the megakaryocyte-specific platelet basic protein.53 The thrombopoietin receptor promoter can be activated by both Ets-1 and Fli-1,12 and the promoter for von Willebrand factor could be transactivated by both Ets-1 and Erg,30 indicating that promoters may be regulated by multiple Ets factors. A recent report described transactivation of the GPIIb promoter by the Ets factor Pu.1 that was dependent on thrombopoietin stimulation.54 These observations suggest that megakaryocyte promoters may be regulated by multiple Ets factors.
The normal target(s) of Fli-1 is uncertain. Fli-1 is expressed in a variety of hematopoietic cell lines and several tissues, including spleen and thymus, and at low levels in embryonic endothelium, heart, lung, and skeletal muscle.55 Fli-1 expression has also been described in cells in embryonic mouse liver that have a characteristic megakaryocytic appearance but the identity of the cells was not definitively established.55 Transgenic studies showed that overexpression of a Fli-1 transgene under control of the H-2Kk promoter caused a lethal autoimmune renal disease, but surprisingly, not an increase in cancer.27 Recently, Melet et al55 described a partial disruption of the amino-terminal 76 amino acids of the Fli-1, leaving intact activation and Ets domains, which led to a reduction in thymic cellularity and an extension in latency of onset of erythroleukemia mediated by Friend Leukemia Virus infection.
The presence of the S2 and S3 complexes shown in Fig 2 indicates that the GPIX Ets site binds, and may be regulated by, other Ets proteins. Whether the GPIX promoter can be regulated by Ets factors other than Fli-1 is currently under investigation. Further experiments are in progress to test whether Ets factors may functionally interact. These experiments may be particularly informative in light of the differential transactivation by Fli-1 of the promoters tested in Fig 3.
Figure 4 shows the presence of Fli-1 protein in lysates derived from purified platelets. It seems unlikely that Fli-1 is translated de novo in platelets. It seems more probable that the presence of Fli-1 in platelets represents residual protein found in the megakaryocyte. Similar assays testing for the presence of Ets-1 were unsuccessful (data not shown). Thus the presence of Fli-1 may reflect the abundance and/or stability of the protein. It is also possible that Fli-1 serves some unknown function in platelet physiology. Figure 5 shows that Fli-1 is expressed in human megakaryocytes and the presence of detectable cytoplasmic Fli-1 may account for the presence of trace Fli-1 in platelets.
In summary, this study provides evidence that the oncogenic Ets factor Fli-1 can regulate several megakaryocyte genes, including GPIX, GPIbα, and GPIIb. Furthermore, Fli-1 is present in the hematopoietic HEL cell line and it binds to the GPIX Ets site. In addition, Fli-1 protein was identified both in platelets and in megakaryocytes. In toto, these data suggest that Fli-1 may be an important gene regulator in megakaryocyte gene expression.
ACKNOWLEDGMENT
We thank Anna Zielinska-Kwiatkowska and Shawn Mohamed for technical assistance, Kin Ritchie for critical reading of the manuscript, and Sally Swedine for assistance with preparation of the figures.
Supported by a National Research Service Award F32 HL09265 and R01-DK49855-01 (L.S.B.). G.R. is supported by a Merit Review Grant from the Veterans Administration and NIH grant HL39947.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Gerald Roth, MD, VA Puget Sound Health Care System (M/S 111), 1660 S. Columbian Way, Seattle, WA 98108.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal