Abstract
We have determined the prognostic significance of hypodiploidy (<46 chromosomes) in a large cohort of children with acute lymphoblastic leukemia (ALL) treated by the Children's Cancer Group. Among 1,880 patients, 110 (5.8%) had hypodiploid karyotypes: 87 had 45 chromosomes, 15 had 33 to 44 chromosomes, none had 29 to 32 chromosomes, and 8 had 24 to 28 chromosomes (near-haploidy). Six-year event-free survival (EFS) estimates for patients with 45 chromosomes, 33 to 44 chromosomes, or 24 to 28 chromosomes were 65% (standard deviation [SD], 8%), 40% (SD, 18%), and 25% (SD, 22%), respectively (log rank, P = .002; test for trend, P = .0009). The combined hypodiploid group had worse outcome than nonhypodiploid patients, with 6-year EFS of 58% (SD, 7%) and 76% (SD, 2%), respectively (P < .0001). EFS for the subgroup with 45 chromosomes was similar to that of patients with pseudodiploidy (P = .43) or 47 to 50 chromosomes (P = .76). None of the patients with 24 to 28 chromosomes had a t(4;11), a t(9;22), or a t(1;19), and most received highly intensive therapy. The adverse risk associated with 33 to 44 and 24 to 28 chromosomes remained significant in multivariate analyses adjusted for important risk factors including age, white blood cell count, and Philadelphia chromosome status. Thus, hypodiploidy with less than 45 chromosomes, particularly 24 to 28 chromosomes, is a significant adverse risk factor despite treatment with contemporary intensive therapies.
CURRENT CHILDREN'S cancer group (CCG) risk group-adjusted intensive therapies for childhood acute lymphoblastic leukemia (ALL) are expected to result in 6-year event-free survival (EFS) and survival of greater than 75% and 84%, respectively (Harland Sather, CCG unpublished data). These intensive therapies have overcome many of the clinical features, including organomegaly and leukemic cell lineage, that previously were associated with poor outcome.1-5 Thus, the present challenge is to improve our ability to identify and treat appropriately those patients who will fail these current intensive therapies. Higher and lower risk patients are now identified primarily by age and white blood cell (WBC) count,6 but early response to therapy, as measured by the bone marrow or peripheral blood leukemic blast content, has emerged as an important prognostic variable.7-10Genetic and biological characteristics, such as the presence of specific chromosomal translocations or fusion transcripts, may provide the tools required for more accurately predicting risk of treatment failure. In addition, further refinement of currently known risk factors may allow better allocation of less or more intensive and potentially toxic therapy to patients with lower or higher risk of failure.
Leukemic cell ploidy is a known risk factor for treatment outcome in pediatric ALL: high hyperdiploidy (>50 chromosomes or DNA index >1.16) is associated with improved outcome, whereas a hypodiploid (<46 chromosomes) karyotype is thought to be an adverse risk factor.11-17 Pui et al18 reported particularly poor outcome for patients with near-haploidy, and similarly, Chessels et al19 reported that hypodiploidy with 24 to 29 chromosomes was a significant independent risk factor for children with ALL. These findings suggested, albeit with small numbers of patients, that subsets of hypodiploid patients may have different treatment outcomes. Therefore, we have examined the prognostic significance of leukemic cell hypodiploidy, including the effect of chromosome number less than 45, in a very large cohort (N = 1,880) of children with ALL treated on contemporary intensive protocols of the CCG. In both univariate and multivariate analyses, our data indicate that hypodiploid ALL patients with 45 chromosomes have an outcome similar to that of ALL patients with pseudodiploid or low hyperdiploid (47 to 50 chromosomes) ALL, whereas patients with less than 45 chromosomes, particularly those with 24 to 28 chromosomes, achieve poor outcomes despite intensive risk-adjusted therapy. Thus, hypodiploid ALL is heterogeneous with respect to treatment outcome, and novel therapies should be explored for those with 24 to 28 chromosomes.
MATERIALS AND METHODS
Patients.
Diagnosis of ALL required determination of lymphoblast morphology by Wright-Giemsa staining of bone marrow smears, negative lymphoblast staining for myeloperoxidase, and cell surface expression of 2 or more lymphoid differentiation antigens.20 Immunophenotyping was performed centrally in the CCG ALL Biology Reference Laboratory by indirect immunofluorescence and flow cytometry, as previously described.2 20 Patients were classified as B-lineage if ≥30% of the leukemic cells were positive for CD19 and/or CD24 and less than 30% were positive for 1 or more of the T-cell–associated antigens CD2, CD3, CD5, or CD7. Likewise, patients were classified as T-lineage if ≥30% of the isolated blasts were positive for 1 or more of the T-cell–associated antigens CD2, CD3, CD5, or CD7 and less than 30% were positive for CD19 and/or CD24.
The current study involved children with newly diagnosed ALL enrolled on CCG risk-adjusted protocols between 1988 and 1995. Children 2 to 9 years of age with WBC counts less than 10,000/μL (low-risk ALL) were enrolled on CCG-1881; children 2 to 9 years of age with WBC counts of 10,000 to 49,999/μL or 12 to 23 months of age with WBC counts less than 50,000/μL (intermediate-risk ALL) were enrolled on CCG-1891. After completion of these studies, patients with low- or intermediate-risk ALL were enrolled on a single protocol, CCG-1922, for standard-risk ALL (1 to 9 years of age and WBC counts <50,000/μL) based on National Cancer Institute (NCI) criteria.6Children 1 to 9 years of age with WBC counts ≥50,000/μL or ≥10 years of age (NCI poor-risk group)6 were assigned to CCG-1882. In addition, children with multiple unfavorable features were enrolled on the CCG-1901 protocol for lymphomatous syndrome ALL.21 Infants less than 12 months of age were excluded from this analysis. All protocols were approved by the NCI and the Institutional Review Boards of the participating CCG-affiliated institutions. Informed consent was obtained from parents, patients, or both, according to the guidelines of the Department of Health and Human Services. EFS and overall survival estimates at 8 years from study entry for the combined group of patients included in this analysis were 74% (standard deviation [SD], 2%) and 82% (SD, 2%), respectively.
Cytogenetic analysis.
Diagnostic karyotyping of leukemic cells was performed by institutional laboratories before the initiation of therapy. Banded chromosomes were prepared from unstimulated peripheral blood or direct and 24-hour–cultured preparations of fresh bone marrow, as described previously.22 Aberrations were designated according to the ISCN (1995).23 Designation as an abnormal clone required the identification of 2 or more metaphase cells with identical structural abnormalities or extra chromosomes or 3 or more metaphase cells with identical missing chromosomes. Designation as normal required complete analysis of a minimum of 20 banded metaphases from bone marrow only. A minimum of 2 original karyotypes of each abnormal clone or of normal cells were reviewed by at least 2 members of the CCG Cytogenetics Committee.
Between 1988 and 1995, a total of 4,986 children were entered on the CCG studies included in this analysis. Among these, 1,880 cases had centrally reviewed and accepted cytogenetic data: 110 cases (6%) were classified as hypodiploid and 1,770 cases (94%) were classified as nonhypodiploid. Classification into ploidy groups was based on the karyotype of the simplest clone.
The current cohort of patients with accepted cytogenetic data was similar to concurrently enrolled patients who did not have accepted cytogenetic data with respect to many presenting features, although patients with accepted data were more likely to be white and to have high WBC counts, high platelet and hemoglobin levels, a large mediastinal mass, lymphadenopathy, a T-lineage immunophenotype, and a non-L1 French-American-British (FAB) morphology. However, the actual percentages of patients with or without accepted cytogenetic data in these categories did not appear to differ; thus, the statistical differences may be due to the large sample size. Importantly, various measures of outcome were similar for concurrently enrolled patients with or without accepted cytogenetic data. For example, similar percentages of patients in the 2 groups had M1 (<5% blasts), M2 (5% to 25% blasts), or M3 (>25% blasts) at day 7 of induction therapy (P = .50). Greater than 97% of each group achieved remission by the end of induction therapy (P = .10), and 6-year EFS estimates were 75% (SD, 2%) and 76% (SD, 1%) for patients with or without accepted data, respectively (P = .36).
Statistical methods.
Analyses were based on patient follow-up through June 20, 1998. Clinical, demographic, and laboratory features of hypodiploid and nonhypodiploid patients were compared using χ2 tests for homogeneity of proportions. Outcome was analyzed using life table methods and associated statistics. The primary endpoint examined was EFS from study entry; events included induction failure (nonresponse to therapy or death during induction), leukemic relapse at any site, death during remission, or second malignant neoplasm, whichever occurred first. Patients not experiencing an event at the time of EFS analysis were censored at the time of their last contact. The Kaplan-Meier24 life table estimate of EFS and its SD25 are provided for selected time points. An approximate 95% confidence interval (CI) can be obtained from the life table estimate ±1.96 SDs. Life table comparisons of EFS outcome pattern for patient groups used the log rank statistic.25 26P values are based on the pattern of outcome across the entire period of patient follow-up; values ≤.05 are referred to as statistically significant and values between .06 and .10 are considered to have borderline statistical significance.
Within the hypodiploid group, we also compared presenting features and outcome for patients with 24 to 28 chromosomes (near-haploidy), 33 to 44 chromosomes, and 45 chromosomes (no patients had 29 to 32 chromosomes). These numerical cutoffs were based on the distinct cytogenetic features found in patients with near-haploidy, as well as on previous reports that such patients have a very poor outcome.18,19 Multivariate analysis of the prognostic effect of hypodiploid status was performed using the Cox regression model.27 Significance levels were based on the likelihood ratio test. The relative risks associated with hypodiploid status were estimated using the exponentiated maximum likelihood coefficient from the multivariate regression analysis.
RESULTS
Karyotypes of children with hypodiploid ALL.
Of the 110 hypodiploid patients, the majority (N = 87) had 45 chromosomes in the simplest clone. Of the patients with less than 45 chromosomes, 8 were near-haploid with 24 to 28 chromosomes (1 with 24; 1 with 25; 2 with 26; 3 with 27; and 1 with 28); none had between 29 and 32 chromosomes; and 15 had 33 to 44 chromosomes (1 with 33; 2 with 34; 1 with 36; 1 with 40; 1 with 42; 1 with 43; and 8 with 44). Six patients with a modal chromosome number of 46 were classified as hypodiploid: 4 cases had Down syndrome with loss of another chromosome; 2 additional cases each had 45 chromosomes in the simplest clone.
Recurrent karyotypic aberrations of the hypodiploid patients are summarized in Table 1. A doubling of the hypodiploid clone was seen in 2 patients with 24 to 28 chromosomes and in 2 patients with 33 to 44 chromosomes. The majority (5/8) of patients with 24 to 28 chromosomes had numerical aberrations only, whereas only 2 of 15 patients with 33 to 44 chromosomes (1 with 33 and 1 with 34 chromosomes) lacked structural aberrations. All patients with 24 to 28 chromosomes had disomy of chromosome 21, 4 had disomy 18, and 2 each had disomy 8, 10, or 14. Four of these patients had 2 sex chromosomes (3 were XY and 1 was XX). Among patients with 33 to 44 chromosomes, 7 had monosomy 7, 6 had monosomy 13, 5 had monosomy 20, 4 had monosomy 14, and 8 had monosomy X (3 females and 5 males).
Recurrent Chromosome Abnormalities Among Hypodiploid Patients
| Abnormality . | Karyotypic Group . | ||||
|---|---|---|---|---|---|
| 24-28 Chromosomes (N = 8) . | 33-44 Chromosomes No Dicentric (N = 13) . | 33-44 Chromosomes With Dicentric (N = 2) . | 45 Chromosomes No Dicentric (N = 59) . | 45 Chromosomes With Dicentric (N = 28) . | |
| Chromosome loss | |||||
| −7 | 8 (100) | 6 (46) | 1 (50) | 10 (17) | 0 (0) |
| −13 | 7 (88) | 6 (46) | 0 (0) | 11 (19) | 0 (0) |
| −14 | 6 (75) | 4 (38) | 0 (0) | 8 (14) | 0 (0) |
| −20 | 8 (100) | 5 (31) | 0 (0) | 8 (14) | 0 (0) |
| −X | 3 (38) | 3 (23) | 0 (0) | 10 (17) | 0 (0) |
| −Y | 1 (13) | 5 (38) | 0 (0) | 3 (5) | 0 (0) |
| Dicentric chromosome | |||||
| dic(9;12) | NA | NA | 1 (50) | NA | 10 (36) |
| dic(9;20) | NA | NA | 0 (0) | NA | 9 (32) |
| dic(7;9) | NA | NA | 0 (0) | NA | 4 (14) |
| dic(9;V) | NA | NA | 1 (50) | NA | 2 (7) |
| Other dicentric | NA | NA | 0 (0) | NA | 3 (11) |
| Other abnormality | |||||
| del(6q) | 8* (100) | 2 (15) | 0 (0) | 7 (12) | 1 (4) |
| Abnormal 9p | 8* (100) | 11 (85) | 2 (100) | 18 (31) | 25 (89) |
| Abnormal 12p | 8* (100) | 6 (46) | 2 (100) | 13 (22) | 11 (39) |
| Philadelphia chromosome | |||||
| Monosomy 7 | 0 (0) | 1 (8) | 0 (0) | 3 (5) | 0 (0) |
| Disomy 7 | 0 (0) | 0 (0) | 0 (0) | 2 (3) | 1 (4) |
| One aberration | NA† | NA† | NA† | 6 (10) | 13 (46) |
| Two aberrations | NA† | 0 (0) | 1 (50) | 16 (27) | 7 (25) |
| ≥Three aberrations | 8 (100) | 13 (100) | 1 (50) | 37 (63) | 8 (29) |
| Doubling of abnormal clone | 2 (25) | 2 (15) | 0 (0) | 0 (0) | 0 (0) |
| Numerical abnormality only | 5 (63) | 2 (15) | 0 (0) | 6 (10) | 0 (0) |
| Abnormality . | Karyotypic Group . | ||||
|---|---|---|---|---|---|
| 24-28 Chromosomes (N = 8) . | 33-44 Chromosomes No Dicentric (N = 13) . | 33-44 Chromosomes With Dicentric (N = 2) . | 45 Chromosomes No Dicentric (N = 59) . | 45 Chromosomes With Dicentric (N = 28) . | |
| Chromosome loss | |||||
| −7 | 8 (100) | 6 (46) | 1 (50) | 10 (17) | 0 (0) |
| −13 | 7 (88) | 6 (46) | 0 (0) | 11 (19) | 0 (0) |
| −14 | 6 (75) | 4 (38) | 0 (0) | 8 (14) | 0 (0) |
| −20 | 8 (100) | 5 (31) | 0 (0) | 8 (14) | 0 (0) |
| −X | 3 (38) | 3 (23) | 0 (0) | 10 (17) | 0 (0) |
| −Y | 1 (13) | 5 (38) | 0 (0) | 3 (5) | 0 (0) |
| Dicentric chromosome | |||||
| dic(9;12) | NA | NA | 1 (50) | NA | 10 (36) |
| dic(9;20) | NA | NA | 0 (0) | NA | 9 (32) |
| dic(7;9) | NA | NA | 0 (0) | NA | 4 (14) |
| dic(9;V) | NA | NA | 1 (50) | NA | 2 (7) |
| Other dicentric | NA | NA | 0 (0) | NA | 3 (11) |
| Other abnormality | |||||
| del(6q) | 8* (100) | 2 (15) | 0 (0) | 7 (12) | 1 (4) |
| Abnormal 9p | 8* (100) | 11 (85) | 2 (100) | 18 (31) | 25 (89) |
| Abnormal 12p | 8* (100) | 6 (46) | 2 (100) | 13 (22) | 11 (39) |
| Philadelphia chromosome | |||||
| Monosomy 7 | 0 (0) | 1 (8) | 0 (0) | 3 (5) | 0 (0) |
| Disomy 7 | 0 (0) | 0 (0) | 0 (0) | 2 (3) | 1 (4) |
| One aberration | NA† | NA† | NA† | 6 (10) | 13 (46) |
| Two aberrations | NA† | 0 (0) | 1 (50) | 16 (27) | 7 (25) |
| ≥Three aberrations | 8 (100) | 13 (100) | 1 (50) | 37 (63) | 8 (29) |
| Doubling of abnormal clone | 2 (25) | 2 (15) | 0 (0) | 0 (0) | 0 (0) |
| Numerical abnormality only | 5 (63) | 2 (15) | 0 (0) | 6 (10) | 0 (0) |
Values are the number of patients with percentages in parentheses.
Abbreviation: NA, not applicable.
All monosomies.
By definition, patients with 30 to 44 chromosomes had at least 2 aberrations and patients with 24 to 28 chromosomes had at least 3 aberrations.
Among patients in the subgroups with 33 to 44 and 45 chromosomes, the frequency of specific recurring aberrations differed among subsets with and without a dicentric chromosome. Among those with 45 chromosomes, dic(9;12), dic(9;20), dic (7;9), and dic(9;V) were particularly common. Six patients with 45 chromosomes had numerical abnormalities only (1 had a constitutional structural abnormality): −X, 1 patient; −18, 1 patient; −7, 1 patient; −22, 1 patient; and −20, 2 patients. In contrast, only 2 patients in the group with 33 to 44 chromosomes had dicentric chromosomes. Thirteen patients with 33 to 44 chromosomes had structural aberrations, and 8 patients were missing a sex chromosome (−X, 3 patients; and −Y, 5 patients).
Among the combined group of patients with 33 to 45 chromosomes, monosomies, particularly of chromosomes 7 (N = 17) and 13 (N = 17), were frequent in patients lacking a dicentric chromosome. Four of the 7 patients with a Philadelphia chromosome (Ph) also had monosomy 7. In addition to the 17 patients with monosomy 13, 6 patients had partial deletions of 13q (data not shown). Monosomy of chromosome 14, which has not been reported previously, and monosomy of chromosome 20 also were relatively frequent, occurring in 12 and 13 patients, respectively.
Other abnormalities that occurred frequently among all hypodiploid patients included an abnormal chromosome arm 9p, an abnormal chromosome arm 12p, a deletion of chromosome arm 6q, and a Ph. Many patients in the 33 to 44 or 45 chromosome group had 2 or more abnormalities. Constitutional structural abnormalities were observed in 7 patients, including the 4 cases with Down syndrome as well as 3 patients with a modal chromosome number of 45: 1 had inv(1)(q23q32), 1 had t(3;17)(q25;q23), and 1 had t(10;13)(q26;q21).
Clinical and biological features of children with hypodiploid ALL.
Distinguishing characteristics of hypodiploid patients at presentation are shown in Table 2. Hypodiploid patients were more likely than nonhypodiploid patients to be ≥10 years of age (P = .007), to be classified as poor risk by NCI criteria (P = .002), to have L2/L1 or L2 FAB morphology (P = .002), or to have a Ph (P = .004). Comparison of the frequencies of WBC count and T-lineage and CD10+immunophenotypes reached borderline significance. No statistically significant differences were observed for other laboratory or clinical parameters, including sex, race, organomegaly, platelet or hemoglobin levels, or presence of central nervous system (CNS) disease, t(4;11), t(1;19), or Down syndrome (data not shown). Within the hypodiploid group, patients in each of the subgroups were generally similar with respect to their presenting features, except that those with 24 to 28 chromosomes were less likely to have hepatomegaly (P = .02) and more likely to have enlarged lymph nodes (P = .02) and be CD10+ (P = .02); those with 45 chromosomes were less likely to have a mediastinal mass (P = .007).
Clinical and Biological Presenting Features for Patients With Hypodiploid ALL
| Variable . | Category . | Hypodiploid . | Nonhypodiploid* . | P Value† . | ||
|---|---|---|---|---|---|---|
| 45 Chromosomes (N = 87) . | 33-44 Chromosomes (N = 15) . | 24-28 Chromosomes (N = 8) . | >45 Chromosomes (N = 1,770) . | |||
| Age (yrs) | 1-9 ≥10 | 60 (69) 27 (31) | 9 (60) 6 (40) | 5 (63) 3 (38) | 1,386 (78) 384 (22) | .007; .76 |
| WBC (×109/L) | <20 20-49 | 45 (52) 17 (20) | 8 (53) 4 (27) | 4 (50) 1 (13) | 1,113 (63) 261 (15) | .07; .88 |
| ≥50 | 25 (29) | 3 (20) | 3 (38) | 396 (22) | ||
| FAB morphology | L1 L1/L2 | 61 (70) 4 (5) | 10 (67) 2 (13) | 4 (50) 1 (13) | 1,368 (78) 152 (9) | .002; .61 |
| L2/L1 L2 | 13 (15) 9 (10) | 1 (7) 2 (13) | 1 (13) 2 (25) | 103 (6) 123 (7) | ||
| Immunophenotype | B-lineage T-lineage | 52 (93) 4 (7) | 8 (80) 2 (20) | 6 (100) 0 (0) | 1,024 (84) 198 (16) | .08; .55 |
| CD10+ CD10− | 63 (9) 6 (91) | 7 (64) 4 (36) | 6 (100) 0 (0) | 1,122 (19) 266 (81) | .08; .02 | |
| NCI Risk Group‡ | Standard Poor | 42 (48) 45 (52) | 6 (40) 9 (60) | 4 (50) 4 (50) | 1,101 (62) 669 (38) | .002; .83 |
| Philadelphia chromosome | Positive Negative | 6 (7) 81 (93) | 1 (7) 14 (93) | 0 (0) 8 (100) | 37 (2) 1,733 (98) | .004; .75 |
| Liver | Normal Moderately enlarged | 42 (48) 44 (51) | 6 (40) 7 (47) | 7 (87) 1 (13) | 874 (50) 825 (47) | .89; .02 |
| Markedly enlarged | 1 (1) | 2 (13) | 0 (0) | 64 (3) | ||
| Mediastinal mass | Absent Moderately enlarged | 85 (98) 0 (0) | 14 (93) 0 (0) | 7 (87) 1 (13) | 1,603 (91) 85 (5) | .10; .007 |
| Markedly enlarged | 2 (2) | 1 (7) | 0 (0) | 2 (5) | ||
| Lymph nodes | Normal Moderately enlarged | 41 (47) 42 (48) | 11 (73) 3 (20) | 1 (13) 5 (63) | 898 (51) 753 (43) | .84; .02 |
| Markedly enlarged | 4 (5) | 1 (7) | 2 (25) | 119 (7) | ||
| Variable . | Category . | Hypodiploid . | Nonhypodiploid* . | P Value† . | ||
|---|---|---|---|---|---|---|
| 45 Chromosomes (N = 87) . | 33-44 Chromosomes (N = 15) . | 24-28 Chromosomes (N = 8) . | >45 Chromosomes (N = 1,770) . | |||
| Age (yrs) | 1-9 ≥10 | 60 (69) 27 (31) | 9 (60) 6 (40) | 5 (63) 3 (38) | 1,386 (78) 384 (22) | .007; .76 |
| WBC (×109/L) | <20 20-49 | 45 (52) 17 (20) | 8 (53) 4 (27) | 4 (50) 1 (13) | 1,113 (63) 261 (15) | .07; .88 |
| ≥50 | 25 (29) | 3 (20) | 3 (38) | 396 (22) | ||
| FAB morphology | L1 L1/L2 | 61 (70) 4 (5) | 10 (67) 2 (13) | 4 (50) 1 (13) | 1,368 (78) 152 (9) | .002; .61 |
| L2/L1 L2 | 13 (15) 9 (10) | 1 (7) 2 (13) | 1 (13) 2 (25) | 103 (6) 123 (7) | ||
| Immunophenotype | B-lineage T-lineage | 52 (93) 4 (7) | 8 (80) 2 (20) | 6 (100) 0 (0) | 1,024 (84) 198 (16) | .08; .55 |
| CD10+ CD10− | 63 (9) 6 (91) | 7 (64) 4 (36) | 6 (100) 0 (0) | 1,122 (19) 266 (81) | .08; .02 | |
| NCI Risk Group‡ | Standard Poor | 42 (48) 45 (52) | 6 (40) 9 (60) | 4 (50) 4 (50) | 1,101 (62) 669 (38) | .002; .83 |
| Philadelphia chromosome | Positive Negative | 6 (7) 81 (93) | 1 (7) 14 (93) | 0 (0) 8 (100) | 37 (2) 1,733 (98) | .004; .75 |
| Liver | Normal Moderately enlarged | 42 (48) 44 (51) | 6 (40) 7 (47) | 7 (87) 1 (13) | 874 (50) 825 (47) | .89; .02 |
| Markedly enlarged | 1 (1) | 2 (13) | 0 (0) | 64 (3) | ||
| Mediastinal mass | Absent Moderately enlarged | 85 (98) 0 (0) | 14 (93) 0 (0) | 7 (87) 1 (13) | 1,603 (91) 85 (5) | .10; .007 |
| Markedly enlarged | 2 (2) | 1 (7) | 0 (0) | 2 (5) | ||
| Lymph nodes | Normal Moderately enlarged | 41 (47) 42 (48) | 11 (73) 3 (20) | 1 (13) 5 (63) | 898 (51) 753 (43) | .84; .02 |
| Markedly enlarged | 4 (5) | 1 (7) | 2 (25) | 119 (7) | ||
Values are the number of patients with percentages in parentheses.
Nonhypodiploid included 578 normal diploid patients, 496 pseudodiploid patients, 204 low hyperdiploid (47 to 50 chromosomes) patients, and 492 high hyperdiploid (>50 chromosomes) patients.
Global χ2 test for homogeneity; the first Pvalue refers to comparison between combined hypodiploid group and nonhypodiploid group and the second P value refers to comparison among hypodiploid subgroups.
NCl standard risk = 1 to 9 years of age with WBC count less than 50,000/μL; NCl poor risk = ≥10 years of age or WBC count ≥50,000/μL.11
Treatment outcome.
Early treatment response was similar for the 101 hypodiploid and 1,408 nonhypodiploid patients with a marrow evaluation on day 7 of induction chemotherapy: 75% of each group achieved a rapid response (M1 or M2 marrow status) and 25% were slow responders (M3 marrow status). Within the hypodiploid subgroups, 80 patients with 45 chromosomes, 15 patients with 33 to 44 chromosomes, and 6 pa- tients with 24 to 28 chromosomes underwent a day-7 marrow evaluation. A rapid early response was achieved by 75% of those with 45 chromosomes, 80% of those with 33 to 44 chromosomes, and 67% of those with 24 to 28 chromo- somes, although the comparison did not reach statistical significance (P = .81). Nearly all hypodiploid and nonhypo- diploid patients (≥97% in each group) achieved remission by the end (day 28) of induction chemotherapy (P = .68). Within the hypodiploid subgroups, 97% of patients with 45 chromosomes and 100% of those with either 33 to 44 chromosomes or 24 to 28 chromosomes achieved remission by day 28 (P = .95).
EFS was significantly different between hypodiploid patients and nonhypodiploid patients, with 6-year estimates of 58% (SD, 7%) and 76% (SD, 2%), respectively (log rank, P < .0001). Overall survival for hypodiploid and nonhypodiploid patients also was significantly different, with 6-year estimates of 67% (SD, 7%) and 84% (SD, 1%), respectively (log rank, P < .0001). Within the hypodiploid group, we observed a significant trend for progressively worse outcome with decreasing chromosome number, with 6-year EFS of 65% (SD, 8%), 40% (SD, 18%), and 25% (SD, 22%) for the 3 groups, respectively (Fig 1; log rank, P = .002; test for trend, P = .0009). By comparison, patients with normal diploidy, pseudodiploidy, low hyperdiploidy (47 to 50 chromosomes), and high hyperdiploidy (>50 chromosomes) had 6-year EFS of 81% (SD, 2%), 70% (SD, 3%), 66% (SD, 6%), and 80% (SD, 3%), respectively (Fig 1). Notably, the subgroup of hypodiploid patients with 45 chromosomes had an EFS outcome similar to that of patients with either pseudodiploidy (P = .43) or low hyperdiploidy (P = .76). Overall survival also was significantly different for the 3 hypodiploid subgroups, with 6-year estimates of 72% (SD, 7%), 47% (SD, 15%), and 50% (SD, 35%), respectively (log rank, P = .02; test for trend,P = .01).
EFS for hypodiploid and nonhypodiploid ALL subgroups. Probabilities for patients with normal diploidy (N = 578); pseudodiploidy (N = 496); low hyperdiploidy (47 to 50 chromosomes, N = 204); high hyperdiploidy (>50 chromosomes, N = 492); 45 chromosomes (N = 87); 33 to 44 chromosomes (N = 12); or ≤28 chromosomes (N = 11).
EFS for hypodiploid and nonhypodiploid ALL subgroups. Probabilities for patients with normal diploidy (N = 578); pseudodiploidy (N = 496); low hyperdiploidy (47 to 50 chromosomes, N = 204); high hyperdiploidy (>50 chromosomes, N = 492); 45 chromosomes (N = 87); 33 to 44 chromosomes (N = 12); or ≤28 chromosomes (N = 11).
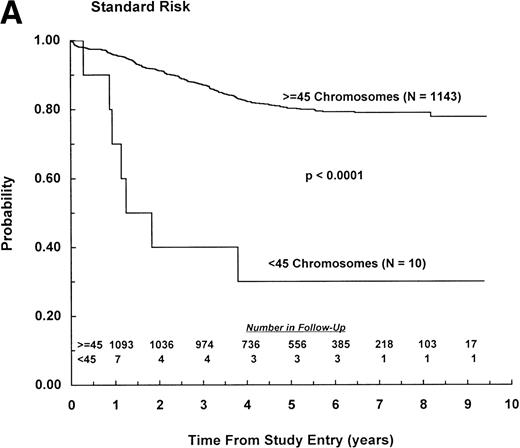
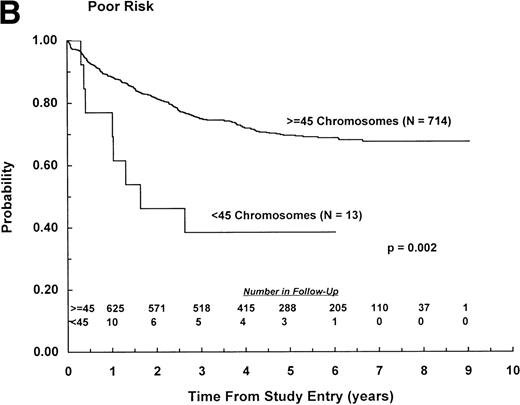
Among patients classified as NCI standard risk (1 to 9 years of age with WBC counts <50,000/μL), those with less than 45 chromosomes had significantly worse EFS outcome than those with ≥45 chromosomes, with estimates of 30% (SD, 15%) and 79% (SD, 2%), respectively (P < .0001; Fig 2A). Similarly, among patients classified as NCI poor risk (age ≥10 years or WBC ≥50,000/μL), those with less than 45 chromosomes had significantly worse EFS outcome than those with ≥45 chromosomes, with estimates of 39% (SD, 21%) and 69% (SD, 3%), respectively (P = .002; Fig2B). Furthermore, among patients with less than 45 chromosomes, the outcome of high-risk patients, all of whom received highly intensive chemotherapy, was not significantly different from that of standard-risk patients (P = .79). Ten hypodiploid patients with 45 chromosomes had a dic(9;12), and there was a trend for improved outcome (relative risk = .55) among this group, with all events occurring within 2.5 years of diagnosis. However, log rank analysis showed similar outcome for these patients and other patients with 45 chromosomes, with 6-year EFS of 78% (SD, 18%) and 63% (SD, 9%), respectively (P = .40). Overall survival also was similar for the 2 subsets (P = .70). Nine hypodiploid patients with 45 chromosomes had a dic(9;20). Outcome for these patients was similar to that of other patients with 45 chromosomes, with 6-year EFS of 63% (SD, 22%) and 65% (SD, 9%), respectively (P = .93). Overall survival also was similar for these 2 groups (P = .83).
EFS for patients with less than 45 chromosomes and ≥45 chromosomes according to NCI risk classification.11 (A) NCI standard risk (1 to 9 years of age with WBC counts <50,000/μL). (B) NCI poor risk (≥10 years of age or WBC counts ≥50,000/μL). (Inset) Number of patients remaining in follow-up.
EFS for patients with less than 45 chromosomes and ≥45 chromosomes according to NCI risk classification.11 (A) NCI standard risk (1 to 9 years of age with WBC counts <50,000/μL). (B) NCI poor risk (≥10 years of age or WBC counts ≥50,000/μL). (Inset) Number of patients remaining in follow-up.
There were a total of 405 events in the nonhypodiploid group, 28 events in the group with 45 chromosomes, and 15 events in the group with less than 45 chromosomes (Table 3). The most common event in all 3 groups was a relapse involving the marrow, either isolated or combined with relapse at another site. Relapses involving the marrow were significantly more frequent among the hypodiploid subgroup with less than 45 chromosomes than in patients with 45 chromosomes (P = .001). Other types of events, including extra- medullary relapses, occurred with similar frequency in these 2 groups.
Frequency and Type of First Events for Hypodiploid and Nonhypodiploid Patients
| Event Type . | Hypodiploid 45 (N = 87) . | Hypodiploid 33-44 (N = 15) . | Hypodiploid 24-28 (N = 8) . | Non- hypodiploid (N = 1,770) . |
|---|---|---|---|---|
| Marrow relapse | ||||
| Isolated | 11 (13) | 8 (53) | 3 (38) | 206 (12) |
| Combined | 3 (3) | 0 (0) | 1 (13) | 23 (1) |
| Total3-150 | 14 (16) | 8 (53) | 4 (50) | 229 (13) |
| Other event3-151 | ||||
| CNS relapse, isolated | 5 (6) | 0 (0) | 1 (13) | 69 (4) |
| Testicular relapse, isolated | 2 (2) | 0 (0) | 0 (0) | 17 (1) |
| Other relapse | 0 (0) | 0 (0) | 0 (0) | 11 (1) |
| Second malignancy | 0 (0) | 0 (0) | 0 (0) | 7 (0) |
| Induction failure or death in induction | 3 (3) | 0 (0) | 0 (0) | 30 (2) |
| Death in remission | 4 (5) | 1 (7) | 1 (13) | 42 (2) |
| Total | 14 (16) | 1 (7) | 2 (18) | 176 (10) |
| Total events | 28 | 9 | 6 | 405 |
| Event Type . | Hypodiploid 45 (N = 87) . | Hypodiploid 33-44 (N = 15) . | Hypodiploid 24-28 (N = 8) . | Non- hypodiploid (N = 1,770) . |
|---|---|---|---|---|
| Marrow relapse | ||||
| Isolated | 11 (13) | 8 (53) | 3 (38) | 206 (12) |
| Combined | 3 (3) | 0 (0) | 1 (13) | 23 (1) |
| Total3-150 | 14 (16) | 8 (53) | 4 (50) | 229 (13) |
| Other event3-151 | ||||
| CNS relapse, isolated | 5 (6) | 0 (0) | 1 (13) | 69 (4) |
| Testicular relapse, isolated | 2 (2) | 0 (0) | 0 (0) | 17 (1) |
| Other relapse | 0 (0) | 0 (0) | 0 (0) | 11 (1) |
| Second malignancy | 0 (0) | 0 (0) | 0 (0) | 7 (0) |
| Induction failure or death in induction | 3 (3) | 0 (0) | 0 (0) | 30 (2) |
| Death in remission | 4 (5) | 1 (7) | 1 (13) | 42 (2) |
| Total | 14 (16) | 1 (7) | 2 (18) | 176 (10) |
| Total events | 28 | 9 | 6 | 405 |
Values are the number of patients. The percentage of all patients for given group is in parentheses.
Any relapse involving the marrow, alone or in combination with relapse at another site.
All events other than a relapse involving the marrow.
The poorer outcome of the 23 patients with less than 45 chromosomes prompted us to examine their clinical and biological characteristics and treatment in more detail. The majority of these patients had B-lineage, CD10+ ALL, and none of the patients in the 24 to 28 modal number subgroup had a Ph, a t(4;11), a t(1;19), CNS disease at diagnosis, or Down syndrome. Only 1 patient in the 33 to 44 modal number group was Ph+. Most patients with 24 to 28 chromosomes lacked marked organomegaly, and only 1 patient had a mediastinal mass. Karyotypes, race, NCI risk group, and outcome for the 23 patients with less than 45 chromosomes are shown in Table 4. Nearly all of these patients were white; 10 of the 23 patients (4 with modal number 24 to 28 and 6 with modal number 33 to 44) were classified as NCI standard risk and 13 patients (4 with modal number 24 to 28 and 9 with modal number 33 to 44) were classified as NCI poor risk. Specific karyotypic features for these 23 patients were summarized above (Table 1) and are shown in Table 4 for comparison with outcome and other features.
Clinical Characteristics, Karyotypes, and Outcome for Patients With Less Than 45 Chromosomes
| Patient No. . | Modal Chromosome Number . | Race . | NCI Risk Group . | Karyotype . | Outcome . |
|---|---|---|---|---|---|
| 1 | 24 | White | Standard | 24,X,+21/48,idem,x2/46,XX | Alive postrelapse |
| 2 | 25 | White | Standard | 25,X,−Y,+14,+21/46,XY | Died postrelapse |
| 3 | 26 | White | Poor | 26,X,+Y,+14,+21/46,XY | Died in first remission |
| 4 | 26 | White | Poor | 26,XY,+del(5)(q13q33),+21/52,idem,x2 | Died after a postrelapse BMT |
| 5 | 27 | White | Standard | 27,X,+X,+18,+21,+mar/46,XX | Alive, event-free |
| 6 | 27 | Black | Poor | 27,X,+Y,+del(13)(q1?4q2?2),+18,+21/27,idem,−del(13)(q1?4q2?2), +dup(13)(q1?3q1?4)/46,XY | Alive, event-free after a BMT in first remission |
| 7 | 27 | Other4-150 | Poor | 27,X,+8,+10,+18,+21/46,XX | Alive postrelapse |
| 8 | 28 | White | Standard | 28,X,+8,+9,+10,+18,+21/46,XX | Died postrelapse |
| 9 | 33 | White | Poor | 33〈1n〉,X,+1,+2,+3,+6,+7,+8,+10,+11,+12,+15, +16,−22/46,XY | Alive, event-free |
| 10 | 34 | Hispanic | Poor | 34〈1n〉,X,+Y,+1,+5,+6,+8,+10,+11,+18,+19,+21, +22/46,XY | Died postrelapse |
| 11 | 34 | White | Poor | 34〈1n〉,X,+1,+5,+6,inv(7)(p1?5q11.2),+8,+10,+11, +13,+14,+19,+21,+22/69〈3n〉,XX,−X,+1,−2, −3,−4,+5,+6, inv(7)(p1?5q11.2),−9,+11,−12, +13,+14,−15, −16,−17,−18,+19,+20,+21, +22/46, XX | Died after a postrelapse BMT |
| 12 | 36 | White | Poor | 36,XX,−3,−4,−5,−7,−9,−13,−15,−16,−17,−17, −20,+mar/72,idemx2/46, XX | Alive, event-free |
| 134-151 | 40 | White | Poor | 40,X,−Y,−3,der(4)t(3;4)(p21;p16),−7,−9,−15,−16, −17,−18,+2mar/46, XY | Alive, event-free |
| 14‡ | 42 | White | Standard | 42,XY,−3,−5,der(6)t(6;7)(p25;q11.2),−7,add(8)(p21),der(9)t(9;12) (p22;p13) t(9;22)(q34;q11),der(12)t(9;12)(p22;p13),−13, der(15;17) (q10;q10),add(22)(p11),+mar/46,XY | Died post-2nd relapse, after a first remission BMT |
| 15 | 43 | White | Poor | 43,X,−X,add(1)(p3?4),t(2;12)(p21;p13),−7,t(7;9)(p13;p22),add(12)(p11), −15/46,XX | Died, postrelapse |
| 16 | 44 | White | Standard | 44,XX,der(2)t(2;7)(q31;p15),−7,psudic(9;16)(p2?2;p1?1.2),der(12)t(7;12)(q1?1.2;p11.2)/45, idem,+mar/46,XX | Alive, postrelapse |
| 17 | 44 | White | Standard | 44,X,−Y,add(12)(p11),−13/46,XY | Alive, event-free |
| 18 | 44 | White | Poor | 44,X,−X,−6,der(9)t(6;9)(p11.1;p13)/46,XX | Alive, event-free |
| 19 | 44 | Hispanic | Standard | 44,XY,add(1)(q4?),del(2)(p1?),add(6)(p21),add(11) (p11.1),?i(12)(p10)x2, −14,−20,−21,+mar/46,XY | Alive, event-free |
| 20 | 44 | White | Poor | 44,X,−Y,der(6)del(6)(p22)del(6)(q12),i(9)(q10), der(12)t(12;14)(p1?3;q11.2 ),−14,add(14)(q32), add(16)(p13)/46,XY | Died in first remission |
| 21 | 44 | White | Poor | 44,XX,der(4)t(4;18)(p1?5;q11∼21),add(9)(p13),−18,−20/46,XX | Died postrelapse |
| 22 | 44 | Black | Standard | 44,X,−Y,add(9)(p22),del(11)(q2?1q2?3),−13,add(17) (p1?3),der(19)t(1;19) (q23;p13)/46,XY | Died postrelapse |
| 23 | 44 | White | Standard | 44,XY,dic(9;12)(p11;p12),−15 | Died postrelapse |
| Patient No. . | Modal Chromosome Number . | Race . | NCI Risk Group . | Karyotype . | Outcome . |
|---|---|---|---|---|---|
| 1 | 24 | White | Standard | 24,X,+21/48,idem,x2/46,XX | Alive postrelapse |
| 2 | 25 | White | Standard | 25,X,−Y,+14,+21/46,XY | Died postrelapse |
| 3 | 26 | White | Poor | 26,X,+Y,+14,+21/46,XY | Died in first remission |
| 4 | 26 | White | Poor | 26,XY,+del(5)(q13q33),+21/52,idem,x2 | Died after a postrelapse BMT |
| 5 | 27 | White | Standard | 27,X,+X,+18,+21,+mar/46,XX | Alive, event-free |
| 6 | 27 | Black | Poor | 27,X,+Y,+del(13)(q1?4q2?2),+18,+21/27,idem,−del(13)(q1?4q2?2), +dup(13)(q1?3q1?4)/46,XY | Alive, event-free after a BMT in first remission |
| 7 | 27 | Other4-150 | Poor | 27,X,+8,+10,+18,+21/46,XX | Alive postrelapse |
| 8 | 28 | White | Standard | 28,X,+8,+9,+10,+18,+21/46,XX | Died postrelapse |
| 9 | 33 | White | Poor | 33〈1n〉,X,+1,+2,+3,+6,+7,+8,+10,+11,+12,+15, +16,−22/46,XY | Alive, event-free |
| 10 | 34 | Hispanic | Poor | 34〈1n〉,X,+Y,+1,+5,+6,+8,+10,+11,+18,+19,+21, +22/46,XY | Died postrelapse |
| 11 | 34 | White | Poor | 34〈1n〉,X,+1,+5,+6,inv(7)(p1?5q11.2),+8,+10,+11, +13,+14,+19,+21,+22/69〈3n〉,XX,−X,+1,−2, −3,−4,+5,+6, inv(7)(p1?5q11.2),−9,+11,−12, +13,+14,−15, −16,−17,−18,+19,+20,+21, +22/46, XX | Died after a postrelapse BMT |
| 12 | 36 | White | Poor | 36,XX,−3,−4,−5,−7,−9,−13,−15,−16,−17,−17, −20,+mar/72,idemx2/46, XX | Alive, event-free |
| 134-151 | 40 | White | Poor | 40,X,−Y,−3,der(4)t(3;4)(p21;p16),−7,−9,−15,−16, −17,−18,+2mar/46, XY | Alive, event-free |
| 14‡ | 42 | White | Standard | 42,XY,−3,−5,der(6)t(6;7)(p25;q11.2),−7,add(8)(p21),der(9)t(9;12) (p22;p13) t(9;22)(q34;q11),der(12)t(9;12)(p22;p13),−13, der(15;17) (q10;q10),add(22)(p11),+mar/46,XY | Died post-2nd relapse, after a first remission BMT |
| 15 | 43 | White | Poor | 43,X,−X,add(1)(p3?4),t(2;12)(p21;p13),−7,t(7;9)(p13;p22),add(12)(p11), −15/46,XX | Died, postrelapse |
| 16 | 44 | White | Standard | 44,XX,der(2)t(2;7)(q31;p15),−7,psudic(9;16)(p2?2;p1?1.2),der(12)t(7;12)(q1?1.2;p11.2)/45, idem,+mar/46,XX | Alive, postrelapse |
| 17 | 44 | White | Standard | 44,X,−Y,add(12)(p11),−13/46,XY | Alive, event-free |
| 18 | 44 | White | Poor | 44,X,−X,−6,der(9)t(6;9)(p11.1;p13)/46,XX | Alive, event-free |
| 19 | 44 | Hispanic | Standard | 44,XY,add(1)(q4?),del(2)(p1?),add(6)(p21),add(11) (p11.1),?i(12)(p10)x2, −14,−20,−21,+mar/46,XY | Alive, event-free |
| 20 | 44 | White | Poor | 44,X,−Y,der(6)del(6)(p22)del(6)(q12),i(9)(q10), der(12)t(12;14)(p1?3;q11.2 ),−14,add(14)(q32), add(16)(p13)/46,XY | Died in first remission |
| 21 | 44 | White | Poor | 44,XX,der(4)t(4;18)(p1?5;q11∼21),add(9)(p13),−18,−20/46,XX | Died postrelapse |
| 22 | 44 | Black | Standard | 44,X,−Y,add(9)(p22),del(11)(q2?1q2?3),−13,add(17) (p1?3),der(19)t(1;19) (q23;p13)/46,XY | Died postrelapse |
| 23 | 44 | White | Standard | 44,XY,dic(9;12)(p11;p12),−15 | Died postrelapse |
Race other than white, black, or hispanic.
Bone marrow transplant in remission.
Ph+ patient.
Of the 8 patients with 24 to 28 chromosomes, half are survivors. Two of the 4 patients are alive after a relapse, and 2 are alive event-free for 5 and 9 years; 1 of the event-free survivors underwent bone marrow transplantation in first remission. Of the 4 patients who died, 1 died after a postrelapse bone marrow transplant, 1 died in remission, and 2 died after a relapse. Because of the small number of patients in the group with modal number of 24 to 28 chromosomes, we were unable to identify differences in treatment that could account for their poor outcome. Half of the 8 patients with modal number of 24 to 28 chromosomes were treated on protocols for higher risk ALL, indicating that, regardless of specific regimen, they would have received relatively intensive therapy. No obvious associations were noted between karyotype and outcome for these patients with modal number 24 to 28.
Among the 15 patients with 33 to 44 chromosomes, 7 are survivors. Six of these 7 patients remain event-free for 3 to 6 years after study entry; 1 of these event-free survivors underwent bone marrow transplantation in first remission. One patient is alive 2 years after a relapse. Of the 8 patients who died, 1 died in first remission and the other 7 died after a relapse. Five of the 7 were in postrelapse remission at the time of death. One of the 7 patients had Ph+ ALL, underwent bone marrow transplant in first remission, and died after a second marrow relapse. Another patient died after a postrelapse bone marrow transplant. No obvious associations were noted between karyotype and outcome for these patients with modal number 33 to 44.
In addition to the 5 hypodiploid patients with less than 45 chromosomes who underwent bone marrow transplantation, 5 hypodiploid patients with 45 chromosomes also had a transplant. Two patients who had a transplant after a relapse remain alive 2.5 and 3 years posttransplant (6.7 and 8.5 years after study entry); the other 3 (1 had a transplant in remission and 2 had transplants after a relapse) died.
Multivariate analysis.
The independent significance of hypodiploidy with less than 45 chromosomes relative to hypodiploidy with 45 chromosomes or nonhypodiploidy was determined using a multivariate analysis. Significant factors identified by stepwise regression for inclusion in the multivariate analysis model included age, WBC count, spleen size, race, and Ph status. Using this model, hypodiploidy for all patients with less than 45 chromosomes remained a significant adverse risk factor for EFS: relative to the nonhypodiploid group, estimated relative risks and their 95% CI were 1.53 (1.04, 2.25), 3.34 (1.65, 6.74), and 5.72 (2.53, 12.95) for patients with 45 chromosomes, 33 to 44 chromosomes, or 24 to 28 chromosomes, respectively (P < .0001).
DISCUSSION
We have assessed the prognostic significance of hypodiploidy in a very large cohort of children with ALL. As has been observed previously,16 the majority (79%) of hypodiploid patients had 45 chromosomes, whereas the remainder had less than 45 chromosomes. Most patients had multiple cytogenetic abnormalities, particularly dicentrics, monosomies, and abnormalities of 6q, 9p, and 12p. In contrast to their counterparts with 33 or more chromosomes, hypodiploid patients with 24 to 28 chromosomes generally had numerical, rather than structural abnormalities (see below). Although hypodiploid patients were more likely than their nonhypodiploid counterparts to have some unfavorable characteristics, presenting features were generally similar within the hypodiploid subgroups; approximately half of all hypodiploid patients were classified as poor risk by NCI criteria.
Similar to previous reports,16 18 our results indicate that patients whose leukemic cells have a hypodiploid karyotype have a poorer EFS and survival compared with nonhypodiploid patients. Notably, however, our data demonstrate that hypodiploid patients with 45 chromosomes have EFS and survival outcomes similar to those of patients with pseudodiploidy or low hyperdiploidy (47 to 50 chromosomes), whereas patients with less than 45 chromosomes have significantly worse outcome. Moreover, there was a significant trend for progressively worse outcome with decreasing chromosome number: patients with 45 chromosomes had the best outcome, patients with 33 to 44 chromosomes had intermediate outcome, and patients with 24 to 28 chromosomes had the worst outcome. Interestingly, the poor outcome of hypodiploid patients with less than 45 chromosomes was due to an excess of marrow relapses rather than to high rates of CNS or other extramedullary relapses. The poorer outcome for patients with less than 45 chromosomes also was observed for patients regardless of NCI risk classification. In addition, the prognostic effects of hypodiploidy with 33 to 44 or 24 to 28 chromosomes were maintained in a multivariate analysis adjusted for important risk factors, including age, WBC count, and Ph status. These findings indicate that leukemic cell hypodiploidy comprises a heterogeneous group of patients with respect to clinical features at diagnosis and treatment outcome.
The 8 patients with 24 to 28 chromosomes (near-hap- loidy) represent a unique subgroup of ALL patients who lack certain adverse risk factors: the majority are white with B-lineage, CD10+ALL, and lack CNS disease, t(9;22), t(1;19), or t(4;11), but have a poor outcome due to high rates of mar- row relapse. Two of these patients had a second abnormal cell line with twice the number of chromosomes seen in the near-haploid clone; 5 had numerical abnormalities only. Disomies of the sex chromosomes as well as autosomes 21, 18, 14, 10, and 8 were frequent. Previous studies also noted a high frequency of numerical abnormalities and, in cases with near-haploidy, the presence of a second abnormal cell line with twice the number of chromosomes found in the near-haploid clone.11
In one of the larger series previously studied, Pui et al16reported that 31 patients with 45 or fewer chromosomes had worse outcome than patients in other ploidy groups, although the significance of this difference was not maintained in a multivariate analysis. In a later study, Pui et al18 reported that hypodiploid patients with fewer than 45 chromosomes, including those with near-haploidy, had significantly worse outcome than patients in all other ploidy groups, suggesting that the precise number of chromosomes may be an important determinant of risk. More recently, Chessells et al19 reported that hypodiploidy with 24 to 29 chromosomes was a signifi- cant and independent risk factor for children treated on the Medical Research Council (MRC) United Kingdom ALL group (UKALL) protocol X. Our data confirm and extend these previous reports by demonstrating that hypodiploidy in ALL comprises a heterogeneous group with respect to treatment outcome, with progressively worse outcome for those with 45, 33 to 44, and 24 to 28 chromosomes. Patients in the subgroup with 24 to 28 chromosomes represent a subset of ALL patients with a particularly poor outcome. These findings suggest that novel treatment programs are warranted for children with ALL and a hypodiploid leukemic cell karyotype with 24 to 28 chromosomes.
ACKNOWLEDGMENT
Contributing Cytogeneticists
| Investigator . | Institution . |
|---|---|
| A.A. Al Saadi | William Beaumont Hospital, Royal Oak, MI |
| D.C. Arthur | University of Minnesota, Minneapolis, MN |
| H. Aviv | University of Medicine and Dentistry, Newark, NJ |
| B.L. Barnoski | University Medical Center, Camden, NJ |
| P. Benn | University of Connecticut Health Center, Farmingham, CT |
| R. Best | Richland Memorial Hospital, Columbia, SC |
| D.S. Borgaonkar | Medical Center of Delaware, Wilmington, DE |
| C. Bradley | Children's Hospital and Medical Center, Seattle, WA |
| A. Brothman | University of Utah, Salt Lake City, UT |
| M.G. Butler | Vanderbilt University, Nashville, TN |
| Z. Chen | Genzyme Genetics, Scottsdale, AZ |
| H. Chen | Louisiana State University Medical Center, Shreveport, LA |
| P.D. Cotter | Children's Hospital, Oakland, CA |
| A.J. Dawson | Cytogenetics/HSC, Winnepeg, MB, Canada |
| G. Dewald | Mayo Clinic, Rochester, MN |
| Y.-S. Fan | London Health Science Centre, London, ON, Canada |
| P.A. Farber | Geisinger Medical Center, Danville, PA |
| A.B. Glassman | M.D. Anderson Cancer Center, Houston, TX |
| T.W. Glover | University of Michigan, Ann Arbor, MI |
| W. Golden | Rainbow Babies & Children's Hospital, Cleveland, OH |
| S. M. Golin | University of Pittsburgh, Pittsburgh, PA |
| D. Harris | Children's Mercy Hospital, Kansas City, MO |
| N.A. Heerema | Indiana University, Indianapolis, IN |
| R. Higgins | Children's Mercy Hospital, Kansas City, MO |
| J.V. Higgins | Butterworth Hospital, Grand Rapids, MI |
| B. Hirsch | University of Minnesota, Minneapolis, MN |
| D.K. Kalousek | British Columbia Children's Hospital, Vancouver, BC, Canada |
| M.M. LeBeau | University of Chicago, IL |
| C. Lee | Izaak Walton Killam Children's Hospital, Halifax, NS, Canada |
| K. Leppig | University of Washington, Seattle, WA |
| S. Lewis | Scottish Rite Children's Hospital, Atlanta, GA |
| W.D. Loughman | Children's Hospital, Oakland, CA |
| C.B. Lozzio | University of Tennessee Medical Center, Knoxville, TN |
| R.E. Magenis | Oregon Health Sciences Center, Portland, OR |
| L. McGavran | Children's Hospital of Denver, CO |
| L.E. McMorrow | University of Medicine & Dentistry, Newark, NJ |
| A. Milatovitch | Children's Hospital and Medical Center, Cincinnati, OH |
| T.K. Mohandas | Harbor/University of California Los Angeles Medical Center, Torrance, CA |
| B. Mouron | Children's Mercy Hospital, Kansas City, MO |
| A. Murch | King Edward Memorial Hospital, Perth, Australia |
| P. Nowell | Children's Hospital of Philadelphia, PA |
| K. Opheim | Children's Hospital and Medical Center, Seattle, WA |
| L. Pasztor | Children's Mercy Hospital, Kansas City, MO |
| S. Patil | University of Iowa, Iowa City, IA |
| C. Pehl | Loma Linda University, Redlands, CA |
| M.A. Perle | New York University Medical Center, New York, NY |
| A. Pettigrew | University of Kentucky, Lexington, KY |
| C. Phillips | Emory University Hospital, Atlanta, GA |
| K. Rao | University of North Carolina, Chapel Hill, NC |
| K.E. Richkind | Genzyme Genetics, Santa Fe, NM |
| K.N. Rosenbaum | Children's Hospital National Medical Center, Washington, DC |
| D. Roulston | University of Chicago, IL |
| C. Sandlin | Memorial Genetics Center, Long Beach, CA |
| W.G. Sanger | University of Nebraska, Omaha, NB |
| K.L. Satya-Prakash | Medical College of Georgia, Augusta, GA |
| S. Schwartz | Rainbow Babies & Children's Hospital, Cleveland, OH |
| G.S. Sekhon | University of Wisconsin, Madison, WI |
| S. Sheldon | University of Michigan, Ann Arbor, MI |
| S. Soukup | Children's Hospital and Medical Center, Cincinnati, OH |
| R. Sparkes | University of California, Los Angeles, CA |
| R. Stallard | Rainbow Babies & Children's Hospital, Cleveland, OH |
| W.S. Stanley | Genetics and IVF Institute, Fairfax, VA and Children's Hospital National Medical Center, Washington, DC |
| J. Stone | Genetrix, Inc, Scottsdale, AZ |
| P. Storto | Michigan State University, Lansing, MI |
| K.S. Theil | Children's Hospital of Columbus, OH |
| B. Torchia | Western Reserve Healthcare, Youngstown, OH |
| D. Van Dyke | Henry Ford Health System, Detroit, MI |
| N.V. Vigfusson | Sacred Heart Medical Center, Spokane, WA |
| D. Warburton | Columbia Presbyterian College of Physicians & Surgeons, New York, NY |
| J.R. Waterson | Children's Hospital Medical Center, Akron, OH |
| S.L. Wenger | University of Pittsburgh, PA |
| G. Williams | Manitoba Cancer Foundation, Winnepeg, MB, Canada |
| K.-L. Yin | Children's Hospital of Los Angeles, CA |
| C.W. Yu | Valley Children's Hospital, Fresno, CA |
| T. Zadeh | Genetics Center, Orange, CA |
| M.C. Zapata | Children's Medical Center, Dayton, OH |
| S. Zneimer | Kaiser-Permanente, San Jose, CA |
| Investigator . | Institution . |
|---|---|
| A.A. Al Saadi | William Beaumont Hospital, Royal Oak, MI |
| D.C. Arthur | University of Minnesota, Minneapolis, MN |
| H. Aviv | University of Medicine and Dentistry, Newark, NJ |
| B.L. Barnoski | University Medical Center, Camden, NJ |
| P. Benn | University of Connecticut Health Center, Farmingham, CT |
| R. Best | Richland Memorial Hospital, Columbia, SC |
| D.S. Borgaonkar | Medical Center of Delaware, Wilmington, DE |
| C. Bradley | Children's Hospital and Medical Center, Seattle, WA |
| A. Brothman | University of Utah, Salt Lake City, UT |
| M.G. Butler | Vanderbilt University, Nashville, TN |
| Z. Chen | Genzyme Genetics, Scottsdale, AZ |
| H. Chen | Louisiana State University Medical Center, Shreveport, LA |
| P.D. Cotter | Children's Hospital, Oakland, CA |
| A.J. Dawson | Cytogenetics/HSC, Winnepeg, MB, Canada |
| G. Dewald | Mayo Clinic, Rochester, MN |
| Y.-S. Fan | London Health Science Centre, London, ON, Canada |
| P.A. Farber | Geisinger Medical Center, Danville, PA |
| A.B. Glassman | M.D. Anderson Cancer Center, Houston, TX |
| T.W. Glover | University of Michigan, Ann Arbor, MI |
| W. Golden | Rainbow Babies & Children's Hospital, Cleveland, OH |
| S. M. Golin | University of Pittsburgh, Pittsburgh, PA |
| D. Harris | Children's Mercy Hospital, Kansas City, MO |
| N.A. Heerema | Indiana University, Indianapolis, IN |
| R. Higgins | Children's Mercy Hospital, Kansas City, MO |
| J.V. Higgins | Butterworth Hospital, Grand Rapids, MI |
| B. Hirsch | University of Minnesota, Minneapolis, MN |
| D.K. Kalousek | British Columbia Children's Hospital, Vancouver, BC, Canada |
| M.M. LeBeau | University of Chicago, IL |
| C. Lee | Izaak Walton Killam Children's Hospital, Halifax, NS, Canada |
| K. Leppig | University of Washington, Seattle, WA |
| S. Lewis | Scottish Rite Children's Hospital, Atlanta, GA |
| W.D. Loughman | Children's Hospital, Oakland, CA |
| C.B. Lozzio | University of Tennessee Medical Center, Knoxville, TN |
| R.E. Magenis | Oregon Health Sciences Center, Portland, OR |
| L. McGavran | Children's Hospital of Denver, CO |
| L.E. McMorrow | University of Medicine & Dentistry, Newark, NJ |
| A. Milatovitch | Children's Hospital and Medical Center, Cincinnati, OH |
| T.K. Mohandas | Harbor/University of California Los Angeles Medical Center, Torrance, CA |
| B. Mouron | Children's Mercy Hospital, Kansas City, MO |
| A. Murch | King Edward Memorial Hospital, Perth, Australia |
| P. Nowell | Children's Hospital of Philadelphia, PA |
| K. Opheim | Children's Hospital and Medical Center, Seattle, WA |
| L. Pasztor | Children's Mercy Hospital, Kansas City, MO |
| S. Patil | University of Iowa, Iowa City, IA |
| C. Pehl | Loma Linda University, Redlands, CA |
| M.A. Perle | New York University Medical Center, New York, NY |
| A. Pettigrew | University of Kentucky, Lexington, KY |
| C. Phillips | Emory University Hospital, Atlanta, GA |
| K. Rao | University of North Carolina, Chapel Hill, NC |
| K.E. Richkind | Genzyme Genetics, Santa Fe, NM |
| K.N. Rosenbaum | Children's Hospital National Medical Center, Washington, DC |
| D. Roulston | University of Chicago, IL |
| C. Sandlin | Memorial Genetics Center, Long Beach, CA |
| W.G. Sanger | University of Nebraska, Omaha, NB |
| K.L. Satya-Prakash | Medical College of Georgia, Augusta, GA |
| S. Schwartz | Rainbow Babies & Children's Hospital, Cleveland, OH |
| G.S. Sekhon | University of Wisconsin, Madison, WI |
| S. Sheldon | University of Michigan, Ann Arbor, MI |
| S. Soukup | Children's Hospital and Medical Center, Cincinnati, OH |
| R. Sparkes | University of California, Los Angeles, CA |
| R. Stallard | Rainbow Babies & Children's Hospital, Cleveland, OH |
| W.S. Stanley | Genetics and IVF Institute, Fairfax, VA and Children's Hospital National Medical Center, Washington, DC |
| J. Stone | Genetrix, Inc, Scottsdale, AZ |
| P. Storto | Michigan State University, Lansing, MI |
| K.S. Theil | Children's Hospital of Columbus, OH |
| B. Torchia | Western Reserve Healthcare, Youngstown, OH |
| D. Van Dyke | Henry Ford Health System, Detroit, MI |
| N.V. Vigfusson | Sacred Heart Medical Center, Spokane, WA |
| D. Warburton | Columbia Presbyterian College of Physicians & Surgeons, New York, NY |
| J.R. Waterson | Children's Hospital Medical Center, Akron, OH |
| S.L. Wenger | University of Pittsburgh, PA |
| G. Williams | Manitoba Cancer Foundation, Winnepeg, MB, Canada |
| K.-L. Yin | Children's Hospital of Los Angeles, CA |
| C.W. Yu | Valley Children's Hospital, Fresno, CA |
| T. Zadeh | Genetics Center, Orange, CA |
| M.C. Zapata | Children's Medical Center, Dayton, OH |
| S. Zneimer | Kaiser-Permanente, San Jose, CA |
Supported in part by research grants including CCG Chairman's Grant No. CA-13539 and CA-60437 from the National Cancer Institute, National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Nyla A. Heerema, PhD, c/o The Children's Cancer Group, Attention Ms. Lucia Noll, PO Box 60012, Arcadia, CA 91066-6012.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal