To the Editor:
Anaplastic large-cell lymphoma (ALCL) is frequently associated with the recurrent translocation t(2;5)(p23;q35) that results in activation of the anaplastic lymphoma kinase (ALK ) gene at 2p23 by fusion to the ubiquitously expressed gene encoding the nucleolar phosphoprotein nucleophosmin (NPM ) at 5q35. This translocation leads to aberrant nuclear (and cytoplasmic) expression ofALK, which is normally silent in hematopoietic tissues.1 2
Approximately 20% of ALK-positive ALCLs do not expressALK in the nucleus but show aberrant expression of ALKrestricted to the cytoplasm.3,4 These “cytoplasmicALK only” ALCL do not contain the t(2;5), suggesting that other genetic abnormalities can result in aberrant ALKexpression. So far, only very limited data on cytogenetic alterations of this subtype of ALCL are available. Nevertheless, in very recent issues of BLOOD, “cytoplasmic ALK only” ALCL cases have been described to contain variant rearrangements of the chromosomal region 2p23. Wlodarska et al5 reported 3 cases with cryptic inv(2)(p23q35). Rosenwald et al6 described 1 case each with t(1;2)(q21;p23) and t(2;3)(p23;q21). Finally, Lamant et al7 reported a t(1;2)(q25;p23) in 1 patient. Cloning of the translocation breakpoint showed a fusion of ALK to theTPM3 gene in 1q25 encoding a nonmuscle tropomyosin. ATPM3-ALK fusion transcript was detectable by reverse transcriptase-polymerase chain reaction (RT-PCR) in 2 of 3 additional ALCL, with ALK staining restricted to the cytoplasm.7
We recently investigated 3 cases of “cytoplasmic ALKonly” ALCL by means of fluorescence in situ hybridization (FISH). Whereas 2 cases turned out to contain the previously described inv(2)(p23q35), we report here a cryptic insertion into the ALKgene observed in the third patient. The lymph node biopsy specimen was obtained from a 5-year-old male patient with stage IVB disease. Histological evaluation showed a CD30(Kil)-positive ALCL of null cell phenotype. Using the ALK1 antibody (Dako, Glostrup, Denmark), only 20% of the tumor cells stained positive, with strong cytoplasmicALK protein expression but lack of any nuclear ALKexpression.
Chromosome analysis of R-banded metaphases derived from unstimulated short-term lymph node cultures showed clonal aberrations in 6 of 16 metaphases. The karyotype was 50 ∼ 51, XY, +X, del(1)(q21), +2, der(2)?dup(2)(p25p21)dup(2)(p11p25) × 2, +6, +7, +17 [cp6]/46, XY [10]. To investigate the case for a cryptic ALKrearrangement, we performed FISH with the LSI ALK assay (Vysis, Downers Grove, IL) according to the manufacturer’s instructions. The LSI ALK assay consists of 2 differentially labeled probes for the centromeric (5′) and telomeric (3′) regions of the ALK gene that showed to be separated in cases with t(2;5) and inv(2). Using this assay, 38 of 100 interphase cells each displayed 2 isolated signals for the centromeric (green) and telomeric (red) ALK probes, indicating 2 rearranged copies of the ALK gene in addition to 1 colocalization of each 1 signal derived from an intact ALKlocus. FISH on metaphases followed by subsequent R-banding analysis showed the short arms of the intact chromosome 2 and both der(2) chromosomes each to contain 1 signal for the centromeric and the telomeric ALK probe. Nevertheless, the spatial separation of the 2ALK probes on the der(2) chromosome suggests an insertion of genomic material into the ALK gene on the der(2) (Fig 1A through D).
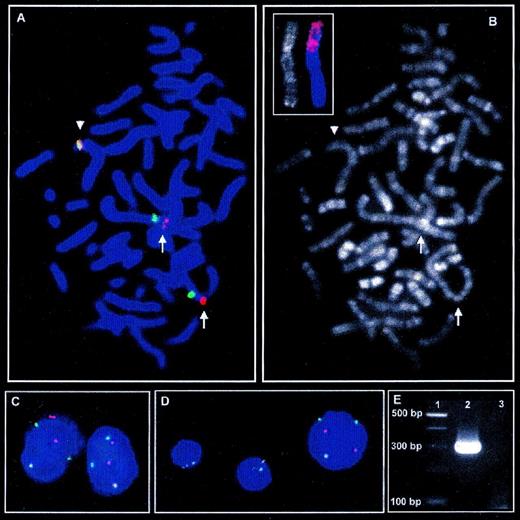
Metaphase FISH with LSI ALK probe (A) and subsequent R-banding analysis (B) shows insertion of genomic material into theALK locus indicated by spatial separation of the red signal for 3′ALK (telomeric) and the green signal for 5′ALK(centromeric) on the short arms of both der(2) chromosomes (arrows) as compared with the intact chromosome 2 (arrowhead). (C) In interphase nuclei of tumor cells, insertion into the ALK locus on both derivative chromosome 2 leads to separation of 3′ and 5′ALKsignals in addition to each 1 colocalized, indicating the intactALK locus. (D) In contrast to the larger nucleus of the tumor cells, the smaller nuclei of nonneoplastic cells show 2 colocalized signals for 3′ and 5′ALK, which is the regular signal constellation for 2 intact chromosomes 2. (B, small picture) FISH with a chromosome 1 painting probe on R-banded chromosomes shows a major part of the short arm of the der(2) chromosomes to derive from chromosome 1. (E) RT-PCR amplifies a PCR product of approximately 300 bp specific for TPM3-ALK fusion (Lane 2). Lane 1, 100-bp ladder; lane 3: H2O control.
Metaphase FISH with LSI ALK probe (A) and subsequent R-banding analysis (B) shows insertion of genomic material into theALK locus indicated by spatial separation of the red signal for 3′ALK (telomeric) and the green signal for 5′ALK(centromeric) on the short arms of both der(2) chromosomes (arrows) as compared with the intact chromosome 2 (arrowhead). (C) In interphase nuclei of tumor cells, insertion into the ALK locus on both derivative chromosome 2 leads to separation of 3′ and 5′ALKsignals in addition to each 1 colocalized, indicating the intactALK locus. (D) In contrast to the larger nucleus of the tumor cells, the smaller nuclei of nonneoplastic cells show 2 colocalized signals for 3′ and 5′ALK, which is the regular signal constellation for 2 intact chromosomes 2. (B, small picture) FISH with a chromosome 1 painting probe on R-banded chromosomes shows a major part of the short arm of the der(2) chromosomes to derive from chromosome 1. (E) RT-PCR amplifies a PCR product of approximately 300 bp specific for TPM3-ALK fusion (Lane 2). Lane 1, 100-bp ladder; lane 3: H2O control.
As for R-banding analysis, there was no evidence for material from chromosomes other than chromosome 2 to be contained in the der(2) chromosomes, suggesting that a complex inv(2) might lead to insertion of 2q35 material into the ALK locus. To test this hypothesis, FISH with YAC probes 884F10, 770F5, and 914E7 flanking and spanning the 2q35 breakpoint of the inv(2), respectively, was applied.5These hybridizations did not provide evidence for the genomic material inserted into the ALK locus to be derived from a complex inv(2).
Considering the loss of chromosome 1 material due to the del(1)(q21) in the tumor cells, it was also tempting to speculate that the material inserted into the ALK locus might be derived from the long arm of chromosome 1. Consequently, FISH on archived R-banded slides was performed with a whole chromosome 1 painting probe (AGS, Heidelberg, Germany). Surprisingly, this analysis showed a major part of the short arms of the der(2) chromosomes to be derived from chromosome 1 (Fig 1B, small picture). Thus, the present case contains a complex t(1;2) translocation that is hardly detectable by banding analyses due to the underlying multiple break events.
Because this complex aberration might be a variant of the recently cloned translocation t(1;2)(q25;p23) leading to a TPM3-ALKfusion, we aimed to investigate the case for this hybrid transcript.7 No fresh material for molecular analyses was available from initial diagnosis and the patient is in continuous complete remission for 2.5 years now after initial treatment according to the German NHL-BFM95 trial (group III-K3).8 Thus, RNA was extracted from cells in Carnoy’s fixative by means of RNAzol (WAK Chemie, Bad Homburg, Germany), and first-strand synthesis was performed with pooled ALK-specific primers (5′-AGC ACA CTT CAG GCA GCG TCT TCA CAG CCA-3′ and 5′-CAT TCC GGA CAC CTG GCC TTC ATA CAC CTC-3′) using the Reverse Transcription System (Promega, Madison, WI). Nested PCR according to Lamant et al7 indeed amplified the characteristic PCR product of approximately 300 bp (Fig 1E). Sequencing of this PCR product confirmed the presence of the TPM3-ALKfusion transcript.
In summary, we identified a cryptic ALK gene rearrangement due to an insertion that, by molecular cytogenetics and RT-PCR analysis, turned out to be a complex variant translocation t(1;2) leading toTPM3-ALK fusion. On the one hand, the present case confirmsTPM3-ALK fusion to be a recurrent mode of ALKactivation in ALCL with ALK expression restricted to the cytoplasm.7 On the other hand, this case again indicates that ALK gene rearrangements might be cytogenetically hidden, eg, due to complex breakage events. Indeed, in the present case as well as in the cases with inv(2), the ALK rearrangements would not have been cytogenetically detected without applying molecular cytogenetics. Thus, in our opinion, FISH with an ALK-specific probe should be generally integrated into the cytogenetic analysis of “cytoplasmic ALK only” ALCL so that cryptic ALKrearrangements are not missed.
ACKNOWLEDGMENT
The authors are grateful to C. Becher, D. Schuster, and F. Jäger for their excellent technical assistance. This work was supported by the Deutsche Krebshilfe, the Wilhelm-Sander-Stiftung, and the IZKF Kiel. S.G. is a scholar of the Hensel-Stiftung (Kiel, Germany).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal